

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.14 - phase: 0
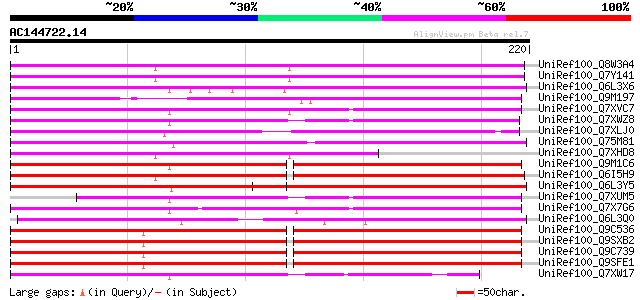
(220 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8W3A4 Putative gag-pol polyprotein [Oryza sativa] 175 9e-43
UniRef100_Q7Y141 Putative polyprotein [Oryza sativa] 174 1e-42
UniRef100_Q6L3X6 Putative polyprotein [Solanum demissum] 156 4e-37
UniRef100_Q9M197 Copia-type reverse transcriptase-like protein [... 149 4e-35
UniRef100_Q7XVC7 OSJNBa0072D21.17 protein [Oryza sativa] 145 1e-33
UniRef100_Q7XWZ8 OSJNBb0072N21.1 protein [Oryza sativa] 137 2e-31
UniRef100_Q7XLJ0 OSJNBa0009K15.20 protein [Oryza sativa] 131 1e-29
UniRef100_Q75M81 Putative integrase [Oryza sativa] 129 5e-29
UniRef100_Q7XHD8 Putative copia-type polyprotein [Oryza sativa] 123 4e-27
UniRef100_Q9M1C6 Hypothetical protein T2O9.150 [Arabidopsis thal... 120 2e-26
UniRef100_Q6I5H9 Putative polyprotein [Oryza sativa] 118 1e-25
UniRef100_Q6L3Y5 Putative gag-pol polyprotein [Solanum demissum] 115 6e-25
UniRef100_Q7XUM5 OSJNBb0002J11.7 protein [Oryza sativa] 115 8e-25
UniRef100_Q7X7G6 OSJNBb0006N15.2 protein [Oryza sativa] 113 4e-24
UniRef100_Q6L3Q0 Putative polyprotein [Solanum demissum] 110 2e-23
UniRef100_Q9C536 Copia-type polyprotein, putative [Arabidopsis t... 107 2e-22
UniRef100_Q9SXB2 T28P6.8 protein [Arabidopsis thaliana] 107 2e-22
UniRef100_Q9C739 Copia-type polyprotein, putative [Arabidopsis t... 107 2e-22
UniRef100_Q9SFE1 T26F17.17 [Arabidopsis thaliana] 107 2e-22
UniRef100_Q7XW17 OSJNBb0013O03.1 protein [Oryza sativa] 107 3e-22
>UniRef100_Q8W3A4 Putative gag-pol polyprotein [Oryza sativa]
Length = 1167
Score = 175 bits (443), Expect = 9e-43
Identities = 101/269 (37%), Positives = 151/269 (55%), Gaps = 51/269 (18%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
MM +EMSDLG + YFL +EV Q + IFI QRKYA +LKKF M+N KS++TP+ P E+
Sbjct: 892 MMHTYEMSDLGLLHYFLGMEVHQSDEGIFISQRKYAENILKKFKMDNCKSVTTPLLPNEK 951
Query: 61 -LCKDVEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL-- 117
+D ++VD T+Y+SL+G L+YLT TR DI++ ++LSR+M+ + F AKRVL
Sbjct: 952 QKARDGADKVDPTIYRSLVGSLLYLTATRPDIMFAASLLSRYMSSPSQLNFTAAKRVLRY 1011
Query: 118 ------------------------------------------------SRVLAATTAVNQ 129
+ +AA+ AV+Q
Sbjct: 1012 IKGTADYGIWYKPVKESKLIGYTDSDWAGCLDDMKSTSGYAFSLGSAEAEYVAASKAVSQ 1071
Query: 130 ALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFFLRDVQKDGDV 189
+WLR+++ DL +Q + T + D+ + IAIS+N V H +TKH +IK ++R+ +V
Sbjct: 1072 VVWLRRIMEDLGEKQYQPTTIYCDSKSAIAISENPVSHDRTKHIAIKYHYIREAVDRQEV 1131
Query: 190 CLKYCKTEDQLSYIFTKALPTGRFELLRE 218
L++C+T +QL+ IFTKAL +F RE
Sbjct: 1132 KLEFCRTNEQLADIFTKALSKEKFVRDRE 1160
>UniRef100_Q7Y141 Putative polyprotein [Oryza sativa]
Length = 1335
Score = 174 bits (441), Expect = 1e-42
Identities = 100/269 (37%), Positives = 151/269 (55%), Gaps = 51/269 (18%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
MM +EMSDLG + YFL +EV Q + IFI QRKYA +LKKF M+N KS++TP+ P E+
Sbjct: 1060 MMHTYEMSDLGLLHYFLGMEVHQSDEGIFISQRKYAENILKKFKMDNCKSVTTPLLPNEK 1119
Query: 61 -LCKDVEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL-- 117
+D ++ D T+Y+SL+G L+YLT TR DI++ ++LSR+M+ + F AKRVL
Sbjct: 1120 QKARDGADKADPTIYRSLVGSLLYLTATRPDIMFAASLLSRYMSSPSQLNFTAAKRVLRY 1179
Query: 118 ------------------------------------------------SRVLAATTAVNQ 129
+ +AA+ AV+Q
Sbjct: 1180 IKGTADYGIWYKPVKESKLIGYTDSDWAGCLDDMKSTSGYAFSLGSAEAEYVAASKAVSQ 1239
Query: 130 ALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFFLRDVQKDGDV 189
+WLR+++ DL +Q + T + D+ + IAIS+N V H +TKH +IK ++R+ +V
Sbjct: 1240 VVWLRRIMEDLGEKQYQPTTIYCDSKSAIAISENPVSHDRTKHIAIKYHYIREAVDRQEV 1299
Query: 190 CLKYCKTEDQLSYIFTKALPTGRFELLRE 218
L++C+T++QL+ IFTKAL +F RE
Sbjct: 1300 KLEFCRTDEQLADIFTKALSKEKFVRDRE 1328
>UniRef100_Q6L3X6 Putative polyprotein [Solanum demissum]
Length = 1758
Score = 156 bits (394), Expect = 4e-37
Identities = 99/289 (34%), Positives = 151/289 (51%), Gaps = 70/289 (24%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
M ++FEM+DLG M YFL +EV Q + IFICQ+KY ++L +F M++ K +STP+ +
Sbjct: 913 MEKIFEMTDLGVMKYFLGMEVLQSSDGIFICQQKYILDILNRFKMQDCKPVSTPISTGVK 972
Query: 61 LCKDVE-EQVDETLYQ--------------------SLIGCLMY------LTTTRSDILY 93
L KD + ++VD+++Y+ SL+ M+ TT + + Y
Sbjct: 973 LGKDEDSKKVDDSMYRSLIGNLLYLTASRLDILFVVSLLSRFMHSPRDTHFTTAKRVLRY 1032
Query: 94 ----------------------------------DVNVLSRFMNCAKESLFKGAKR---- 115
D + S ++ C S F + R
Sbjct: 1033 IKGTSKFGTFFPTSAEVTMNLIGYSDSDWGGGVDDSRITSGYLFCLGTSWFSWSSRKQET 1092
Query: 116 -----VLSRVLAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKT 170
V + +AA + VNQA+WLR +L DL EQ + T++M DN + ++ISKN VFHG+T
Sbjct: 1093 TAQPIVEAEYIAAASTVNQAIWLRNMLKDLGYEQTKATKIMCDNSSAVSISKNPVFHGRT 1152
Query: 171 KHFSIKLFFLRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLREK 219
KH IK F+R+VQ+ +V L +C +E+QL+ IFTK+LP RFE LR++
Sbjct: 1153 KHIKIKFHFIREVQQSNEVLLVHCSSENQLADIFTKSLPKERFEDLRQR 1201
Score = 38.1 bits (87), Expect = 0.17
Identities = 53/221 (23%), Positives = 86/221 (37%), Gaps = 17/221 (7%)
Query: 7 MSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKERL-CKDV 65
+ DL ++YFL IEV + I + Q Y ++L++ NM + K TPM E L D
Sbjct: 1528 VKDLEALTYFLGIEVIRSVDGIIMTQSTYKRDILREENMADCKLAKTPMSTTEVLKVNDG 1587
Query: 66 EEQVDETLYQSL-----IGCLMYL--TTTRSDILYDVNVLSRFMNCAKESLFKGAKRVLS 118
VD T Y + L YL T L N S M + + R +
Sbjct: 1588 GLLVDATRYMRIHWKIVKRVLRYLQGTAHLGLRLMRHNDFSLHMYSDADWVGDVNDRAST 1647
Query: 119 RVLAATTAVNQALWLRK-----VLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHF 173
N W K + + E R T + + + N + + KH
Sbjct: 1648 IGYLLFIGQNPVSWSSKKQRTIARSSIEAEYRAVTSALTETNWV----TNLLAELRMKHI 1703
Query: 174 SIKLFFLRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFE 214
++ F+R+ + V + + ++ DQL+ TK+L F+
Sbjct: 1704 AVDFHFVRNQVQLKQVQVVHIQSLDQLADTLTKSLSKSAFD 1744
>UniRef100_Q9M197 Copia-type reverse transcriptase-like protein [Arabidopsis thaliana]
Length = 1272
Score = 149 bits (377), Expect = 4e-35
Identities = 93/230 (40%), Positives = 129/230 (55%), Gaps = 38/230 (16%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
M + FEM+D+G MSY+L IEV+Q++ IFI Q YA E+LKKF M++ S P
Sbjct: 1052 MTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDD----SNP------ 1101
Query: 61 LCKDVEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVLSRV 120
SL+G L YLT TR DILY V V+SR+M + FK AKR+L +
Sbjct: 1102 ---------------SLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYI 1146
Query: 121 LA--------ATTA-----VNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFH 167
+TT+ V A+WLR +L +L L Q E T++ VDN + IA++KN VFH
Sbjct: 1147 KGTVNFGLHYSTTSDYKLVVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFH 1206
Query: 168 GKTKHFSIKLFFLRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLR 217
++KH + ++R+ DV L+Y KT DQ++ IFTK L F +R
Sbjct: 1207 DRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADIFTKPLKREDFIKMR 1256
>UniRef100_Q7XVC7 OSJNBa0072D21.17 protein [Oryza sativa]
Length = 1452
Score = 145 bits (365), Expect = 1e-33
Identities = 89/230 (38%), Positives = 131/230 (56%), Gaps = 14/230 (6%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
M + FEMS +GE+SYFL ++++Q F+ Q KY ++LK+F MEN K +STP+
Sbjct: 1212 MRREFEMSMMGELSYFLGLQIKQTPQGTFVHQTKYTKDLLKRFKMENYKPISTPIGSTAV 1271
Query: 61 LCKDVE-EQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL-- 117
L D + E VD+ Y+S+IG L+YLT +R +I + V + +RF + S + KR++
Sbjct: 1272 LDPDEDGEAVDQKEYRSMIGSLLYLTASRPNIQFVVCLCARFQASPRASHRQAVKRIMRK 1331
Query: 118 ----------SRVLAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFH 167
S +AA + +Q LWL L D L E + DN + I I+KN V H
Sbjct: 1332 QSSVAQSTAESEYVAAASCCSQILWLLSTLKDYGLTF-EKVHLFCDNTSAINIAKNPVQH 1390
Query: 168 GKTKHFSIKLFFLRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLR 217
+TKH I+ FLRD + GDV L++ T+ Q++ IFTK L + RF LR
Sbjct: 1391 SRTKHIDIRFHFLRDHVEKGDVELQFLDTKLQIADIFTKPLDSNRFAFLR 1440
>UniRef100_Q7XWZ8 OSJNBb0072N21.1 protein [Oryza sativa]
Length = 420
Score = 137 bits (345), Expect = 2e-31
Identities = 84/217 (38%), Positives = 125/217 (56%), Gaps = 9/217 (4%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
M + FEMS +GE+SYFL ++++Q F+ Q KY ++L++F MEN K +STP+
Sbjct: 199 MCREFEMSMMGELSYFLGLQIKQTPQRTFVHQTKYTKDLLRRFKMENCKPISTPIGSTAV 258
Query: 61 LCKDVE-EQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVLSR 119
L D + E VD+ Y+S+IG L+YLT +R DI + V + +RF + S + KR++S
Sbjct: 259 LDPDEDGEAVDQKEYRSMIGSLLYLTASRPDIHFAVCLCARFQASRRVSHRQAVKRIMS- 317
Query: 120 VLAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFF 179
+Q LWL L D L E + DN + I I+KN + H + H I+ F
Sbjct: 318 ------CCSQILWLLSTLKDYGLTF-EKIPLFCDNTSAINIAKNPIQHSRINHIDIRFHF 370
Query: 180 LRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELL 216
LRD + GDV L++ +T+ QL+ IFTK L + RF L
Sbjct: 371 LRDHVEKGDVELQFLETKLQLADIFTKPLDSNRFAFL 407
>UniRef100_Q7XLJ0 OSJNBa0009K15.20 protein [Oryza sativa]
Length = 1592
Score = 131 bits (329), Expect = 1e-29
Identities = 75/217 (34%), Positives = 119/217 (54%), Gaps = 15/217 (6%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
M F+MSDLG +S++L IEV Q + I + Q YA +++ + + TPM + +
Sbjct: 1371 MKATFQMSDLGPLSFYLGIEVHQDDSGITLRQTAYAKRVVELAGLTDCNPALTPMEERLK 1430
Query: 61 LCKD-VEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVLSR 119
L +D E+VD T Y+ L+G L YLT TR D+ + V + +C E
Sbjct: 1431 LSRDSTAEEVDATQYRRLVGSLRYLTHTRPDLAFSVGYVVALSSCEAE------------ 1478
Query: 120 VLAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFF 179
+AA+ QALWL ++L+DL T E+ VD+ + +A++KN VFH ++KH ++ F
Sbjct: 1479 YMAASATSTQALWLARLLSDLLGRDTGTVELRVDSKSALALAKNPVFHERSKHIRVRYHF 1538
Query: 180 LRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELL 216
+R ++G + Y T+DQL+ + TK P GR + L
Sbjct: 1539 IRSYLEEGSIKASYINTKDQLADLLTK--PLGRIKFL 1573
>UniRef100_Q75M81 Putative integrase [Oryza sativa]
Length = 1094
Score = 129 bits (324), Expect = 5e-29
Identities = 76/220 (34%), Positives = 123/220 (55%), Gaps = 4/220 (1%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
M +F +SDLG +SY+L IEV Q + DI I Q YA ++LK + + S PM P+ +
Sbjct: 866 MHSLFCISDLGLLSYYLGIEVHQSDDDITINQSAYAQKLLKIGGLADCNSTCIPMEPRLK 925
Query: 61 LCKDVEEQ-VDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVLSR 119
L K+ + VD TLY+SL+G L YL R +I + V+ +SRFM ++ K +L
Sbjct: 926 LTKNSDRMAVDSTLYRSLVGSLRYLVHARPNIAFAVSYVSRFMETPHDNHMAAVKHILQY 985
Query: 120 VLAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFF 179
V A +WL ++L ++ + E + VDN + I++ KN VFH ++KH + F
Sbjct: 986 VAGAACL---GMWLARLLAEVLGTEPEMAILKVDNKSAISVCKNPVFHDRSKHIDTRYHF 1042
Query: 180 LRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLREK 219
+R ++ V +++ T++Q++ I TK L +F+ R K
Sbjct: 1043 IRQCIEEKKVEVEFISTDEQVADILTKPLARIKFQEFRNK 1082
>UniRef100_Q7XHD8 Putative copia-type polyprotein [Oryza sativa]
Length = 1350
Score = 123 bits (308), Expect = 4e-27
Identities = 73/207 (35%), Positives = 109/207 (52%), Gaps = 51/207 (24%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
MM +EMSDLG + YFL +EV Q + IFI QRKYA +LKKF M+N KS++TP+ P E+
Sbjct: 973 MMHTYEMSDLGLLHYFLGMEVHQSDEGIFISQRKYAENILKKFKMDNYKSVTTPLLPNEK 1032
Query: 61 -LCKDVEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL-- 117
+D ++ D T+Y+SL+G L+YLT TR DI++ ++LSR+M+ + F AKRVL
Sbjct: 1033 QKARDGADKADPTIYRSLVGSLLYLTATRPDIMFAASLLSRYMSSPSQLNFTAAKRVLRY 1092
Query: 118 ------------------------------------------------SRVLAATTAVNQ 129
+ +AA+ AV+Q
Sbjct: 1093 IKGTADYGIWYKPVKESKLIGYTDSDWAGCLDDMKSTSGYAFSLGSAEAEYVAASKAVSQ 1152
Query: 130 ALWLRKVLTDLHLEQRETTEVMVDNHA 156
+WLR+++ DL +Q + T + D+ A
Sbjct: 1153 VVWLRRIMEDLGEKQYQPTTIYCDDLA 1179
>UniRef100_Q9M1C6 Hypothetical protein T2O9.150 [Arabidopsis thaliana]
Length = 1339
Score = 120 bits (302), Expect = 2e-26
Identities = 60/118 (50%), Positives = 85/118 (71%), Gaps = 1/118 (0%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
MM FEMSDLG+M +FL IEV+Q +G IFICQR+YA E+L +F M+ ++ P+ P +
Sbjct: 1041 MMLEFEMSDLGKMKHFLGIEVKQSDGGIFICQRRYAREVLARFGMDESNAVKNPIVPGTK 1100
Query: 61 LCKDVE-EQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL 117
L KD E+VDET+++ L+G LMYLT TR D++Y V ++SRFM+ + S + AKR+L
Sbjct: 1101 LTKDENGEKVDETMFKQLVGSLMYLTVTRPDLMYGVCLISRFMSNPRMSHWLAAKRIL 1158
Score = 88.2 bits (217), Expect = 1e-16
Identities = 44/97 (45%), Positives = 61/97 (62%)
Query: 121 LAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFFL 180
+AA Q +WLRKVL L E++ T + DN +TI +SK+ V HGK+KH ++ +L
Sbjct: 1231 IAAAFCACQCVWLRKVLEKLGAEEKSATVINCDNSSTIQLSKHPVLHGKSKHIEVRFHYL 1290
Query: 181 RDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLR 217
RD+ V L+YC TEDQ++ IFTK L +FE LR
Sbjct: 1291 RDLVNGDVVKLEYCPTEDQVADIFTKPLKLEQFEKLR 1327
>UniRef100_Q6I5H9 Putative polyprotein [Oryza sativa]
Length = 1136
Score = 118 bits (295), Expect = 1e-25
Identities = 59/118 (50%), Positives = 83/118 (70%), Gaps = 1/118 (0%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
MM +EMSDLG + YFL +EV Q + IFI QRKYA +LKKF M+N KS++TP+ P E+
Sbjct: 844 MMHTYEMSDLGLLHYFLGMEVHQSDEGIFISQRKYAENILKKFKMDNCKSVTTPLLPNEK 903
Query: 61 -LCKDVEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL 117
+D ++ D T+Y+SL+G L+YLT TR DI++ ++LSR+M+ + F AKRVL
Sbjct: 904 QKARDGADKADPTIYRSLVGSLLYLTATRPDIMFAASLLSRYMSSPSQLNFTAAKRVL 961
Score = 83.2 bits (204), Expect = 5e-15
Identities = 40/98 (40%), Positives = 67/98 (67%)
Query: 121 LAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFFL 180
+AA+ AV+Q +WLR+++ DL +Q + T + D+ + IAI++N V H +TKH +IK ++
Sbjct: 1032 VAASKAVSQVVWLRRIMEDLGEKQYQPTTIYCDSKSAIAINENPVSHDRTKHIAIKYHYI 1091
Query: 181 RDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLRE 218
R+ +V L++C+T++QL+ IFTKAL +F RE
Sbjct: 1092 REAVDRQEVKLEFCRTDEQLADIFTKALSKEKFICDRE 1129
>UniRef100_Q6L3Y5 Putative gag-pol polyprotein [Solanum demissum]
Length = 1133
Score = 115 bits (289), Expect = 6e-25
Identities = 60/118 (50%), Positives = 85/118 (71%), Gaps = 1/118 (0%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
M +VFEM+DLG+M+YFL +EV Q + IF+ Q+KYA E+LKKF ++ K +STP E+
Sbjct: 835 MKKVFEMNDLGKMTYFLGMEVNQSSQGIFVSQKKYATEILKKFCLDKCKPVSTPAVQGEK 894
Query: 61 LCKDVEE-QVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL 117
L K+ E VD ++Y+SLIG L+YL+ TR DI+Y +VLSRFM+ ++ K AKR+L
Sbjct: 895 LMKEDESGLVDASIYRSLIGSLLYLSATRPDIMYATSVLSRFMHLPTKTHLKAAKRIL 952
Score = 108 bits (270), Expect = 1e-22
Identities = 55/116 (47%), Positives = 74/116 (63%)
Query: 104 CAKESLFKGAKRVLSRVLAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKN 163
C K+ + +AA+ AVNQALWLRK+L DL + TEV+ DN + +A+ KN
Sbjct: 1006 CTKKQGSVAQSTAEAEYVAASGAVNQALWLRKILIDLGFHPSKATEVLCDNKSVVAMVKN 1065
Query: 164 HVFHGKTKHFSIKLFFLRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLREK 219
VFHG+TKH IK +R+ +++ +V + YC EDQL+ IFTKAL RFELLR K
Sbjct: 1066 PVFHGRTKHIKIKYHSIREAERENEVHILYCCGEDQLADIFTKALQKQRFELLRAK 1121
>UniRef100_Q7XUM5 OSJNBb0002J11.7 protein [Oryza sativa]
Length = 752
Score = 115 bits (288), Expect = 8e-25
Identities = 74/190 (38%), Positives = 106/190 (54%), Gaps = 9/190 (4%)
Query: 29 FICQRKYANEMLKKFNMENRKSMSTPMCPKERLCKDVE-EQVDETLYQSLIGCLMYLTTT 87
F+ Q KY ++L++F MEN K +STP+ L D + E VD+ Y+S+I L+YLT +
Sbjct: 559 FVHQMKYTKDLLRRFKMENCKPISTPIGSTAVLDPDEDGEAVDQKEYRSMIRSLLYLTAS 618
Query: 88 RSDILYDVNVLSRFMNCAKESLFKGAKRVLSRVLAATTAVNQALWLRKVLTDLHLEQRET 147
R DI + V + +RF + S + KR++S +Q LWL L D L E
Sbjct: 619 RPDIQFLVCLCARFQASPRASHRRAIKRIIS-------CCSQILWLLSTLKDYCLTF-EK 670
Query: 148 TEVMVDNHATIAISKNHVFHGKTKHFSIKLFFLRDVQKDGDVCLKYCKTEDQLSYIFTKA 207
+ DN + I I+KN V H +TKH I+ FLRD + GDV L++ + QL+ IFTK
Sbjct: 671 VPLFCDNTSVINIAKNPVQHSRTKHIDIRFHFLRDRVEKGDVELQFLDIKLQLADIFTKP 730
Query: 208 LPTGRFELLR 217
L + RF LR
Sbjct: 731 LDSNRFAFLR 740
>UniRef100_Q7X7G6 OSJNBb0006N15.2 protein [Oryza sativa]
Length = 1134
Score = 113 bits (282), Expect = 4e-24
Identities = 79/227 (34%), Positives = 117/227 (50%), Gaps = 12/227 (5%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
M + FEMS +GE+SYFL ++++Q F+ Q KY ++L++F MEN K +STP+
Sbjct: 898 MRREFEMSMMGELSYFLGLQIKQTPQGTFVHQTKYTKDLLRRFKMENCKPISTPIGSTAV 957
Query: 61 LCKDVE-EQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVLSR 119
L D + E VD+ Y++ Y + + D S + SL + R S
Sbjct: 958 LDPDEDGETVDQKEYRTSPRA-SYRQAVKRIMRIDRKSTSGTCHFLGTSLIAWSSRKQSS 1016
Query: 120 V---------LAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKT 170
V +AA ++ +Q WL L D L E + DN + I I+KNHV H +T
Sbjct: 1017 VAQSTTESEYVAAASSCSQIFWLLSTLKDYGLTF-EKVPLFCDNTSAINIAKNHVQHSRT 1075
Query: 171 KHFSIKLFFLRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLR 217
KH I+ FLRD + GDV L++ T+ Q++ IFTK L + F LR
Sbjct: 1076 KHIDIRFHFLRDHVEKGDVELQFLDTKLQIADIFTKPLDSNHFAFLR 1122
>UniRef100_Q6L3Q0 Putative polyprotein [Solanum demissum]
Length = 1336
Score = 110 bits (276), Expect = 2e-23
Identities = 75/237 (31%), Positives = 116/237 (48%), Gaps = 31/237 (13%)
Query: 4 VFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKERLCK 63
+F DLG++ YFL IEV + IF+ QRKY ++L++ K STPM P +L
Sbjct: 1100 MFHTKDLGQLKYFLGIEVSRSKKGIFLSQRKYILDLLEETGKSAAKPCSTPMVPNTQLTN 1159
Query: 64 DVEEQVDE-TLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVLSRVLA 122
D + D+ Y+ L+G L YLT TR DI + V++ +L R++ +L
Sbjct: 1160 DDGDPFDDPERYRRLVGKLNYLTVTRPDISFAVSI----------TLIGLDPRLIEGLLL 1209
Query: 123 ATTAVNQALW----LRKVLTDLHLEQRETTE----------------VMVDNHATIAISK 162
AT ++ +W +R + L L Q +TE + DN A + I+
Sbjct: 1210 ATVSLWVEIWYHGEVRNKMLYLDLVQNPSTELWHNPQVRLCGYFNPKLWCDNQAALHIAS 1269
Query: 163 NHVFHGKTKHFSIKLFFLRDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLREK 219
N V+H +TKH + F+R+ ++ + Y KT +QL+ +FTKAL R L K
Sbjct: 1270 NPVYHERTKHIEVDCHFIREKIQENLISTSYVKTGEQLADLFTKALTGARVNYLCNK 1326
>UniRef100_Q9C536 Copia-type polyprotein, putative [Arabidopsis thaliana]
Length = 1320
Score = 107 bits (268), Expect = 2e-22
Identities = 59/118 (50%), Positives = 79/118 (66%), Gaps = 1/118 (0%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPM-CPKE 59
M + FEM+D+G MSY+L IEV+Q++ IFI Q YA E+LKKF M++ + TPM C +
Sbjct: 1020 MTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIK 1079
Query: 60 RLCKDVEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL 117
K+ E VD T ++SL+G L YLT TR DILY V V+SR+M + FK AKR+L
Sbjct: 1080 LSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRIL 1137
Score = 85.1 bits (209), Expect = 1e-15
Identities = 42/97 (43%), Positives = 62/97 (63%)
Query: 121 LAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFFL 180
+AAT+ V A+WLR +L +L L Q E T++ VDN + IA++KN VFH ++KH + ++
Sbjct: 1208 VAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYI 1267
Query: 181 RDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLR 217
R+ DV L+Y KT DQ++ IFTK L F +R
Sbjct: 1268 RECVSKKDVQLEYVKTHDQVADIFTKPLKREDFIKMR 1304
>UniRef100_Q9SXB2 T28P6.8 protein [Arabidopsis thaliana]
Length = 1352
Score = 107 bits (268), Expect = 2e-22
Identities = 59/118 (50%), Positives = 79/118 (66%), Gaps = 1/118 (0%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPM-CPKE 59
M + FEM+D+G MSY+L IEV+Q++ IFI Q YA E+LKKF M++ + TPM C +
Sbjct: 1052 MTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIK 1111
Query: 60 RLCKDVEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL 117
K+ E VD T ++SL+G L YLT TR DILY V V+SR+M + FK AKR+L
Sbjct: 1112 LSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRIL 1169
Score = 84.3 bits (207), Expect = 2e-15
Identities = 41/97 (42%), Positives = 61/97 (62%)
Query: 121 LAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFFL 180
+AAT+ V A+WLR +L +L L Q E T++ VDN + IA++KN VFH ++KH + ++
Sbjct: 1240 VAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYI 1299
Query: 181 RDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLR 217
R+ DV L+Y KT DQ++ FTK L F +R
Sbjct: 1300 RECVSKKDVQLEYVKTHDQVADFFTKPLKRENFIKMR 1336
>UniRef100_Q9C739 Copia-type polyprotein, putative [Arabidopsis thaliana]
Length = 1352
Score = 107 bits (268), Expect = 2e-22
Identities = 59/118 (50%), Positives = 79/118 (66%), Gaps = 1/118 (0%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPM-CPKE 59
M + FEM+D+G MSY+L IEV+Q++ IFI Q YA E+LKKF M++ + TPM C +
Sbjct: 1052 MTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIK 1111
Query: 60 RLCKDVEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL 117
K+ E VD T ++SL+G L YLT TR DILY V V+SR+M + FK AKR+L
Sbjct: 1112 LSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRIL 1169
Score = 85.1 bits (209), Expect = 1e-15
Identities = 42/97 (43%), Positives = 62/97 (63%)
Query: 121 LAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFFL 180
+AAT+ V A+WLR +L +L L Q E T++ VDN + IA++KN VFH ++KH + ++
Sbjct: 1240 VAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYI 1299
Query: 181 RDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLR 217
R+ DV L+Y KT DQ++ IFTK L F +R
Sbjct: 1300 RECVSKKDVQLEYVKTHDQVADIFTKPLKREDFIKMR 1336
>UniRef100_Q9SFE1 T26F17.17 [Arabidopsis thaliana]
Length = 1291
Score = 107 bits (267), Expect = 2e-22
Identities = 59/118 (50%), Positives = 79/118 (66%), Gaps = 1/118 (0%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPM-CPKE 59
M + FEM+D+G MSY+L IEV+Q++ IFI Q YA E+LKKF M++ + TPM C +
Sbjct: 991 MTKEFEMTDIGLMSYYLGIEVKQEDNRIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIK 1050
Query: 60 RLCKDVEEQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVL 117
K+ E VD T ++SL+G L YLT TR DILY V V+SR+M + FK AKR+L
Sbjct: 1051 LSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRIL 1108
Score = 85.1 bits (209), Expect = 1e-15
Identities = 42/97 (43%), Positives = 62/97 (63%)
Query: 121 LAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFFL 180
+AAT+ V A+WLR +L +L L Q E T++ VDN + IA++KN VFH ++KH + ++
Sbjct: 1179 VAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYI 1238
Query: 181 RDVQKDGDVCLKYCKTEDQLSYIFTKALPTGRFELLR 217
R+ DV L+Y KT DQ++ IFTK L F +R
Sbjct: 1239 RECVSKKDVQLEYVKTHDQVADIFTKPLKREDFIKMR 1275
>UniRef100_Q7XW17 OSJNBb0013O03.1 protein [Oryza sativa]
Length = 832
Score = 107 bits (266), Expect = 3e-22
Identities = 70/200 (35%), Positives = 105/200 (52%), Gaps = 15/200 (7%)
Query: 1 MMQVFEMSDLGEMSYFLVIEVEQKNGDIFICQRKYANEMLKKFNMENRKSMSTPMCPKER 60
M + FEMS +GE+SYFL ++++Q F+ Q KY ++L+ F MEN K +STP+
Sbjct: 609 MHKEFEMSMMGELSYFLGLQIKQTPQGTFVHQTKYTKDLLRWFKMENCKPISTPIGSTAV 668
Query: 61 LCKDVE-EQVDETLYQSLIGCLMYLTTTRSDILYDVNVLSRFMNCAKESLFKGAKRVLSR 119
L D + E VD+ Y+S+IG L+YL DI +DV + +RF S + KR++S
Sbjct: 669 LDPDEDSEAVDQKEYRSMIGSLLYLIAYMPDIQFDVCLCARFQASPHASHRQAVKRIMS- 727
Query: 120 VLAATTAVNQALWLRKVLTDLHLEQRETTEVMVDNHATIAISKNHVFHGKTKHFSIKLFF 179
+Q LWL L D L E + DN + I I+KN V H +TKH F
Sbjct: 728 ------CCSQILWLLSTLKDYGL-TFEKVPLFCDNTSAINIAKNPVQHSRTKHMIFVFTF 780
Query: 180 LRDVQKDGDVCLKYCKTEDQ 199
+G + +C+ E++
Sbjct: 781 ------EGSFLIAHCEKENK 794
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 326,772,678
Number of Sequences: 2790947
Number of extensions: 12075164
Number of successful extensions: 40500
Number of sequences better than 10.0: 737
Number of HSP's better than 10.0 without gapping: 629
Number of HSP's successfully gapped in prelim test: 109
Number of HSP's that attempted gapping in prelim test: 38781
Number of HSP's gapped (non-prelim): 1256
length of query: 220
length of database: 848,049,833
effective HSP length: 122
effective length of query: 98
effective length of database: 507,554,299
effective search space: 49740321302
effective search space used: 49740321302
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 72 (32.3 bits)
Medicago: description of AC144722.14


