

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144720.7 + phase: 2 /pseudo/partial
(1007 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
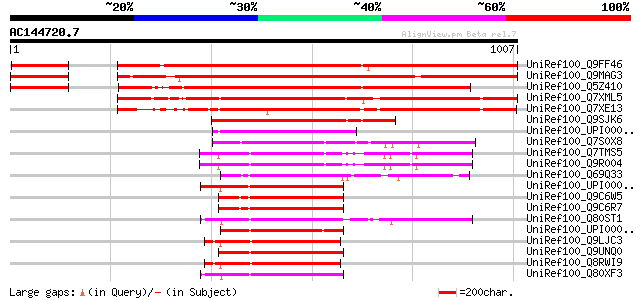
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FF46 ABC transporter-like protein [Arabidopsis thali... 1046 0.0
UniRef100_Q9MAG3 F12M16.28 [Arabidopsis thaliana] 987 0.0
UniRef100_Q5Z410 ABC transporter-like [Oryza sativa] 858 0.0
UniRef100_Q7XML5 OSJNBa0040D17.13 protein [Oryza sativa] 857 0.0
UniRef100_Q7XE13 Putative ABC transporter [Oryza sativa] 747 0.0
UniRef100_Q9SJK6 Putative ABC transporter [Arabidopsis thaliana] 507 e-142
UniRef100_UPI000028D6BC UPI000028D6BC UniRef100 entry 234 8e-60
UniRef100_Q7S0X8 Hypothetical protein [Neurospora crassa] 233 3e-59
UniRef100_Q7TMS5 ATP-binding cassette, sub-family G, member 2 [M... 211 7e-53
UniRef100_Q9R004 Breast cancer resistance protein 1 [Mus musculus] 211 1e-52
UniRef100_Q69Q33 Putative ATP-binding cassette transporter1 [Ory... 210 2e-52
UniRef100_UPI000027C604 UPI000027C604 UniRef100 entry 209 3e-52
UniRef100_Q9C6W5 Hypothetical protein F27M3_2 [Arabidopsis thali... 207 1e-51
UniRef100_Q9C6R7 ABC transporter, putative [Arabidopsis thaliana] 207 1e-51
UniRef100_Q80ST1 ATP-binding cassette protein G2 transcript vari... 206 2e-51
UniRef100_UPI000024A2F0 UPI000024A2F0 UniRef100 entry 205 5e-51
UniRef100_Q9LJC3 ABC transporter-like protein [Arabidopsis thali... 205 6e-51
UniRef100_Q9UNQ0 ATP-binding cassette, sub-family G, member 2 [H... 205 6e-51
UniRef100_Q8RWI9 Hypothetical protein At3g21090 [Arabidopsis tha... 204 8e-51
UniRef100_Q80XF3 ATP-binding cassette transporter ABCG2 [Rattus ... 204 1e-50
>UniRef100_Q9FF46 ABC transporter-like protein [Arabidopsis thaliana]
Length = 1109
Score = 1046 bits (2705), Expect = 0.0
Identities = 524/805 (65%), Positives = 636/805 (78%), Gaps = 21/805 (2%)
Query: 215 CSDQVLTTRERRVAKSRESAARSARKTANAHQRWKVAKDAAKKGATGLQAQLSRKFSRKK 274
CSDQVL TRERR AKSRE A +S R + + ++WK AKD AKK AT LQ SR FSR+K
Sbjct: 313 CSDQVLATRERRQAKSREKAVQSVRDS-QSREKWKSAKDIAKKHATELQQSFSRTFSRRK 371
Query: 275 DEENLEKVKILNQETSETDVELLPHSQPSNMVASSSAVPTEKGKTPSGLMHMMHEIENDP 334
+ + ++ L+Q +D L P M+ SSS K K + L M+H+IE +P
Sbjct: 372 SMKQPDLMRGLSQAKPGSDAALPP------MLGSSSDTKKGKKKEKNKLTEMLHDIEQNP 425
Query: 335 H--VNYNPNTGKETRHKSATKEKQPQTNTQIFKYAYAQLEKEKAQQQENKNLTFSGVLKM 392
+N G + K A K K T +Q+F+YAY Q+EKEKA Q++NKNLTFSGV+ M
Sbjct: 426 EDPEGFNLEIGDKNIKKHAPKGKALHTQSQMFRYAYGQIEKEKAMQEQNKNLTFSGVISM 485
Query: 393 ATNTEKSKRPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSA 452
A + + KRP IE++F+DL++TLK +NKH++R VTGK+ PGR++A+MGPSGAGKTTFL+A
Sbjct: 486 ANDIDIRKRPMIEVAFKDLSITLKGKNKHLMRCVTGKLSPGRVSAVMGPSGAGKTTFLTA 545
Query: 453 LAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSAD 512
L GKA GC++TG IL+NG+ ESI S+KKIIGFVPQDD+VHGNLTVEENLWFSA+CRL AD
Sbjct: 546 LTGKAPGCIMTGMILVNGKVESIQSYKKIIGFVPQDDIVHGNLTVEENLWFSARCRLPAD 605
Query: 513 LSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDE 572
L KPEKVLVVERVIE LGLQ VR+S+VGTVEKRG+SGGQRKRVNVGLEMVMEPSLL+LDE
Sbjct: 606 LPKPEKVLVVERVIESLGLQHVRDSLVGTVEKRGISGGQRKRVNVGLEMVMEPSLLILDE 665
Query: 573 PTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSA 632
PTSGLDS+SSQLLLRALRREALEGVNICMVVHQPSY LF MFDDLILL KGGL+ Y G
Sbjct: 666 PTSGLDSSSSQLLLRALRREALEGVNICMVVHQPSYTLFRMFDDLILLAKGGLICYQGPV 725
Query: 633 KKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYPIPLD 692
KKVEEYFS LGI VPER+NPPDYYIDILEGI P SSG++Y+ LPV+WMLHN YP+P+D
Sbjct: 726 KKVEEYFSSLGIVVPERVNPPDYYIDILEGILKPSTSSGVTYKQLPVRWMLHNGYPVPMD 785
Query: 693 MRQHAAQFGIPQSVNSAND----------LGEVGKTFAGELWNDVRSNVELRGEKIRLNF 742
M + G+ S + N +G+ G +FAGE W DV++NVE++ + ++ NF
Sbjct: 786 MLKSIE--GMASSASGENSAHGGSAHGSVVGDDGTSFAGEFWQDVKANVEIKKDNLQNNF 843
Query: 743 LKSKDLSNRKTPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKSSDQT 802
S DLS R+ PGV++QY+YFL R+GKQRLREAR AVDYLILLLAG CLG++ K SD+T
Sbjct: 844 SSSGDLSEREVPGVYQQYRYFLGRLGKQRLREARTLAVDYLILLLAGICLGTLAKVSDET 903
Query: 803 FGASGYTYTVIAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDHFNTVIK 862
FGA GYTYTVIAVSLLCKI ALRSFSLDKLHYWRES +GMSSLAYFL+KDT+DHFNT++K
Sbjct: 904 FGAMGYTYTVIAVSLLCKITALRSFSLDKLHYWRESRAGMSSLAYFLAKDTVDHFNTIVK 963
Query: 863 PVVYLSMFYFLTNPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPGAAQLWSVLLPVVST 922
P+VYLSMFYF NPRST TDNY+VL+CLVYCVTGIAY L+I+FEPG AQLWSVLLPVV T
Sbjct: 964 PLVYLSMFYFFNNPRSTVTDNYVVLICLVYCVTGIAYTLAILFEPGPAQLWSVLLPVVLT 1023
Query: 923 LIATQQKDSKILKAIANLCYSKWALQALVIANAERYQGVWLITRCGSLLKSGYNLHDWSL 982
LIAT D+KI+ +I+ LCY++WAL+A V++NA+RY+GVWLITRCGSL+++GYN+ +
Sbjct: 1024 LIATSTNDNKIVDSISELCYTRWALEAFVVSNAQRYKGVWLITRCGSLMENGYNIKHFPR 1083
Query: 983 CISILILMGVIGRAIAFFCMVTFKK 1007
C+ L L G++ R AFFCMVTF+K
Sbjct: 1084 CLVFLTLTGILSRCAAFFCMVTFQK 1108
Score = 160 bits (405), Expect = 2e-37
Identities = 70/115 (60%), Positives = 85/115 (73%), Gaps = 1/115 (0%)
Query: 3 DWNQAFNFSSDLRFLSSCIKKTKGDISNRLCTAAEVKFYLNSLMEKSSSA-NYLKPNRNC 61
D+N+AFNFS+ FL++C K TKGD+ R+CTAAEV+ Y N L+ + A NYLKPN+NC
Sbjct: 79 DYNEAFNFSTKPDFLNACGKTTKGDMMQRICTAAEVRIYFNGLLGGAKRATNYLKPNKNC 138
Query: 62 NLTSWVSGCEPGWACSVPSGQKIDLKDSKEMPARTSNCRACCEGFFCPHGITCMI 116
NL+SW+SGCEPGWAC K+DLKD K +P RT C CC GFFCP GITCMI
Sbjct: 139 NLSSWMSGCEPGWACRTAKDVKVDLKDDKNVPVRTQQCAPCCAGFFCPRGITCMI 193
>UniRef100_Q9MAG3 F12M16.28 [Arabidopsis thaliana]
Length = 1096
Score = 987 bits (2551), Expect = 0.0
Identities = 508/798 (63%), Positives = 620/798 (77%), Gaps = 33/798 (4%)
Query: 215 CSDQVLTTRERRVAKSRESAARSARKTANAHQRWKVAKDAAKKGATGLQAQLSRKFSRKK 274
CSDQ+LTTRERR AKSRE+A + AR AH RWK A++AAKK +G++AQ++R FS K+
Sbjct: 300 CSDQILTTRERRQAKSREAAVKKAR----AHHRWKAAREAAKKHVSGIRAQITRTFSGKR 355
Query: 275 DEENLEKVKILNQETSETDVELLPHSQPSNMVASSSAVPTEKGKTPSGLMHMMHEIENDP 334
++ + K+L + S E + S S+ +SS+A EN+
Sbjct: 356 ANQDGDTNKMLGRGDSSEIDEAIDMSTCSSPASSSAA---------------QSSYENED 400
Query: 335 HV----NYNPNTGKE-TRHKSATKEKQPQTNTQIFKYAYAQLEKEKAQQQENKNLTFSGV 389
H N + G E R K T K +T +QIFKYAY ++EKEKA +QENKNLTFSG+
Sbjct: 401 HAAAGSNGRASLGIEGKRVKGQTLAKIKKTQSQIFKYAYDRIEKEKAMEQENKNLTFSGI 460
Query: 390 LKMATNTEKSKRPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTF 449
+KMATN+E KR +E+SF+DLTLTLK+ K +LR VTG +KPGRITA+MGPSGAGKT+
Sbjct: 461 VKMATNSETRKRHLMELSFKDLTLTLKSNGKQVLRCVTGSMKPGRITAVMGPSGAGKTSL 520
Query: 450 LSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRL 509
LSALAGKA+GC ++G ILING+ ESIHS+KKIIGFVPQDDVVHGNLTVEENLWF A+CRL
Sbjct: 521 LSALAGKAVGCKLSGLILINGKQESIHSYKKIIGFVPQDDVVHGNLTVEENLWFHAKCRL 580
Query: 510 SADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLM 569
ADLSK +KVLVVER+I+ LGLQ+VR+S+VGTVEKRG+SGGQRKRVNVGLEMVMEPS+L
Sbjct: 581 PADLSKADKVLVVERIIDSLGLQAVRSSLVGTVEKRGISGGQRKRVNVGLEMVMEPSVLF 640
Query: 570 LDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYH 629
LDEPTSGLDSASSQLLLRALR EALEGVNICMVVHQPSY LF F+DL+LL KGGL VYH
Sbjct: 641 LDEPTSGLDSASSQLLLRALRHEALEGVNICMVVHQPSYTLFKTFNDLVLLAKGGLTVYH 700
Query: 630 GSAKKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYPI 689
GS KVEEYFSGLGI+VP+RINPPDYYID+LEG+ G+SG+ Y++LP +WMLH Y +
Sbjct: 701 GSVNKVEEYFSGLGIHVPDRINPPDYYIDVLEGVVISMGNSGIGYKELPQRWMLHKGYSV 760
Query: 690 PLDMRQHAAQFGIPQSVNSANDLGEVGKTFAGELWNDVRSNVELRGEKIRLNFLKSKDLS 749
PLDMR ++A N +TFA ELW DV+SN LR +KIR NFLKS+DLS
Sbjct: 761 PLDMRNNSAAGLETNPDLGTNSPDNAEQTFARELWRDVKSNFRLRRDKIRHNFLKSRDLS 820
Query: 750 NRKTPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKSSDQTFGASGYT 809
+R+TP + QYKYFL R+ KQR+REA++QA DYLILLLAGACLGS+ K+SD++FGA
Sbjct: 821 HRRTPSTWLQYKYFLGRIAKQRMREAQLQATDYLILLLAGACLGSLIKASDESFGAP--- 877
Query: 810 YTVIAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDHFNTVIKPVVYLSM 869
+LLCKIAALRSFSLDKLHYWRES SGMSS A FL+KDT+D FN ++KP+VYLSM
Sbjct: 878 ------ALLCKIAALRSFSLDKLHYWRESASGMSSSACFLAKDTIDIFNILVKPLVYLSM 931
Query: 870 FYFLTNPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPGAAQLWSVLLPVVSTLIATQQK 929
FYF TNPRSTF DNYIVL+CLVYCVTGIAYAL+I +P AQL+SVLLPVV TL+ATQ K
Sbjct: 932 FYFFTNPRSTFFDNYIVLVCLVYCVTGIAYALAIFLQPSTAQLFSVLLPVVLTLVATQPK 991
Query: 930 DSKILKAIANLCYSKWALQALVIANAERYQGVWLITRCGSLLKSGYNLHDWSLCISILIL 989
+S++++ IA+L Y KWAL+A VI NA++Y GVW+ITRCGSL+KSGY+++ WSLCI IL+L
Sbjct: 992 NSELIRIIADLSYPKWALEAFVIGNAQKYYGVWMITRCGSLMKSGYDINKWSLCIMILLL 1051
Query: 990 MGVIGRAIAFFCMVTFKK 1007
+G+ R +AF M+ +K
Sbjct: 1052 VGLTTRGVAFVGMLILQK 1069
Score = 185 bits (469), Expect = 7e-45
Identities = 78/117 (66%), Positives = 94/117 (79%)
Query: 1 DSDWNQAFNFSSDLRFLSSCIKKTKGDISNRLCTAAEVKFYLNSLMEKSSSANYLKPNRN 60
D+DWN+AFNFSS+L FLSSCIKKT+G I R+CTAAE+KFY N K+++ YLKPN N
Sbjct: 80 DADWNRAFNFSSNLNFLSSCIKKTQGSIGKRICTAAEMKFYFNGFFNKTNNPGYLKPNVN 139
Query: 61 CNLTSWVSGCEPGWACSVPSGQKIDLKDSKEMPARTSNCRACCEGFFCPHGITCMIR 117
CNLTSWVSGCEPGW CSV +++DL++SK+ P R NC CCEGFFCP G+TCMIR
Sbjct: 140 CNLTSWVSGCEPGWGCSVDPTEQVDLQNSKDFPERRRNCMPCCEGFFCPRGLTCMIR 196
>UniRef100_Q5Z410 ABC transporter-like [Oryza sativa]
Length = 995
Score = 858 bits (2218), Expect = 0.0
Identities = 448/702 (63%), Positives = 534/702 (75%), Gaps = 28/702 (3%)
Query: 216 SDQVLTTRERRVAKSRESAARSARKTANAHQRWKVAKDAAKKGATGLQAQLSRKFSRKKD 275
S Q+LT RE++ AKSRE+AAR A++TA A +RWK AKD AKK A GLQ+ LSR FSRKK
Sbjct: 313 SGQLLTNREKKQAKSREAAARHAKETAMARERWKSAKDVAKKHAVGLQSSLSRTFSRKKT 372
Query: 276 EENLEKVKILNQETSETDVELLPHSQPSNMVASSSAVPTEKGKTPSGLMHMMHEIENDPH 335
E K ETDVE PS G+ S L MM +E +P
Sbjct: 373 LRTHEPSK----GAVETDVE------PSK----------GSGEKKSNLTDMMRSLEENPE 412
Query: 336 VNYNPNTGKETRHKSATKEKQPQTNTQIFKYAYAQLEKEKAQQQENKNLTFSGVLKMATN 395
N E K TK + T +QIFKYAY Q+EKEKA +Q+NKNLTFSGV+ MAT+
Sbjct: 413 KGEGFNV--EIGEKKKTKGRHAHTQSQIFKYAYGQIEKEKAMEQQNKNLTFSGVISMATD 470
Query: 396 TEKSKRPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAG 455
+ RP IEI+F+DLTLTLK K +LR+VTGK+ PGR+ A+MGPSGAGKTTFLSA+AG
Sbjct: 471 EDIRTRPRIEIAFKDLTLTLKGSKKKLLRSVTGKLMPGRVAAVMGPSGAGKTTFLSAIAG 530
Query: 456 KALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSK 515
KA GC TG +LING+ E I ++KKIIGFVPQDD+VHGNLTV+ENLWF+A+CRLSAD+SK
Sbjct: 531 KATGCETTGMVLINGKMEPIRAYKKIIGFVPQDDIVHGNLTVQENLWFNARCRLSADMSK 590
Query: 516 PEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTS 575
+KVLVVERVIE LGLQ+VR+S+VGTVE+RG+SGGQRKRVNVGLEMVMEPS+L+LDEPTS
Sbjct: 591 ADKVLVVERVIEALGLQAVRDSLVGTVEQRGISGGQRKRVNVGLEMVMEPSVLILDEPTS 650
Query: 576 GLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKV 635
GLDS+SS LLLRALRREALEGVNI MVVHQPSY L+ MFDDLILL KGGL VYHG KKV
Sbjct: 651 GLDSSSSLLLLRALRREALEGVNISMVVHQPSYTLYKMFDDLILLAKGGLTVYHGPVKKV 710
Query: 636 EEYFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYPIPLDMRQ 695
EEYFSGLGI VP+R+NPPDYYIDILEGI P + ++ +DLP++WMLHN Y +P DM Q
Sbjct: 711 EEYFSGLGIVVPDRVNPPDYYIDILEGIVKPNANVAVNAKDLPLRWMLHNGYEVPRDMLQ 770
Query: 696 HAAQFGIPQSVNSANDL---GEVGKTFAGELWNDVRSNVELRGEKIRLNFLKSKDLSNRK 752
+ S DL G+ G++ AGE+W +V+ V + ++ N S++LSNR
Sbjct: 771 SGSD--AESSFRGGGDLTPGGDTGQSIAGEVWGNVKDIVGQKKDEYDYN-KSSQNLSNRC 827
Query: 753 TPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKSSDQTFGASGYTYTV 812
TPG+ +QYKY+L R GKQRLREARIQ VDYLIL LAG CLG++ K SD+TFGA GYTYTV
Sbjct: 828 TPGILRQYKYYLGRCGKQRLREARIQGVDYLILGLAGICLGTLAKVSDETFGALGYTYTV 887
Query: 813 IAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDHFNTVIKPVVYLSMFYF 872
IAVSLLCKI ALRSFSL+K+HYWRE SGMSSLAYF+SKDT+DHFNT+IKP+VYLSMFYF
Sbjct: 888 IAVSLLCKIGALRSFSLEKIHYWRERASGMSSLAYFMSKDTIDHFNTIIKPIVYLSMFYF 947
Query: 873 LTNPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPGAAQLWS 914
NPRS+ +NY+VL+ LVYCVTGI Y +I F+PG+AQL S
Sbjct: 948 FNNPRSSIWENYVVLVALVYCVTGIGYTFAIFFQPGSAQLVS 989
Score = 166 bits (420), Expect = 3e-39
Identities = 67/116 (57%), Positives = 88/116 (75%)
Query: 1 DSDWNQAFNFSSDLRFLSSCIKKTKGDISNRLCTAAEVKFYLNSLMEKSSSANYLKPNRN 60
D +WN AFNFS+D FLS+C++ T GD+ R+CTAAE+KFY S ++ + NY++PN+N
Sbjct: 77 DDEWNIAFNFSTDPTFLSNCMQATDGDVPQRVCTAAEMKFYFESFLDSNGRKNYVRPNKN 136
Query: 61 CNLTSWVSGCEPGWACSVPSGQKIDLKDSKEMPARTSNCRACCEGFFCPHGITCMI 116
CNLTSW+ GCE GWACS Q I+L+D+ P+RT +CR CC GFFCPHG+TCMI
Sbjct: 137 CNLTSWMDGCEAGWACSAGPDQNINLQDAVNFPSRTLDCRGCCAGFFCPHGLTCMI 192
>UniRef100_Q7XML5 OSJNBa0040D17.13 protein [Oryza sativa]
Length = 964
Score = 857 bits (2215), Expect = 0.0
Identities = 451/803 (56%), Positives = 578/803 (71%), Gaps = 41/803 (5%)
Query: 215 CSDQVLTTRERRVAKSRESAARSARKTANAHQRWKVAKDAAKKGATGLQAQLSRKFSRKK 274
CS Q LT RE+R A+SRE+A + AR+ AH+ WK AK A+K +Q+ LSR FSR++
Sbjct: 192 CSGQFLTIREKRKARSRENAIQLARQQLKAHEGWKAAKRLARKHVNDMQSHLSRTFSRRR 251
Query: 275 D-EENLEKVKILNQETSETDVELLPHSQPSNMVASSSAVPTEKGKTPSGLMHMMHEIEND 333
++L+ N + L + + S+ A S+ T G+ + ++ ++ +D
Sbjct: 252 SFRQHLDSE---NSGHRLQEAPLFMNQELSDSAAFSAHQST--GEISEVMPSVVVDVSDD 306
Query: 334 PHVNYNPNTGKETRHKSATKEKQPQTNTQIFKYAYAQLEKEKAQQQENKNLTFSGVLKMA 393
+ GK+ +SA K K T+TQIFKYAY ++EKEK +QQENKNLTF+GVL M
Sbjct: 307 GEIV----AGKD---RSAPKGKHRSTHTQIFKYAYGEIEKEKVRQQENKNLTFTGVLSMV 359
Query: 394 TNTEKS-KRPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSA 452
+ +K RP +++ F+DLTL+L K +LR++ G+++PGR+TA+MGPSGAGKTTFL+A
Sbjct: 360 SEQQKEITRPLLKVEFKDLTLSLG--KKKLLRSINGELRPGRVTAVMGPSGAGKTTFLNA 417
Query: 453 LAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSAD 512
+ GK G V+GS+L+NGR+++I S+KKIIGFVPQDDVVHGNLTVEENLWFSA+CRLSA
Sbjct: 418 VTGKVAGYKVSGSVLVNGRHDNIRSYKKIIGFVPQDDVVHGNLTVEENLWFSAKCRLSAT 477
Query: 513 LSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDE 572
+ KVL VERVI+ L LQ VR+S+VGTVEKRG+SGGQRKRVNVG+EMVMEPSLL+LDE
Sbjct: 478 TAHRHKVLTVERVIDSLDLQGVRSSLVGTVEKRGISGGQRKRVNVGIEMVMEPSLLILDE 537
Query: 573 PTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSA 632
PTSGLDS+SSQLLLRALR EALEGVN+C VVHQPSY L+NMFDDLILL KGGL+VY+G
Sbjct: 538 PTSGLDSSSSQLLLRALRHEALEGVNVCAVVHQPSYTLYNMFDDLILLAKGGLIVYNGPV 597
Query: 633 KKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYPIPLD 692
K VE+YFS LGI VPER+NPPD+YIDILEGI P SG++ + P+ WML+N Y +P D
Sbjct: 598 KSVEDYFSTLGITVPERVNPPDHYIDILEGIVKP--ESGINAKHFPLHWMLYNGYEVPND 655
Query: 693 MRQHAAQFG--------IPQSVNSANDLGEVGKTFAGELWNDVRSNVELRGEKIRLNFLK 744
M+ G P + ++ + L V FA E ++I + K
Sbjct: 656 MKDDLKAIGEQRPHLGSSPSAGSTPHCLPHVRNAFAEE------------RDRIEHHLSK 703
Query: 745 SKDLSNRKTPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKSSDQTFG 804
KDLS+R+TPGV +QYKY+L RV KQRLREAR+ AVD+LIL LAG CLG+I K SD FG
Sbjct: 704 PKDLSSRRTPGVIRQYKYYLGRVTKQRLREARLLAVDFLILGLAGICLGTIAKLSDPNFG 763
Query: 805 ASGYTYTVIAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDHFNTVIKPV 864
GY YT+IAVSLLCKIAALRSFSL++L Y RE +SGMSSLAYFL++DT+DHF+T++KP+
Sbjct: 764 MPGYIYTIIAVSLLCKIAALRSFSLERLQYLRERESGMSSLAYFLARDTIDHFSTIVKPI 823
Query: 865 VYLSMFYFLTNPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPGAAQLWSVLLPVVSTLI 924
VYLSMFY+ NPRST TDNYI+LL LVYCVTGI Y +I F PG+AQL S L+PVV TL+
Sbjct: 824 VYLSMFYYFNNPRSTITDNYIILLALVYCVTGIGYTFAICFNPGSAQLCSALIPVVLTLL 883
Query: 925 ATQQKDSKILKAIANLCYSKWALQALVIANAERYQGVWLITRCGSLLKSGYNLHDWSLCI 984
+TQ IL LCY KWAL+ +I NA+RY GVWLITRCG L +S +++H + LCI
Sbjct: 884 STQNNTPAILN---RLCYPKWALEGFIIVNAKRYPGVWLITRCGLLFRSRFDIHHYMLCI 940
Query: 985 SILILMGVIGRAIAFFCMVTFKK 1007
+L + G+ R +AF ++ KK
Sbjct: 941 LVLFMYGLFFRIVAFVALILVKK 963
Score = 38.1 bits (87), Expect = 1.4
Identities = 15/36 (41%), Positives = 23/36 (63%)
Query: 3 DWNQAFNFSSDLRFLSSCIKKTKGDISNRLCTAAEV 38
DWN+AFN++SDL F+ +C+ +T+ C A V
Sbjct: 86 DWNEAFNYTSDLGFVQNCLAETRACPLGSYCPRATV 121
>UniRef100_Q7XE13 Putative ABC transporter [Oryza sativa]
Length = 908
Score = 747 bits (1929), Expect = 0.0
Identities = 406/803 (50%), Positives = 527/803 (65%), Gaps = 91/803 (11%)
Query: 215 CSDQVLTTRERRVAKSRESAARSARKTANAHQRWKVAKDAAKKGATGLQAQLSRKFSRKK 274
CSDQ + R + ++KSR AA A+++A A RWK+AK+
Sbjct: 186 CSDQFIKIRAKILSKSRRKAATIAQESATARGRWKLAKEL-------------------- 225
Query: 275 DEENLEKVKILNQETSETDVELLPHSQPSNMVASSSAVPTEKGKTPSGLMHMMHEIENDP 334
+L+ E L S+ + ASS+ H E +
Sbjct: 226 ---------VLSHE--------LEMSESDQLAASSN--------------EARHATEGN- 253
Query: 335 HVNYNPNTGKETRHKSATKEKQPQTNTQIFKYAYAQLEKEKAQQQENKNLTFSGVLKMAT 394
GK +++ ++K T+ F+ AY+Q+ +E+ Q +N +T SGV+ +A
Sbjct: 254 --------GKRSKN----RKKLAHARTERFRRAYSQIGRERVLQPDNDKITLSGVVALAA 301
Query: 395 NTEKSKRPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALA 454
+S+RP E+ F+ LTL++ K +L+ VTGK+ PGR+TAIMGPSGAGKTTFL+A+
Sbjct: 302 E-NRSRRPMFEVVFKGLTLSI--GKKKLLQCVTGKLSPGRVTAIMGPSGAGKTTFLNAVL 358
Query: 455 GKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRL----- 509
GK G G +LING++ S+ S+KKIIGFVPQDD+VHGNLTVEENLWFSA C L
Sbjct: 359 GKTTGYKKDGLVLINGKSGSMQSYKKIIGFVPQDDIVHGNLTVEENLWFSACCSLKFSFA 418
Query: 510 ------SADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVM 563
S +SK +K++V+ERVI LGLQ +RNS+VGTVEKRG+SGGQRKRVNVG+EMVM
Sbjct: 419 NPHPWSSKGMSKSDKIIVLERVIGSLGLQEIRNSLVGTVEKRGISGGQRKRVNVGIEMVM 478
Query: 564 EPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKG 623
EPSLL+LDEPT+GLDSASSQLLLRALR EAL+GVN+C V+HQPSY LFNMFDD +LL +G
Sbjct: 479 EPSLLILDEPTTGLDSASSQLLLRALRHEALQGVNVCAVIHQPSYTLFNMFDDFVLLARG 538
Query: 624 GLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWML 683
GL+ Y G +VE YFS LGI VPER NPPDYYIDILEGI + + LP+ WML
Sbjct: 539 GLIAYLGPISEVETYFSSLGIKVPERENPPDYYIDILEGITKTKMRGHAAPKHLPLLWML 598
Query: 684 HNEYPIPLDMRQHAAQFGIPQSVNSANDLGEVGKTFAGELWNDVRSNVELRGEKIRLNFL 743
N Y +P M++ + +N+ ++L VG E + D N + + N
Sbjct: 599 RNGYEVPEYMQKDL------EDINNVHELYTVGSMSREESFGDQSEN----ADSVHQNVR 648
Query: 744 KSKDLSNRKTPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKSSDQTF 803
+ L +RKTPGV QYKY+L RV KQRLREA +QAVDYLIL +AG C+G+I K D TF
Sbjct: 649 EPYSLLDRKTPGVLAQYKYYLGRVTKQRLREATLQAVDYLILCIAGICIGTIAKVKDDTF 708
Query: 804 GASGYTYTVIAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDHFNTVIKP 863
G + Y YT+IAVSLLC++AALRSFS ++L YWRE +SGMS+LAYFL++DT+DHFNT++KP
Sbjct: 709 GVASYGYTIIAVSLLCQLAALRSFSPERLQYWRERESGMSTLAYFLARDTIDHFNTLVKP 768
Query: 864 VVYLSMFYFLTNPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPGAAQLWSVLLPVVSTL 923
V +LS FYF NPRS F DNY+V L LVYCVTGI Y +I FE G AQL S L+PVV L
Sbjct: 769 VAFLSTFYFFNNPRSEFKDNYLVFLALVYCVTGIGYTFAIWFELGLAQLCSALIPVVLVL 828
Query: 924 IATQQKDSKILKAIANLCYSKWALQALVIANAERYQGVWLITRCGSLLKSGYNLHDWSLC 983
+ TQ +K LCY KWAL+AL+IA A++Y GVWLITRCG+LLK GY+++++ LC
Sbjct: 829 VGTQPNIPNFIK---GLCYPKWALEALIIAGAKKYSGVWLITRCGALLKGGYDINNFVLC 885
Query: 984 ISILILMGVIGRAIAFFCMVTFK 1006
I I++LMGV+ R IA ++ K
Sbjct: 886 IVIVMLMGVLFRFIALLSLLKLK 908
Score = 45.8 bits (107), Expect = 0.007
Identities = 23/62 (37%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query: 3 DWNQAFNFSSDLRFLSSCIKKTKGDISNRLCTAAEVKFYLNSLMEKSSSANYLKPNRNCN 62
D+ QAF+FS+ F+S C+++T+G ++ LC AE++ Y+ SL +K S+ + N
Sbjct: 91 DFTQAFSFSN-ASFVSDCMEETQGQMTGMLCGKAEIEIYVKSLGKKPSTRGITVQQQPKN 149
Query: 63 LT 64
LT
Sbjct: 150 LT 151
>UniRef100_Q9SJK6 Putative ABC transporter [Arabidopsis thaliana]
Length = 386
Score = 507 bits (1305), Expect = e-142
Identities = 249/366 (68%), Positives = 305/366 (83%), Gaps = 4/366 (1%)
Query: 401 RPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGC 460
RP IE++F+DLTLTLK ++KHILR+VTGKI PGR++A+MGPSGAGKTTFLSALAGKA GC
Sbjct: 4 RPVIEVAFKDLTLTLKGKHKHILRSVTGKIMPGRVSAVMGPSGAGKTTFLSALAGKATGC 63
Query: 461 LVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVL 520
TG ILINGRN+SI+S+KKI GFVPQDDVVHGNLTVEENL FSA+CRLSA +SK +KVL
Sbjct: 64 TRTGLILINGRNDSINSYKKITGFVPQDDVVHGNLTVEENLRFSARCRLSAYMSKADKVL 123
Query: 521 VVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSA 580
++ERVIE LGLQ VR+S+VGT+EKRG+SGGQRKRVNVG+EMVMEPSLL+LDEPT+GLDSA
Sbjct: 124 IIERVIESLGLQHVRDSLVGTIEKRGISGGQRKRVNVGVEMVMEPSLLILDEPTTGLDSA 183
Query: 581 SSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFS 640
SSQLLLRALRREALEGVNICMVVHQPSY ++ MFDD+I+L KGGL VYHGS KK+EEYF+
Sbjct: 184 SSQLLLRALRREALEGVNICMVVHQPSYTMYKMFDDMIILAKGGLTVYHGSVKKIEEYFA 243
Query: 641 GLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYPIPLDMRQHAAQF 700
+GI VP+R+NPPD+YIDILEGI P G ++ + LPV+WMLHN YP+P DM +
Sbjct: 244 DIGITVPDRVNPPDHYIDILEGIVKPDGD--ITIEQLPVRWMLHNGYPVPHDMLKFCD-- 299
Query: 701 GIPQSVNSANDLGEVGKTFAGELWNDVRSNVELRGEKIRLNFLKSKDLSNRKTPGVFKQY 760
G+P S + +F+ +LW DV++NVE+ ++++ N+ S D SNR TP V +QY
Sbjct: 300 GLPSSSTGSAQEDSTHNSFSNDLWQDVKTNVEITKDQLQHNYSNSHDNSNRVTPTVGRQY 359
Query: 761 KYFLIR 766
+YF+ R
Sbjct: 360 RYFVGR 365
>UniRef100_UPI000028D6BC UPI000028D6BC UniRef100 entry
Length = 301
Score = 234 bits (598), Expect = 8e-60
Identities = 132/285 (46%), Positives = 168/285 (58%), Gaps = 1/285 (0%)
Query: 404 IEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVT 463
I I FR L L L A + L V+ G + AIMGPSG GK+T L+ L GKA ++
Sbjct: 6 ITIEFRKLGLRL-ASGEVALAGVSATFAAGTVVAIMGPSGCGKSTLLNVLCGKATYGNMS 64
Query: 464 GSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVE 523
G +LING ESI S +GFVPQDD+VH LTV EN+ FSA R S + E +V+
Sbjct: 65 GQLLINGEEESISSIAHKVGFVPQDDIVHEKLTVLENIAFSAHLRNSRSKKEDEFASIVD 124
Query: 524 RVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQ 583
+ L L +R SVVG+VE+RG+SGGQRKRVN+GLE+ PSLL+LDEPTSGLD+ SS
Sbjct: 125 DCLSVLQLHHIRMSVVGSVEERGISGGQRKRVNIGLELASSPSLLLLDEPTSGLDATSSL 184
Query: 584 LLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLG 643
++ AL A GV + MV+HQP +LF +FD ++LL KGG V+ GS YF LG
Sbjct: 185 MVCTALSNLAKLGVTVVMVIHQPRRSLFQLFDKVLLLAKGGHAVFEGSPDTAILYFRSLG 244
Query: 644 INVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYP 688
+ P +P D +DI+ G G DL W E P
Sbjct: 245 FHFPPNESPSDVLLDIISGSVPREGYPEFKAADLISLWKKETESP 289
>UniRef100_Q7S0X8 Hypothetical protein [Neurospora crassa]
Length = 1812
Score = 233 bits (593), Expect = 3e-59
Identities = 164/557 (29%), Positives = 272/557 (48%), Gaps = 45/557 (8%)
Query: 404 IEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVT 463
+ + LT LK + IL+NVTG I G +TA+MG SGAGK+TF++ L GKA
Sbjct: 426 LSFGYDQLTFALKNSKRPILQNVTGSINSGSLTAVMGGSGAGKSTFINVLMGKAQ--YTK 483
Query: 464 GSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVE 523
GS+ +NG + +KK+IG+VPQDD+V LTV EN+ SA+ RL + V
Sbjct: 484 GSVTVNGIPGKLKQYKKLIGYVPQDDIVLPELTVYENIMHSAKIRLPRTWKDSDVETHVN 543
Query: 524 RVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQ 583
VI+ L L VR+S+VG+V K +SGGQRKRV++G+E+ P + LDEPTSGLD+ S+
Sbjct: 544 VVIDCLELSHVRDSLVGSVGKPVISGGQRKRVSIGMELAAAPMAIFLDEPTSGLDATSAS 603
Query: 584 LLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLG 643
++ L+ G+ + +++HQP +F M DD+ILLG G L +Y G + + +F +G
Sbjct: 604 SIMSTLKALGKLGITVIVIIHQPRVEIFEMLDDMILLGNGQL-IYEGPQSQAKTFFEQMG 662
Query: 644 INVPERINPPDYYIDILEGIAAPGGSSG-LSYQDLPVKWMLHNEYPIPLDMRQHAAQFGI 702
P N D DI+ G G +S L +W + + L + F
Sbjct: 663 FVFPPGANCSDVITDIITGNGRRYKEEGDISKDALIAQWAQGSR--VSLLTNESVDTFWQ 720
Query: 703 PQSVNSANDLGEVGKTFAGELWNDVRSNVELRGEKIRLNFLKS------KDLSNRKTPGV 756
+ +D+ T AG +D ++ R + ++ LK+ +SN + +
Sbjct: 721 SSHSRNPSDM-----TLAGSESSDADTSRHNRSSLMSISKLKAGARISMLSISNPQLSRI 775
Query: 757 FK-----QYKYFLIRVGKQRLREARIQA---VDYLILLLAGACLGSITKSSDQTFGASGY 808
K +++ L+ + + L++ R ++ + + LAG LG K + F Y
Sbjct: 776 LKHRGAPRWRQTLLCLSRSILQQYRTRSNLWFEIGLSTLAGVLLGLAQKPKNGAFFRGQY 835
Query: 809 T--------------------YTVIAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYF 848
+A+ L ++ FS D L Y RE+++G S +AYF
Sbjct: 836 NKPYEILSLSADYVAAPQMAMLITLAIGLSSAAPGVKLFSEDMLVYRREAEAGHSRVAYF 895
Query: 849 LSKDTMDHFNTVIKPVVYLSMFYFLTNPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPG 908
L+K + + + + + ++ P F ++ L VYC+ G+A +S+V
Sbjct: 896 LAKTISVLPRMALACLHFSAPLFLISMPVIPFWTAFLANLLYVYCIYGLASCMSMVVRRE 955
Query: 909 AAQLWSVLLPVVSTLIA 925
A L + ++ ++ +++
Sbjct: 956 DAPLLATMVSLIVGILS 972
>UniRef100_Q7TMS5 ATP-binding cassette, sub-family G, member 2 [Mus musculus]
Length = 657
Score = 211 bits (538), Expect = 7e-53
Identities = 171/566 (30%), Positives = 271/566 (47%), Gaps = 60/566 (10%)
Query: 378 QQENKNLTFSGVLKMATNTEKSKRPFIEISFRDLT----LTLKAQNKHILRNVTGKIKPG 433
Q+ N L + + T E F I++R L K K IL ++ G +KPG
Sbjct: 14 QRNNNGLPRTNSRAVRTLAEGDVLSFHHITYRVKVKSGFLVRKTVEKEILSDINGIMKPG 73
Query: 434 RITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHG 493
+ AI+GP+G GK++ L LA + ++G +LING + H FK G+V QDDVV G
Sbjct: 74 -LNAILGPTGGGKSSLLDVLAARKDPKGLSGDVLINGAPQPAH-FKCCSGYVVQDDVVMG 131
Query: 494 NLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRK 553
LTV ENL FSA RL + EK + +I+ LGL+ V +S VGT RG+SGG+RK
Sbjct: 132 TLTVRENLQFSAALRLPTTMKNHEKNERINTIIKELGLEKVADSKVGTQFIRGISGGERK 191
Query: 554 RVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNM 613
R ++G+E++ +PS+L LDEPT+GLDS+++ +L L+R + +G I +HQP Y++F +
Sbjct: 192 RTSIGMELITDPSILFLDEPTTGLDSSTANAVLLLLKRMSKQGRTIIFSIHQPRYSIFKL 251
Query: 614 FDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLS 673
FD L LL G L V+HG A+K EYF+ G + NP D+++D++ G SS +
Sbjct: 252 FDSLTLLASGKL-VFHGPAQKALEYFASAGYHCEPYNNPADFFLDVIN-----GDSSAV- 304
Query: 674 YQDLPVKWMLHNEYPIPLDMRQHAAQFGIPQSVNSANDLGEVGKTFAGELWN--DVRSNV 731
ML+ E + N AN E K + N + N
Sbjct: 305 --------MLNRE-----------------EQDNEANKTEEPSKGEKPVIENLSEFYINS 339
Query: 732 ELRGE-KIRLNFL----KSKDLSNRKTP----GVFKQYKYFLIRVGKQRLREARIQAVDY 782
+ GE K L+ L + K S K P Q ++ R K L +
Sbjct: 340 AIYGETKAELDQLPGAQEKKGTSAFKEPVYVTSFCHQLRWIARRSFKNLLGNPQASVAQL 399
Query: 783 LILLLAGACLGSITKSSDQTFGA------SGYTYTVIAVSLLCKIAALRSFSLDKLHYWR 836
++ ++ G +G+I D + A +G + + ++A+ F ++K +
Sbjct: 400 IVTVILGLIIGAI--YFDLKYDAAGMQNRAGVLFFLTTNQCFSSVSAVELFVVEKKLFIH 457
Query: 837 ESDSGMSSL-AYFLSKDTMDHFNTVIKP-VVYLSMFYFLTNPRSTFTDNYIVLLCLV-YC 893
E SG + +YF K D P V++ + YF+ + T +I++ L+
Sbjct: 458 EYISGYYRVSSYFFGKVMSDLLPMRFLPSVIFTCVLYFMLGLKKTVDAFFIMMFTLIMVA 517
Query: 894 VTGIAYALSIVFEPGAAQLWSVLLPV 919
T + AL+I + ++L+ +
Sbjct: 518 YTASSMALAIATGQSVVSVATLLMTI 543
>UniRef100_Q9R004 Breast cancer resistance protein 1 [Mus musculus]
Length = 657
Score = 211 bits (536), Expect = 1e-52
Identities = 171/566 (30%), Positives = 270/566 (47%), Gaps = 60/566 (10%)
Query: 378 QQENKNLTFSGVLKMATNTEKSKRPFIEISFRDLT----LTLKAQNKHILRNVTGKIKPG 433
Q+ N L + T E F I++R L K K IL ++ G +KPG
Sbjct: 14 QRNNNGLPRMNSRAVRTLAEGDVLSFHHITYRVKVKSGFLVRKTVEKEILSDINGIMKPG 73
Query: 434 RITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHG 493
+ AI+GP+G GK++ L LA + ++G +LING + H FK G+V QDDVV G
Sbjct: 74 -LNAILGPTGGGKSSLLDVLAARKDPKGLSGDVLINGAPQPAH-FKCCSGYVVQDDVVMG 131
Query: 494 NLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRK 553
LTV ENL FSA RL + EK + +I+ LGL+ V +S VGT RG+SGG+RK
Sbjct: 132 TLTVRENLQFSAALRLPTTMKNHEKNERINTIIKELGLEKVADSKVGTQFIRGISGGERK 191
Query: 554 RVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNM 613
R ++G+E++ +PS+L LDEPT+GLDS+++ +L L+R + +G I +HQP Y++F +
Sbjct: 192 RTSIGMELITDPSILFLDEPTTGLDSSTANAVLLLLKRMSKQGRTIIFSIHQPRYSIFKL 251
Query: 614 FDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLS 673
FD L LL G L V+HG A+K EYF+ G + NP D+++D++ G SS +
Sbjct: 252 FDSLTLLASGKL-VFHGPAQKALEYFASAGYHCEPYNNPADFFLDVIN-----GDSSAV- 304
Query: 674 YQDLPVKWMLHNEYPIPLDMRQHAAQFGIPQSVNSANDLGEVGKTFAGELWN--DVRSNV 731
ML+ E + N AN E K + N + N
Sbjct: 305 --------MLNRE-----------------EQDNEANKTEEPSKGEKPVIENLSEFYINS 339
Query: 732 ELRGE-KIRLNFL----KSKDLSNRKTP----GVFKQYKYFLIRVGKQRLREARIQAVDY 782
+ GE K L+ L + K S K P Q ++ R K L +
Sbjct: 340 AIYGETKAELDQLPGAQEKKGTSAFKEPVYVTSFCHQLRWIARRSFKNLLGNPQASVAQL 399
Query: 783 LILLLAGACLGSITKSSDQTFGA------SGYTYTVIAVSLLCKIAALRSFSLDKLHYWR 836
++ ++ G +G+I D + A +G + + ++A+ F ++K +
Sbjct: 400 IVTVILGLIIGAI--YFDLKYDAAGMQNRAGVLFFLTTNQCFSSVSAVELFVVEKKLFIH 457
Query: 837 ESDSGMSSL-AYFLSKDTMDHFNTVIKP-VVYLSMFYFLTNPRSTFTDNYIVLLCLV-YC 893
E SG + +YF K D P V++ + YF+ + T +I++ L+
Sbjct: 458 EYISGYYRVSSYFFGKVMSDLLPMRFLPSVIFTCILYFMLGLKKTVDAFFIMMFTLIMVA 517
Query: 894 VTGIAYALSIVFEPGAAQLWSVLLPV 919
T + AL+I + ++L+ +
Sbjct: 518 YTASSMALAIATGQSVVSVATLLMTI 543
>UniRef100_Q69Q33 Putative ATP-binding cassette transporter1 [Oryza sativa]
Length = 688
Score = 210 bits (534), Expect = 2e-52
Identities = 165/509 (32%), Positives = 251/509 (48%), Gaps = 45/509 (8%)
Query: 420 KHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFK 479
KHIL+ + G + PG I A+MGPSG+GKTT L L G+ G V G I N S K
Sbjct: 89 KHILKGIGGSVDPGEILALMGPSGSGKTTLLKILGGRLSGG-VKGQITYNDTPYS-PCLK 146
Query: 480 KIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVV 539
+ IGFV QDDV+ LTVEE L F+A RL A +SK +K V+ +I L L+ R++ +
Sbjct: 147 RRIGFVTQDDVLFPQLTVEETLVFAAFLRLPARMSKQQKRDRVDAIITELNLERCRHTKI 206
Query: 540 GTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGV-- 597
G RGVSGG+RKR ++G E++++PSLL+LDEPTSGLDS S+ LL LRR A
Sbjct: 207 GGAFVRGVSGGERKRTSIGYEILVDPSLLLLDEPTSGLDSTSAAKLLVVLRRLARSAARR 266
Query: 598 NICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYI 657
+ +HQPS +F+MFD L+L+ +G +YHG A+ +F+ LG + +NP ++ +
Sbjct: 267 TVITTIHQPSSRMFHMFDKLLLVAEGH-AIYHGGARGCMRHFAALGFSPGIAMNPAEFLL 325
Query: 658 DI----LEGIAAPGG----SSGLSYQDLPVKWMLHNEYPIPLDMRQHAAQFGIPQSVNSA 709
D+ L+GI++P S+ + D P + I +H A + +A
Sbjct: 326 DLATGNLDGISSPASLLLPSAAAASPDSPE----FRSHVIKYLQARHRAAGEEEAAAAAA 381
Query: 710 NDLGEVGKTFAGELWNDVRSNVELRGEKIRLNFLKSKDLSNRKTPGVFKQYKYFLIRVGK 769
+ G G E +R V +R ++ R G +Q+ R +
Sbjct: 382 REGGGGGGAGRDEAAKQLRMAVRMRKDR-------------RGGIGWLEQFTVLSRRTFR 428
Query: 770 QR----LREARIQAVDYLILLLAGACLGSITKSSDQTFGASGYT-YTVIAVSLLCKIAAL 824
+R L + R+ + LLL S T + Q G Y I + ++
Sbjct: 429 ERAADYLDKMRLAQSVGVALLLGLLWWKSQTSNEAQLRDQVGLIFYICIFWTSSSLFGSV 488
Query: 825 RSFSLDKLHYWRESDSGMSSL-AYFLSKDTMDHFNTVIKPVVYLSMFYFLTNPRSTFTDN 883
F +KL+ +E + M L AY+ S D V+ PV++ ++ YF+ + R T
Sbjct: 489 YVFPFEKLYLVKERKADMYRLSAYYASSTVCDAVPHVVYPVLFTAILYFMADLRRTVP-- 546
Query: 884 YIVLLCLVYCVTGIAYALSIVFEPGAAQL 912
+C+T +A L ++ G +L
Sbjct: 547 -------CFCLTLLATLLIVLTSQGTGEL 568
>UniRef100_UPI000027C604 UPI000027C604 UniRef100 entry
Length = 354
Score = 209 bits (532), Expect = 3e-52
Identities = 121/294 (41%), Positives = 179/294 (60%), Gaps = 13/294 (4%)
Query: 379 QENKNLTFSGVLKMATNTEKSKRPFIEISFRDLTLTLK----------AQNKHILRNVTG 428
Q N L +G K S++ +SF ++ +K +K IL ++ G
Sbjct: 5 QVNIELELNGASKQQPAASASQQQGATVSFHNIHYKVKEGGSCLCRRKTSSKAILIDLNG 64
Query: 429 KIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQD 488
+KPG + AIMG +G+GK++FL LA + +TG +LI+G + +FK + G+V QD
Sbjct: 65 IMKPG-LNAIMGATGSGKSSFLDILAARKDPAGLTGEVLIDGAPQP-PNFKCLSGYVVQD 122
Query: 489 DVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVS 548
DVV G LTV ENL FSA RL A + + EK V ++IE LGL V +S VGT RGVS
Sbjct: 123 DVVMGTLTVRENLNFSAALRLPAHVPQKEKEQKVNKLIEELGLGRVADSRVGTQLIRGVS 182
Query: 549 GGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSY 608
GG+RKR N+G+E++++PS+L LDEPT+GLD++++ +L L+R A G I + +HQP Y
Sbjct: 183 GGERKRTNIGMELIIDPSVLFLDEPTTGLDASTANSVLLLLKRMANNGRTIILSIHQPRY 242
Query: 609 ALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDILEG 662
+++ +FD L LL G VYHG A++ EYFS +G NP D+++D++ G
Sbjct: 243 SIYRLFDSLTLL-VNGKQVYHGPAQRALEYFSDIGYTCETHNNPADFFLDVING 295
>UniRef100_Q9C6W5 Hypothetical protein F27M3_2 [Arabidopsis thaliana]
Length = 648
Score = 207 bits (528), Expect = 1e-51
Identities = 110/248 (44%), Positives = 163/248 (65%), Gaps = 3/248 (1%)
Query: 416 KAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESI 475
K++ K IL +TG + PG A++GPSG+GKTT LSAL G+ L +G ++ NG+ S
Sbjct: 75 KSKEKTILNGITGMVCPGEFLAMLGPSGSGKTTLLSALGGR-LSKTFSGKVMYNGQPFS- 132
Query: 476 HSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVR 535
K+ GFV QDDV++ +LTV E L+F+A RL + L++ EK V+RVI LGL
Sbjct: 133 GCIKRRTGFVAQDDVLYPHLTVWETLFFTALLRLPSSLTRDEKAEHVDRVIAELGLNRCT 192
Query: 536 NSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALE 595
NS++G RG+SGG++KRV++G EM++ PSLL+LDEPTSGLDS ++ ++ ++R A
Sbjct: 193 NSMIGGPLFRGISGGEKKRVSIGQEMLINPSLLLLDEPTSGLDSTTAHRIVTTIKRLASG 252
Query: 596 GVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDY 655
G + +HQPS +++MFD ++LL +G +Y+G+A EYFS LG + +NP D
Sbjct: 253 GRTVVTTIHQPSSRIYHMFDKVVLLSEGS-PIYYGAASSAVEYFSSLGFSTSLTVNPADL 311
Query: 656 YIDILEGI 663
+D+ GI
Sbjct: 312 LLDLANGI 319
>UniRef100_Q9C6R7 ABC transporter, putative [Arabidopsis thaliana]
Length = 646
Score = 207 bits (528), Expect = 1e-51
Identities = 110/248 (44%), Positives = 163/248 (65%), Gaps = 3/248 (1%)
Query: 416 KAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESI 475
K++ K IL +TG + PG A++GPSG+GKTT LSAL G+ L +G ++ NG+ S
Sbjct: 73 KSKEKTILNGITGMVCPGEFLAMLGPSGSGKTTLLSALGGR-LSKTFSGKVMYNGQPFS- 130
Query: 476 HSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVR 535
K+ GFV QDDV++ +LTV E L+F+A RL + L++ EK V+RVI LGL
Sbjct: 131 GCIKRRTGFVAQDDVLYPHLTVWETLFFTALLRLPSSLTRDEKAEHVDRVIAELGLNRCT 190
Query: 536 NSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALE 595
NS++G RG+SGG++KRV++G EM++ PSLL+LDEPTSGLDS ++ ++ ++R A
Sbjct: 191 NSMIGGPLFRGISGGEKKRVSIGQEMLINPSLLLLDEPTSGLDSTTAHRIVTTIKRLASG 250
Query: 596 GVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDY 655
G + +HQPS +++MFD ++LL +G +Y+G+A EYFS LG + +NP D
Sbjct: 251 GRTVVTTIHQPSSRIYHMFDKVVLLSEGS-PIYYGAASSAVEYFSSLGFSTSLTVNPADL 309
Query: 656 YIDILEGI 663
+D+ GI
Sbjct: 310 LLDLANGI 317
>UniRef100_Q80ST1 ATP-binding cassette protein G2 transcript variant B [Rattus
norvegicus]
Length = 657
Score = 206 bits (525), Expect = 2e-51
Identities = 162/562 (28%), Positives = 270/562 (47%), Gaps = 53/562 (9%)
Query: 379 QENKNLTFSGVLKMATNTEKSKRPFIEISFRDLTLTLKAQN---------KHILRNVTGK 429
Q NKN G+ M++ ++ +SF +T +K ++ K IL ++ G
Sbjct: 14 QRNKN----GLPGMSSRGARTLAEGDVLSFHHITYRVKVKSGFLVRKTAEKEILSDINGI 69
Query: 430 IKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDD 489
+KPG + AI+GP+G GK++ L LA + ++G +LING + + FK G+V QDD
Sbjct: 70 MKPG-LNAILGPTGGGKSSLLDVLAARKDPRGLSGDVLINGAPQPAN-FKCSSGYVVQDD 127
Query: 490 VVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSG 549
VV G LTV ENL FSA RL + EK + +I+ LGL V +S VGT RG+SG
Sbjct: 128 VVMGTLTVRENLQFSAALRLPKAMKTHEKNERINTIIKELGLDKVADSKVGTQFTRGISG 187
Query: 550 GQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYA 609
G+RKR ++G+E++ +PS+L LDEPT+GLDS+++ +L L+R + +G I +HQP Y+
Sbjct: 188 GERKRTSIGMELITDPSILFLDEPTTGLDSSTANAVLLLLKRMSKQGRTIIFSIHQPRYS 247
Query: 610 LFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGS 669
+F +FD L LL G LM +HG A+K EYF+ G + NP D+++D++ G
Sbjct: 248 IFKLFDSLTLLASGKLM-FHGPAQKALEYFASAGYHCEPYNNPADFFLDVING------- 299
Query: 670 SGLSYQDLPVKWMLHNEYPIPLDMRQHAAQFGIPQSVNSANDLGEVGKTFAGELWNDVRS 729
D + E + + ++ P N A + T G
Sbjct: 300 ------DSSAVMLNRGEQDHEANKTEEPSKREKPIIENLAEFY--INSTIYG-------- 343
Query: 730 NVELRGEKIRLNFLKSKDLSNRKTPGVF-----KQYKYFLIRVGKQRLREARIQAVDYLI 784
E + E +L + K S+ V+ Q ++ R K L + ++
Sbjct: 344 --ETKAELDQLPVAQKKKGSSAFREPVYVTSFCHQLRWIARRSFKNLLGNPQASVAQLIV 401
Query: 785 LLLAGACLGSI--TKSSDQT--FGASGYTYTVIAVSLLCKIAALRSFSLDKLHYWRESDS 840
++ G +G++ +D T +G + + ++A+ F ++K + E S
Sbjct: 402 TVILGLIIGALYFGLKNDPTGMQNRAGVFFFLTTNQCFTSVSAVELFVVEKKLFIHEYIS 461
Query: 841 GMSSL-AYFLSKDTMDHFNTVIKP-VVYLSMFYFLTNPRSTFTDNYIVLLCLV-YCVTGI 897
G + +YF K D P V+Y + YF+ + T +I++ L+ T
Sbjct: 462 GYYRVSSYFFGKLVSDLLPMRFLPSVIYTCLLYFMLGLKRTVEAFFIMMFTLIMVAYTAS 521
Query: 898 AYALSIVFEPGAAQLWSVLLPV 919
+ AL+I + ++L+ +
Sbjct: 522 SMALAIAAGQSVVSVATLLMTI 543
>UniRef100_UPI000024A2F0 UPI000024A2F0 UniRef100 entry
Length = 613
Score = 205 bits (522), Expect = 5e-51
Identities = 107/243 (44%), Positives = 163/243 (67%), Gaps = 3/243 (1%)
Query: 420 KHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFK 479
K IL ++ G ++PG + AI+GP+G+GK++FL LAG+ ++G +L+NG + + FK
Sbjct: 64 KEILLDLNGIMRPG-LNAILGPTGSGKSSFLDVLAGRKDPAGLSGEVLVNGALQPAN-FK 121
Query: 480 KIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVV 539
+ G+V QDD+V G LTV ENL FSA RLS+ +S EK V +I LGL V +S V
Sbjct: 122 CLSGYVVQDDIVMGTLTVRENLSFSAALRLSSHVSPREKEARVNHLISELGLNKVADSKV 181
Query: 540 GTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNI 599
GT RG+SGG+RKR ++G+E++++PS+L LDEPT+GLD++++ +L L+R A +G I
Sbjct: 182 GTQIIRGISGGERKRTSIGMELIIDPSVLFLDEPTTGLDASTAHSVLLLLKRMAGQGRTI 241
Query: 600 CMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDI 659
M +HQP ++++ +FD L LL G VYHG A+ +YFS +G NP D+++D+
Sbjct: 242 IMSIHQPRFSIYRLFDSLTLLA-NGKQVYHGPAQDALDYFSNIGYACEAHNNPADFFLDV 300
Query: 660 LEG 662
+ G
Sbjct: 301 ING 303
>UniRef100_Q9LJC3 ABC transporter-like protein [Arabidopsis thaliana]
Length = 594
Score = 205 bits (521), Expect = 6e-51
Identities = 115/276 (41%), Positives = 174/276 (62%), Gaps = 10/276 (3%)
Query: 387 SGVLKMATNTEKSKRPFIEISFRDLTLTLK----AQNKHILRNVTGKIKPGRITAIMGPS 442
SG ++ + E S+ ++ ++ DLT+ + + +L+ + G +PGRI AIMGPS
Sbjct: 8 SGRRQLPSKLEMSRGAYL--AWEDLTVVIPNFSDGPTRRLLQRLNGYAEPGRIMAIMGPS 65
Query: 443 GAGKTTFLSALAGK-ALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENL 501
G+GK+T L +LAG+ A ++TG++L+NG+ + ++ +V Q+DV+ G LTV E +
Sbjct: 66 GSGKSTLLDSLAGRLARNVVMTGNLLLNGKKARLDY--GLVAYVTQEDVLLGTLTVRETI 123
Query: 502 WFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEM 561
+SA RL +D+SK E +VE I LGLQ + V+G RGVSGG+RKRV++ LE+
Sbjct: 124 TYSAHLRLPSDMSKEEVSDIVEGTIMELGLQDCSDRVIGNWHARGVSGGERKRVSIALEI 183
Query: 562 VMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLG 621
+ P +L LDEPTSGLDSAS+ +++ALR A +G + VHQPS +F +FDDL LL
Sbjct: 184 LTRPQILFLDEPTSGLDSASAFFVIQALRNIARDGRTVISSVHQPSSEVFALFDDLFLLS 243
Query: 622 KGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYI 657
G VY G AK E+F+ G P++ NP D+++
Sbjct: 244 SGE-SVYFGEAKSAVEFFAESGFPCPKKRNPSDHFL 278
>UniRef100_Q9UNQ0 ATP-binding cassette, sub-family G, member 2 [Homo sapiens]
Length = 655
Score = 205 bits (521), Expect = 6e-51
Identities = 112/247 (45%), Positives = 158/247 (63%), Gaps = 3/247 (1%)
Query: 416 KAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESI 475
K K IL N+ G +KPG + AI+GP+G GK++ L LA + ++G +LING
Sbjct: 57 KPVEKEILSNINGIMKPG-LNAILGPTGGGKSSLLDVLAARKDPSGLSGDVLINGAPRPA 115
Query: 476 HSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVR 535
+ FK G+V QDDVV G LTV ENL FSA RL+ ++ EK + RVIE LGL V
Sbjct: 116 N-FKCNSGYVVQDDVVMGTLTVRENLQFSAALRLATTMTNHEKNERINRVIEELGLDKVA 174
Query: 536 NSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALE 595
+S VGT RGVSGG+RKR ++G+E++ +PS+L LDEPT+GLDS+++ +L L+R + +
Sbjct: 175 DSKVGTQFIRGVSGGERKRTSIGMELITDPSILFLDEPTTGLDSSTANAVLLLLKRMSKQ 234
Query: 596 GVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDY 655
G I +HQP Y++F +FD L LL G LM +HG A++ YF G + NP D+
Sbjct: 235 GRTIIFSIHQPRYSIFKLFDSLTLLASGRLM-FHGPAQEALGYFESAGYHCEAYNNPADF 293
Query: 656 YIDILEG 662
++DI+ G
Sbjct: 294 FLDIING 300
>UniRef100_Q8RWI9 Hypothetical protein At3g21090 [Arabidopsis thaliana]
Length = 691
Score = 204 bits (520), Expect = 8e-51
Identities = 115/276 (41%), Positives = 174/276 (62%), Gaps = 10/276 (3%)
Query: 387 SGVLKMATNTEKSKRPFIEISFRDLTLTLK----AQNKHILRNVTGKIKPGRITAIMGPS 442
SG ++ + E S+ ++ ++ DLT+ + + +L+ + G +PGRI AIMGPS
Sbjct: 8 SGRRQLPSKLEMSRGAYL--AWEDLTVVIPNFSDGPTRRLLQRLNGYAEPGRIMAIMGPS 65
Query: 443 GAGKTTFLSALAGK-ALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENL 501
G+GK+T L +LAG+ A ++TG++L+NG+ + ++ +V Q+DV+ G LTV E +
Sbjct: 66 GSGKSTLLDSLAGRLARNVVMTGNLLLNGKKARLDY--GLVAYVTQEDVLLGTLTVRETI 123
Query: 502 WFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEM 561
+SA RL +D+SK E +VE I LGLQ + V+G RGVSGG+RKRV++ LE+
Sbjct: 124 TYSAHLRLPSDMSKEEVSDIVEGTIIELGLQDCSDRVIGNWHARGVSGGERKRVSIALEI 183
Query: 562 VMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLG 621
+ P +L LDEPTSGLDSAS+ +++ALR A +G + VHQPS +F +FDDL LL
Sbjct: 184 LTRPQILFLDEPTSGLDSASAFFVIQALRNIARDGRTVISSVHQPSSEVFALFDDLFLLS 243
Query: 622 KGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYI 657
G VY G AK E+F+ G P++ NP D+++
Sbjct: 244 SGE-SVYFGEAKSAVEFFAESGFPCPKKRNPSDHFL 278
>UniRef100_Q80XF3 ATP-binding cassette transporter ABCG2 [Rattus norvegicus]
Length = 657
Score = 204 bits (518), Expect = 1e-50
Identities = 117/293 (39%), Positives = 177/293 (59%), Gaps = 16/293 (5%)
Query: 379 QENKNLTFSGVLKMATNTEKSKRPFIEISFRDLTLTLKAQN---------KHILRNVTGK 429
Q NKN G+ M++ ++ +SF +T +K ++ K IL ++ G
Sbjct: 14 QRNKN----GLPGMSSRGARTLAEGDVLSFHHITYRVKVKSGFLVRKTAEKEILSDINGI 69
Query: 430 IKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDD 489
+KPG + AI+GP+G GK++ L LA + ++G +LING + + FK G+V QDD
Sbjct: 70 MKPG-LNAILGPTGGGKSSLLDVLAARKDPRGLSGDVLINGAPQPAN-FKCSSGYVVQDD 127
Query: 490 VVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSG 549
VV G LTV ENL FSA RL + EK + +I+ LGL V +S VGT RG+SG
Sbjct: 128 VVMGTLTVRENLQFSAALRLPKAMKTHEKNERINTIIKELGLDKVADSKVGTQFTRGISG 187
Query: 550 GQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYA 609
G+RKR ++G+E++ +PS+L LDEPT+GLDS+++ +L L+R + +G I +HQP Y+
Sbjct: 188 GERKRTSIGMELITDPSILFLDEPTTGLDSSTANAVLLLLKRMSKQGRTIIFSIHQPRYS 247
Query: 610 LFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDILEG 662
+F +FD L LL G LM +HG A+K EYF+ G + NP D+++D++ G
Sbjct: 248 IFKLFDSLTLLASGKLM-FHGPAQKALEYFASAGYHCEPYNNPADFFLDVING 299
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,605,923,834
Number of Sequences: 2790947
Number of extensions: 66382029
Number of successful extensions: 314099
Number of sequences better than 10.0: 24950
Number of HSP's better than 10.0 without gapping: 17820
Number of HSP's successfully gapped in prelim test: 7136
Number of HSP's that attempted gapping in prelim test: 263427
Number of HSP's gapped (non-prelim): 34334
length of query: 1007
length of database: 848,049,833
effective HSP length: 137
effective length of query: 870
effective length of database: 465,690,094
effective search space: 405150381780
effective search space used: 405150381780
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 80 (35.4 bits)
Medicago: description of AC144720.7


