

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.12 - phase: 0
(160 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
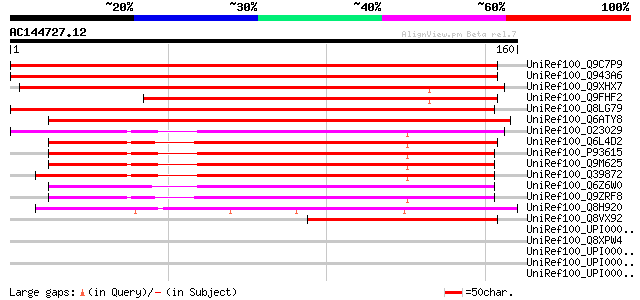
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C7P9 Plasma membrane associated protein, putative; 6... 243 1e-63
UniRef100_Q943A6 Putative plasma membrane associated protein [Or... 213 1e-54
UniRef100_Q9XHX7 Putative ABA-induced plasma membrane protein [O... 187 8e-47
UniRef100_Q9FHF2 Similarity to ABA induced plasma membrane prote... 167 1e-40
UniRef100_Q8LG79 Plasma membrane associated protein-like [Arabid... 139 2e-32
UniRef100_Q6ATY8 Hypothetical protein OSJNBa0035J16.5 [Oryza sat... 136 2e-31
UniRef100_O23029 T1G11.19 protein [Arabidopsis thaliana] 122 4e-27
UniRef100_Q6L4D2 Putative plasma membrane associated protein [Or... 119 3e-26
UniRef100_P93615 ABA induced plasma membrane protein PM 19 [Trit... 118 4e-26
UniRef100_Q9M625 Plasma membrane associated protein [Hordeum vul... 117 7e-26
UniRef100_Q39872 PGmPM3 [Glycine max] 117 1e-25
UniRef100_Q6Z6W0 Putative ABA-induced plasma membrane protein [O... 113 2e-24
UniRef100_Q9ZRF8 Hydrophobic LEA-like protein [Oryza sativa] 110 9e-24
UniRef100_Q8H920 Hypothetical protein OSJNBa0071K18.4 [Oryza sat... 86 2e-16
UniRef100_Q8VX92 Putative hydrophobic LEA-like protein [Pinus pi... 78 7e-14
UniRef100_UPI0000308606 UPI0000308606 UniRef100 entry 39 0.057
UniRef100_Q8XPW4 PROBABLE TRANSMEMBRANE PROTEIN [Ralstonia solan... 38 0.075
UniRef100_UPI0000333B0D UPI0000333B0D UniRef100 entry 37 0.17
UniRef100_UPI00002776F1 UPI00002776F1 UniRef100 entry 37 0.17
UniRef100_UPI000034C36D UPI000034C36D UniRef100 entry 37 0.22
>UniRef100_Q9C7P9 Plasma membrane associated protein, putative; 66162-66952
[Arabidopsis thaliana]
Length = 158
Score = 243 bits (620), Expect = 1e-63
Identities = 117/154 (75%), Positives = 133/154 (85%)
Query: 1 MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMG 60
MA EQ+KP+A+ LL LNFCMYVIV+ IGGWAMNRAIDHGFE+GP +LPAHFSPIYFPMG
Sbjct: 1 MAGEQIKPVASGLLVLNFCMYVIVLGIGGWAMNRAIDHGFEVGPNLELPAHFSPIYFPMG 60
Query: 61 NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK 120
NAATGFFV FAL+AGVVG S ISGL+HIRSWT SLP+AA+ A IAW LTVLAMGF K
Sbjct: 61 NAATGFFVIFALLAGVVGAASTISGLSHIRSWTVGSLPAAATAATIAWTLTVLAMGFAWK 120
Query: 121 EIQLNIRNSRLKTMEAFLIILTATQLFYIAAIHG 154
EI+L RN++L+TMEAFLIIL+ TQL YIAA+HG
Sbjct: 121 EIELQGRNAKLRTMEAFLIILSVTQLIYIAAVHG 154
>UniRef100_Q943A6 Putative plasma membrane associated protein [Oryza sativa]
Length = 156
Score = 213 bits (542), Expect = 1e-54
Identities = 103/154 (66%), Positives = 125/154 (80%)
Query: 1 MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMG 60
MAN +KP+A LLL LNFCMYVIV ++GGWA+N AI G+ IG G LPA+FSPIYFPMG
Sbjct: 1 MANAGLKPVAGLLLVLNFCMYVIVAAVGGWAINHAIHTGYFIGSGMALPANFSPIYFPMG 60
Query: 61 NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK 120
NAATGFFV FA+IAGVVG + ++G +H+R+W+ ESLP+AAS IAW LT+LAMG K
Sbjct: 61 NAATGFFVIFAVIAGVVGAAAALAGFHHVRAWSHESLPAAASSGFIAWTLTLLAMGLAVK 120
Query: 121 EIQLNIRNSRLKTMEAFLIILTATQLFYIAAIHG 154
EI L+ RN+RLKTME+F IIL+ATQLFY+ AIHG
Sbjct: 121 EIDLHGRNARLKTMESFTIILSATQLFYLLAIHG 154
>UniRef100_Q9XHX7 Putative ABA-induced plasma membrane protein [Oryza sativa]
Length = 189
Score = 187 bits (475), Expect = 8e-47
Identities = 95/171 (55%), Positives = 117/171 (67%), Gaps = 18/171 (10%)
Query: 4 EQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAA 63
E +KP+A LLL LN CMYVI+ IGGWA+N +ID GF +GPG LPAHF P++FP+GN A
Sbjct: 3 ETLKPVALLLLILNLCMYVILAIIGGWAVNISIDRGFILGPGLRLPAHFHPMFFPIGNWA 62
Query: 64 TGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQ 123
TGFFV F+L+AGVVGI S + G +HIR W SL AA+ +AWALTVLAMG C+EI
Sbjct: 63 TGFFVVFSLLAGVVGIASGLVGFSHIRHWNYYSLQPAATTGLLAWALTVLAMGLACQEIS 122
Query: 124 LNIRNSRL------------------KTMEAFLIILTATQLFYIAAIHGAA 156
L+ RN++L TMEAF I+LTATQLFY+ AIH +
Sbjct: 123 LDRRNAKLINSFRMVCELKPWEFGIQGTMEAFTIVLTATQLFYVLAIHSGS 173
>UniRef100_Q9FHF2 Similarity to ABA induced plasma membrane protein [Arabidopsis
thaliana]
Length = 138
Score = 167 bits (422), Expect = 1e-40
Identities = 81/118 (68%), Positives = 99/118 (83%), Gaps = 6/118 (5%)
Query: 43 GPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAAS 102
G + LPAHFSPI+FPMGNAATGFF+ FALIAGV G S+ISG++H++SWT+ SLP+A S
Sbjct: 18 GADYSLPAHFSPIHFPMGNAATGFFIMFALIAGVAGAASVISGISHLQSWTTTSLPAAVS 77
Query: 103 VAAIAWALTVLAMGFGCKEIQLNIRNSRL------KTMEAFLIILTATQLFYIAAIHG 154
A IAW+LT+LAMGFGCKEI+L +RN+RL +TMEAFLIIL+ATQL YIAAI+G
Sbjct: 78 AATIAWSLTLLAMGFGCKEIELGMRNARLVSKPLMRTMEAFLIILSATQLLYIAAIYG 135
>UniRef100_Q8LG79 Plasma membrane associated protein-like [Arabidopsis thaliana]
Length = 171
Score = 139 bits (350), Expect = 2e-32
Identities = 70/153 (45%), Positives = 97/153 (62%)
Query: 1 MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMG 60
MA+ K A +LL LN +Y ++ I WA+N I+ E LPA PIYFP+G
Sbjct: 1 MASGGSKSAAFMLLMLNLGLYFVITIIASWAVNHGIERTRESASTLSLPAKIFPIYFPVG 60
Query: 61 NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK 120
N ATGFFV F LIAGVVG+ + ++G+ ++ W S +L SAA+ + I+W+LT+LAMG CK
Sbjct: 61 NMATGFFVIFTLIAGVVGMATSLTGIINVLQWDSPNLHSAAASSLISWSLTLLAMGLACK 120
Query: 121 EIQLNIRNSRLKTMEAFLIILTATQLFYIAAIH 153
EI + + L+T+E II++ATQL AIH
Sbjct: 121 EINIGWTEANLRTLEVLTIIVSATQLLCTGAIH 153
>UniRef100_Q6ATY8 Hypothetical protein OSJNBa0035J16.5 [Oryza sativa]
Length = 169
Score = 136 bits (343), Expect = 2e-31
Identities = 67/146 (45%), Positives = 90/146 (60%)
Query: 13 LLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFAL 72
LL LN +YV+V I GWA+N +ID F G P PIYFP+GN ATGFFV FAL
Sbjct: 13 LLFLNLVLYVVVAVIAGWAINYSIDESFNSLQGVSPPVRLFPIYFPIGNLATGFFVIFAL 72
Query: 73 IAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQLNIRNSRLK 132
+AGVVG+ + ++GL+ + S+ SAA+ + + W LT+LAMG CKEI + R L+
Sbjct: 73 LAGVVGVSTSLTGLHDVSQGYPASMMSAAAASIVTWTLTLLAMGLACKEISIGWRPPSLR 132
Query: 133 TMEAFLIILTATQLFYIAAIHGAAAI 158
+E F IIL TQL ++H A +
Sbjct: 133 ALETFTIILAGTQLLCAGSLHAGAHV 158
>UniRef100_O23029 T1G11.19 protein [Arabidopsis thaliana]
Length = 186
Score = 122 bits (305), Expect = 4e-27
Identities = 68/157 (43%), Positives = 94/157 (59%), Gaps = 14/157 (8%)
Query: 1 MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMG 60
MA + IA LL LN MY+IV+ W +N+ I+ G P F G
Sbjct: 1 MATTVGRNIAAPLLFLNLVMYLIVLGFASWCLNKYIN-GQTNHPSFG------------G 47
Query: 61 NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK 120
N AT FF+TF+++A VVG+ S ++G NHIR W ++SL +A + + +AWA+T LAMG CK
Sbjct: 48 NGATPFFLTFSILAAVVGVASKLAGANHIRFWRNDSLAAAGASSIVAWAITALAMGLACK 107
Query: 121 EIQL-NIRNSRLKTMEAFLIILTATQLFYIAAIHGAA 156
+I + R RLK +EAF+IILT TQL Y+ IH +
Sbjct: 108 QINIGGWRGWRLKMIEAFIIILTFTQLLYLMLIHAGS 144
>UniRef100_Q6L4D2 Putative plasma membrane associated protein [Oryza sativa]
Length = 173
Score = 119 bits (297), Expect = 3e-26
Identities = 65/143 (45%), Positives = 87/143 (60%), Gaps = 14/143 (9%)
Query: 13 LLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFAL 72
LL LN MY+IVI W +N I+ G PG GN AT +F+ FA+
Sbjct: 12 LLVLNLIMYLIVIGFASWNLNHYIN-GETNHPGV------------AGNGATFYFLVFAI 58
Query: 73 IAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQL-NIRNSRL 131
+AGVVG S ++G++H+RSW + SL + A+ A IAWA+T LA G CKEI + R RL
Sbjct: 59 LAGVVGAASKLAGVHHVRSWGAHSLAAGAASALIAWAITALAFGLACKEIHIGGYRGWRL 118
Query: 132 KTMEAFLIILTATQLFYIAAIHG 154
+ +EAF+IIL TQL Y+A +HG
Sbjct: 119 RVLEAFVIILAFTQLLYVAMLHG 141
>UniRef100_P93615 ABA induced plasma membrane protein PM 19 [Triticum aestivum]
Length = 182
Score = 118 bits (296), Expect = 4e-26
Identities = 62/142 (43%), Positives = 87/142 (60%), Gaps = 14/142 (9%)
Query: 13 LLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFAL 72
LL LN MY++VI W +N I+ G PG GN AT +F+ FA+
Sbjct: 12 LLVLNLIMYIVVIGFASWNLNHFIN-GLTNRPGVG------------GNGATFYFLVFAI 58
Query: 73 IAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQL-NIRNSRL 131
+AGVVG S ++G++H+R+W +SL ++AS A +AWA+T LA G CKEI + R RL
Sbjct: 59 LAGVVGAASKLAGVHHVRTWRGDSLATSASSALVAWAITALAFGLACKEIHIGGYRGWRL 118
Query: 132 KTMEAFLIILTATQLFYIAAIH 153
+ +EAF+IIL TQL Y+ A+H
Sbjct: 119 RVLEAFVIILMFTQLLYVLALH 140
>UniRef100_Q9M625 Plasma membrane associated protein [Hordeum vulgare]
Length = 181
Score = 117 bits (294), Expect = 7e-26
Identities = 62/142 (43%), Positives = 87/142 (60%), Gaps = 14/142 (9%)
Query: 13 LLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFAL 72
LL LN MY+IVI W +N I+ G PG GN AT +F+ FA+
Sbjct: 12 LLVLNLIMYIIVIGFASWNLNHFIN-GITNRPGVG------------GNGATFYFLVFAI 58
Query: 73 IAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQL-NIRNSRL 131
+AGVVG S ++G++H+R+W +SL ++AS + +AWA+T LA G CKEI + R RL
Sbjct: 59 LAGVVGAASKLAGVHHVRTWRGDSLATSASSSLVAWAITALAFGLACKEIHVGGYRGWRL 118
Query: 132 KTMEAFLIILTATQLFYIAAIH 153
+ +EAF+IIL TQL Y+ A+H
Sbjct: 119 RVLEAFVIILAFTQLLYVLALH 140
>UniRef100_Q39872 PGmPM3 [Glycine max]
Length = 181
Score = 117 bits (293), Expect = 1e-25
Identities = 66/146 (45%), Positives = 88/146 (60%), Gaps = 14/146 (9%)
Query: 9 IATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFV 68
+A LL LN MY IV+ W +NR I+ G P F GN AT FF+
Sbjct: 8 VAAPLLFLNLIMYFIVLGFASWCLNRFIN-GQTYHPSFG------------GNGATMFFL 54
Query: 69 TFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQL-NIR 127
TF+++A V+GI S + G NH+R+W S+SL SA + + +AWA+T LA G CK+I L R
Sbjct: 55 TFSILAAVLGIVSKLLGGNHMRTWRSDSLASAGATSMVAWAVTALAFGLACKQIHLGGHR 114
Query: 128 NSRLKTMEAFLIILTATQLFYIAAIH 153
RL+ +EAF+IILT TQL Y+ IH
Sbjct: 115 GWRLRVVEAFIIILTFTQLLYLILIH 140
>UniRef100_Q6Z6W0 Putative ABA-induced plasma membrane protein [Oryza sativa]
Length = 185
Score = 113 bits (282), Expect = 2e-24
Identities = 56/141 (39%), Positives = 84/141 (58%), Gaps = 14/141 (9%)
Query: 13 LLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFAL 72
LL +N M+ V+ + GW++N+ ID G GN ++G+ + F+L
Sbjct: 14 LLCVNLVMHAAVLGLAGWSLNKFIDGETHHHLG--------------GNTSSGYLLVFSL 59
Query: 73 IAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQLNIRNSRLK 132
+AGVVG+ S++ L H+R+W E+L +AAS ++WALT L+ G CK I L R RL+
Sbjct: 60 MAGVVGVCSVLPRLLHVRAWRGETLAAAASTGLVSWALTALSFGLACKHITLGNRGRRLR 119
Query: 133 TMEAFLIILTATQLFYIAAIH 153
T+EAF+ ILT TQL Y+ +H
Sbjct: 120 TLEAFIAILTLTQLLYLILLH 140
>UniRef100_Q9ZRF8 Hydrophobic LEA-like protein [Oryza sativa]
Length = 173
Score = 110 bits (276), Expect = 9e-24
Identities = 61/142 (42%), Positives = 85/142 (58%), Gaps = 14/142 (9%)
Query: 13 LLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFAL 72
LL LN MY+IVI W +N I+ G PG GN AT +F+ FA+
Sbjct: 12 LLVLNLIMYLIVIGFASWNLNHFIN-GQTNYPGV------------AGNGATFYFLVFAI 58
Query: 73 IAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQL-NIRNSRL 131
+AGVVG S ++G++H+R+W +SL + A+ + IAWA+T LA G CKEI + R RL
Sbjct: 59 LAGVVGAASKLAGVHHVRAWRHDSLATNAASSLIAWAITALAFGLACKEIHIGGHRGWRL 118
Query: 132 KTMEAFLIILTATQLFYIAAIH 153
+ +EAF+IIL TQL Y+ +H
Sbjct: 119 RVLEAFVIILAFTQLLYVLMLH 140
>UniRef100_Q8H920 Hypothetical protein OSJNBa0071K18.4 [Oryza sativa]
Length = 184
Score = 86.3 bits (212), Expect = 2e-16
Identities = 57/166 (34%), Positives = 83/166 (49%), Gaps = 15/166 (9%)
Query: 9 IATLLLALNFCMYVIVISIGGWAMNRAIDH-GFEIGPGFDLPAHFSPIYFPMGNAATGFF 67
+A LL LN MYV ++ GWA+N +I + G ++G G+ +SP Y A+ F
Sbjct: 11 VAGSLLLLNLLMYVFLLGFAGWALNSSIKNAGADVGVGWG-EQPWSPYYRQSAWFASRFH 69
Query: 68 V-TFALIAGVVGIGSLISGLNHI----RSWTSESLPSAASVAAIAWALTVLAMGFGCKEI 122
+ TFA +AG +G+ + S H SW + L +AAS+ AWA T LA G C+EI
Sbjct: 70 LATFAALAGALGVAAKASAAYHGGRSGASWRPQGLAAAASLGTAAWAATALAFGVACREI 129
Query: 123 Q--------LNIRNSRLKTMEAFLIILTATQLFYIAAIHGAAAIRR 160
R R++ +E + L TQL Y+ +H A A R
Sbjct: 130 HDAAAAGPAGAARGWRMRALEGLTVTLAFTQLLYVLLLHAAVAGER 175
>UniRef100_Q8VX92 Putative hydrophobic LEA-like protein [Pinus pinaster]
Length = 93
Score = 78.2 bits (191), Expect = 7e-14
Identities = 39/60 (65%), Positives = 46/60 (76%)
Query: 95 ESLPSAASVAAIAWALTVLAMGFGCKEIQLNIRNSRLKTMEAFLIILTATQLFYIAAIHG 154
ESL S+AS A AW LT+LAMG CKEI + RNS+L T+E+FLIIL+ TQLFYI IHG
Sbjct: 1 ESLASSASSAMTAWGLTLLAMGLACKEIHVGGRNSKLITLESFLIILSGTQLFYILVIHG 60
>UniRef100_UPI0000308606 UPI0000308606 UniRef100 entry
Length = 186
Score = 38.5 bits (88), Expect = 0.057
Identities = 32/101 (31%), Positives = 51/101 (49%), Gaps = 10/101 (9%)
Query: 44 PGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLIS-GLNHIRSWTSESLPSAAS 102
P F L + PIYF G+ T F FALI+ +V SLI+ GL W + + + S
Sbjct: 34 PAFVLQKVYQPIYFARGDTKTPF--RFALISMIV--NSLIAFGLMPFYGWIAAPIATTVS 89
Query: 103 VAAIAWALTVLAMGFGCKEIQLNIRNSRLKTMEAFLIILTA 143
AWA+ +L G K+ + + S++ + LII+++
Sbjct: 90 ----AWAMIILLFA-GTKKFGASGQLSKISRSKFLLIIISS 125
>UniRef100_Q8XPW4 PROBABLE TRANSMEMBRANE PROTEIN [Ralstonia solanacearum]
Length = 883
Score = 38.1 bits (87), Expect = 0.075
Identities = 24/64 (37%), Positives = 35/64 (54%), Gaps = 4/64 (6%)
Query: 58 PMGNAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAW---ALTVLA 114
P G AT F+ +AL +G+GSL +G R +T+ L +A+ AA+A+ AL V
Sbjct: 117 PPGTVATTSFIAYAL-GNTIGLGSLTAGSVRARLYTAAGLDAASVTAAVAFDASALGVST 175
Query: 115 MGFG 118
FG
Sbjct: 176 TAFG 179
>UniRef100_UPI0000333B0D UPI0000333B0D UniRef100 entry
Length = 472
Score = 37.0 bits (84), Expect = 0.17
Identities = 31/101 (30%), Positives = 51/101 (49%), Gaps = 10/101 (9%)
Query: 44 PGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLIS-GLNHIRSWTSESLPSAAS 102
P F L + PIYF G+ T F FA+I+ +V SLI+ GL W + + + S
Sbjct: 320 PAFVLQKVYQPIYFARGDTKTPF--RFAVISMIV--NSLIAFGLMPFYGWIAAPIATTVS 375
Query: 103 VAAIAWALTVLAMGFGCKEIQLNIRNSRLKTMEAFLIILTA 143
AWA+ +L G K+ + + S++ + LII+++
Sbjct: 376 ----AWAMIILLFA-GSKKFGASGQLSKISKSKFVLIIISS 411
>UniRef100_UPI00002776F1 UPI00002776F1 UniRef100 entry
Length = 435
Score = 37.0 bits (84), Expect = 0.17
Identities = 30/127 (23%), Positives = 53/127 (41%), Gaps = 22/127 (17%)
Query: 21 YVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIG 80
+ + + G+ + ID G + F LP + F L+ +VG G
Sbjct: 141 FSLFLFASGFVVANGIDKGIDKAMRFLLP------------------MLFVLVIAMVGYG 182
Query: 81 SLI----SGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCKEIQLNIRNSRLKTMEA 136
++ SGLN++ SW +E + AAI A +++G G + S K +A
Sbjct: 183 AVAGDFSSGLNYLFSWKAEDFSAGTIQAAIGQAFFSMSIGMGTLMAFGSYMPSDQKVSKA 242
Query: 137 FLIILTA 143
+++TA
Sbjct: 243 SAVVVTA 249
>UniRef100_UPI000034C36D UPI000034C36D UniRef100 entry
Length = 206
Score = 36.6 bits (83), Expect = 0.22
Identities = 41/160 (25%), Positives = 76/160 (46%), Gaps = 16/160 (10%)
Query: 6 MKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYF--PMGNAA 63
M P+ ++L+AL F V + +G W + AI GF+ + + + IY + N
Sbjct: 1 MPPLISILMALLFRSVVPALFLGIWIGSYAIT-GFQKDSILESLLNITSIYVKDALANPD 59
Query: 64 TGFFVTFAL-IAGVVGIGSLISGL----NHIRSWTSESLPSAASVAAIAWAL------TV 112
+ F+L I G+VGI S G+ N++ + S S + + +++ A+
Sbjct: 60 HAAIIIFSLMIGGLVGIISKNGGMQGIVNNLSRFVSSSNRAQLATSSLGVAIFFDDYANT 119
Query: 113 LAMGFGCKEIQLNIRNSRLKTMEAFLIILTATQLFYIAAI 152
L +G +++ +++ SR K AFL+ TA + +A I
Sbjct: 120 LVVGNTMRKVTDSLKISRAKL--AFLVDATAAPIACVALI 157
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.328 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 242,399,721
Number of Sequences: 2790947
Number of extensions: 9315388
Number of successful extensions: 33561
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 33518
Number of HSP's gapped (non-prelim): 70
length of query: 160
length of database: 848,049,833
effective HSP length: 117
effective length of query: 43
effective length of database: 521,509,034
effective search space: 22424888462
effective search space used: 22424888462
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC144727.12


