

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.5 - phase: 0
(458 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
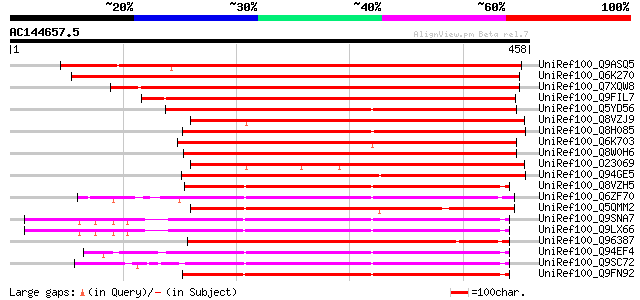
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ASQ5 At2g11520/F14P14.15 [Arabidopsis thaliana] 523 e-147
UniRef100_Q6K270 Nodulation receptor kinase-like protein [Oryza ... 490 e-137
UniRef100_Q7XQW8 OSJNBa0014K14.7 protein [Oryza sativa] 470 e-131
UniRef100_Q9FIL7 Similarity to protein kinase [Arabidopsis thali... 341 3e-92
UniRef100_Q5YD56 Calcium/calmodulin-regulated receptor-like kina... 332 1e-89
UniRef100_Q8VZJ9 Hypothetical protein At4g00330 [Arabidopsis tha... 330 4e-89
UniRef100_Q8H085 Hypothetical protein OSJNBb0050N02.6 [Oryza sat... 329 9e-89
UniRef100_Q6K703 Receptor protein kinase-like [Oryza sativa] 326 1e-87
UniRef100_Q8W0H6 P0529E05.2 protein [Oryza sativa] 320 4e-86
UniRef100_O23069 A_IG005I10.8 protein [Arabidopsis thaliana] 318 2e-85
UniRef100_Q94GE5 Putative receptor kinase [Oryza sativa] 266 1e-69
UniRef100_Q8VZH5 Receptor protein kinase-like protein [Capsicum ... 244 5e-63
UniRef100_Q6ZF70 Putative PTH-2, resistance gene (PTO kinase) ho... 241 2e-62
UniRef100_Q5QMM2 Protein kinase-like [Oryza sativa] 241 3e-62
UniRef100_Q9SNA7 Receptor-like protein kinase homolog [Arabidops... 240 5e-62
UniRef100_Q9LX66 Receptor protein kinase-like [Arabidopsis thali... 240 5e-62
UniRef100_Q96387 Receptor-like protein kinase precursor [Cathara... 240 5e-62
UniRef100_Q94EF4 Hypothetical protein P0665A11.8 [Oryza sativa] 240 7e-62
UniRef100_Q9SC72 L1332.5 protein [Oryza sativa] 240 7e-62
UniRef100_Q9FN92 Receptor-like protein kinase [Arabidopsis thali... 239 9e-62
>UniRef100_Q9ASQ5 At2g11520/F14P14.15 [Arabidopsis thaliana]
Length = 510
Score = 523 bits (1348), Expect = e-147
Identities = 265/414 (64%), Positives = 323/414 (78%), Gaps = 9/414 (2%)
Query: 46 STAGRKLLQKDLNNNSTSQGDPKQVSSSQVGIFAGGALLVCCAVVCPCFYGKRRKATSHA 105
S GR+ L++ +S + + S +V I G LL+CCA+ CPCF+ K RKA SH
Sbjct: 96 SLLGRRFLEEKTVKDSKNSKPKTEYSHVKVSIAGSGFLLLCCALCCPCFH-KERKANSHE 154
Query: 106 VLEKDPNSMELGSSFEPSVS-DKIPASPLRVPPSPSR-------FSMSPKLSRLQSLHLN 157
VL K+ NS+ SSFE S S +KIP SP R PPSPSR ++MSP+ SRL L+L
Sbjct: 155 VLPKESNSVHQVSSFEMSPSSEKIPQSPFRAPPSPSRVPQSPSRYAMSPRPSRLGPLNLT 214
Query: 158 LSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELL 217
+SQ++ AT NF+++ QIGEGGFG V+K LDDG VVA+KRAK+EHFE+LRTEF SEV+LL
Sbjct: 215 MSQINTATGNFADSHQIGEGGFGVVFKGVLDDGQVVAIKRAKKEHFENLRTEFKSEVDLL 274
Query: 218 AKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHG 277
+KI HRNLVKLLGY+DKG+ER++ITE+V NGTLR+HLDG RG L+FNQRLEI IDV HG
Sbjct: 275 SKIGHRNLVKLLGYVDKGDERLIITEYVRNGTLRDHLDGARGTKLNFNQRLEIVIDVCHG 334
Query: 278 LTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGY 337
LTYLH YAE+QIIHRD+KSSNILLT+SMRAKVADFGFA+ GP +++ THI T+VKGTVGY
Sbjct: 335 LTYLHSYAERQIIHRDIKSSNILLTDSMRAKVADFGFARGGPTDSNQTHILTQVKGTVGY 394
Query: 338 LDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVA 397
LDPEYMKTYHLT KSDVYSFGILL+EILTGRRPVE K+ +ER+T+RWAF KYNEG V
Sbjct: 395 LDPEYMKTYHLTAKSDVYSFGILLVEILTGRRPVEAKRLPDERITVRWAFDKYNEGRVFE 454
Query: 398 LLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLKSS 451
L+DP +E V + KMF LAF CAAP + +RPDM+ VG+QLWAIR+ YL+ S
Sbjct: 455 LVDPNARERVDEKILRKMFSLAFQCAAPTKKERPDMEAVGKQLWAIRSSYLRRS 508
>UniRef100_Q6K270 Nodulation receptor kinase-like protein [Oryza sativa]
Length = 526
Score = 490 bits (1262), Expect = e-137
Identities = 238/397 (59%), Positives = 311/397 (77%), Gaps = 1/397 (0%)
Query: 55 KDLNNNSTSQGDPKQVSSSQVGIFAGGALLVCCAVVCPCFYGKRRKATSHAVLEKDPNSM 114
K++ + +Q D + + G +L+CC ++ PCF+ ++++ + H N++
Sbjct: 126 KNMGIHGHNQDDNDSLEGQDHLLAVPGVILLCCGLMIPCFHAEKKEVSRHNTTSIQRNAV 185
Query: 115 ELGSSFEPSVS-DKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQ 173
E +S + S S +K+P +P R+PPSPSRF+ SP+++R+ S++L + Q+ +AT+NFS + +
Sbjct: 186 ESIASLDVSTSSEKVPPTPHRIPPSPSRFAQSPQIARVGSVNLTVQQILRATQNFSPSFK 245
Query: 174 IGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYID 233
+GEGGFGTVY+A L DG VVAVKRAK++ F R EFS+EVELLAKIDHRNLV+LLG+ D
Sbjct: 246 LGEGGFGTVYRAVLPDGQVVAVKRAKKDQFAGPRDEFSNEVELLAKIDHRNLVRLLGFTD 305
Query: 234 KGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRD 293
KG+ERI+ITE+V NGTLREHLDG G+ LDFNQRLEIAIDVAH LTYLHLYAEK IIHRD
Sbjct: 306 KGHERIIITEYVPNGTLREHLDGQYGRTLDFNQRLEIAIDVAHALTYLHLYAEKTIIHRD 365
Query: 294 VKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSD 353
VKSSNILLTES RAKV+DFGFA+ GP + + THISTKVKGT GYLDPEY++TY LTPKSD
Sbjct: 366 VKSSNILLTESYRAKVSDFGFARSGPSDTEKTHISTKVKGTAGYLDPEYLRTYQLTPKSD 425
Query: 354 VYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAV 413
V+SFGILL+EIL+ RRPVELK++AEER+T+RW F+K+NEG+ +LDPL+++ V +V
Sbjct: 426 VFSFGILLVEILSARRPVELKRAAEERITIRWTFKKFNEGNRREILDPLLEDPVDDEVLE 485
Query: 414 KMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLKS 450
++ +LAF CAAP R DRP MK VGEQLW IR +Y KS
Sbjct: 486 RLLNLAFQCAAPTREDRPTMKEVGEQLWEIRKEYGKS 522
>UniRef100_Q7XQW8 OSJNBa0014K14.7 protein [Oryza sativa]
Length = 367
Score = 470 bits (1209), Expect = e-131
Identities = 232/361 (64%), Positives = 282/361 (77%), Gaps = 2/361 (0%)
Query: 90 VCPCFYGKRRKATSHAVLEKDPNSMELGSSFEPSVSDKIPASPLRVPPSPSRFSMSPKLS 149
+CPCF +R+ + VL +D NS L SS S+SD++P SPLRVP SPSRFS+S S
Sbjct: 1 MCPCFGSRRKDGSEDPVLGRDGNS--LNSSELRSMSDRVPPSPLRVPASPSRFSLSSSPS 58
Query: 150 RLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTE 209
R + L+L+L QV K T NF+ L IGEG FG VY+A L DG +VA+KRAK EHF SLR E
Sbjct: 59 RNEPLNLSLEQVIKLTHNFAPDLMIGEGYFGKVYRAQLRDGHIVAIKRAKMEHFASLRAE 118
Query: 210 FSSEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLE 269
FS+E+ LL KI+HRNLV+LLGYIDK NERI+ITE+V NGTLREHLDG RG +L FNQRLE
Sbjct: 119 FSNEIALLKKIEHRNLVQLLGYIDKRNERIVITEYVPNGTLREHLDGQRGLVLSFNQRLE 178
Query: 270 IAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHIST 329
IAIDVAHGLTYLHLYAEK IIHRDVKSSNILL E RAKVADFGFA+ GP D + I T
Sbjct: 179 IAIDVAHGLTYLHLYAEKPIIHRDVKSSNILLNEGFRAKVADFGFARTGPTEPDQSQIQT 238
Query: 330 KVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRK 389
V+GT GY+DPEY++T HLT KSDV+S+G+LLLEIL+GRRP+E++++A ER+T+RWAF K
Sbjct: 239 DVRGTAGYVDPEYLRTNHLTVKSDVFSYGVLLLEILSGRRPIEVRRAARERITVRWAFEK 298
Query: 390 YNEGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLK 449
YN G V +LDP++ E+V D+ K+FD+AF C AP R+DRP MK V E+LW IR DY K
Sbjct: 299 YNRGDVKEILDPMLTESVNEDILNKIFDVAFQCVAPTRADRPTMKEVAERLWKIRRDYAK 358
Query: 450 S 450
+
Sbjct: 359 T 359
>UniRef100_Q9FIL7 Similarity to protein kinase [Arabidopsis thaliana]
Length = 470
Score = 341 bits (874), Expect = 3e-92
Identities = 172/332 (51%), Positives = 235/332 (69%), Gaps = 3/332 (0%)
Query: 117 GSSFEPSVSDKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGE 176
G S E VS + S R SP + S S K + + ++ +AT NFS QIGE
Sbjct: 97 GRSTERKVSGQYRFSGSRFQ-SPGKDSSSSKSWHQGPVIFSFGELQRATANFSSVHQIGE 155
Query: 177 GGFGTVYKAHLDDGLVVAVKRAKREHF-ESLRTEFSSEVELLAKIDHRNLVKLLGYIDKG 235
GGFGTV+K LDDG +VA+KRA++ ++ +S EF +E+ L+KI+H NLVKL G+++ G
Sbjct: 156 GGFGTVFKGKLDDGTIVAIKRARKNNYGKSWLLEFKNEIYTLSKIEHMNLVKLYGFLEHG 215
Query: 236 NERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVK 295
+E++++ E+VANG LREHLDGLRG L+ +RLEIAIDVAH LTYLH Y + IIHRD+K
Sbjct: 216 DEKVIVVEYVANGNLREHLDGLRGNRLEMAERLEIAIDVAHALTYLHTYTDSPIIHRDIK 275
Query: 296 SSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVY 355
+SNIL+T +RAKVADFGFA+L + THIST+VKG+ GY+DP+Y++T+ LT KSDVY
Sbjct: 276 ASNILITNKLRAKVADFGFARLVSEDLGATHISTQVKGSAGYVDPDYLRTFQLTDKSDVY 335
Query: 356 SFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDP-LMQEAVKTDVAVK 414
SFG+LL+EILTGRRP+ELK+ ++R+T++WA R+ + V ++DP L + +VA K
Sbjct: 336 SFGVLLVEILTGRRPIELKRPRKDRLTVKWALRRLKDDEAVLIMDPFLKRNRAAIEVAEK 395
Query: 415 MFDLAFNCAAPVRSDRPDMKTVGEQLWAIRAD 446
M LA C P R+ RP MK + E+LWAIR +
Sbjct: 396 MLRLASECVTPTRATRPAMKGIAEKLWAIRRE 427
>UniRef100_Q5YD56 Calcium/calmodulin-regulated receptor-like kinase [Medicago sativa]
Length = 456
Score = 332 bits (852), Expect = 1e-89
Identities = 165/311 (53%), Positives = 219/311 (70%), Gaps = 2/311 (0%)
Query: 138 SPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKR 197
S S S S +L + + ++ K+T FS QIGEGGFGTVY+ L+DG +VAVKR
Sbjct: 99 SSSYASSSTTSEQLGTGNFTFEEIYKSTAKFSSDNQIGEGGFGTVYRGKLNDGTIVAVKR 158
Query: 198 AKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGL 257
AK+E +S EF +E+ L+KI+H NLV+L GY++ G+E+++I E+V NG L EHLDG+
Sbjct: 159 AKKEALQSHLYEFKNEIYTLSKIEHLNLVRLYGYLEHGDEKLIIVEYVGNGNLTEHLDGI 218
Query: 258 RGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKL 317
RG L+ +RL+IAID+AH +TYLH+Y + IIHRD+K+SNIL++E++RAKVADFGFA+L
Sbjct: 219 RGDGLEIGERLDIAIDIAHAITYLHMYTDNPIIHRDIKASNILISENLRAKVADFGFARL 278
Query: 318 GPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSA 377
+ THIST+VKGT GY+DPEY++TY LT KSDVYSFG+LL+E++TGR PVE KK
Sbjct: 279 SE-DPGATHISTQVKGTAGYMDPEYLRTYQLTEKSDVYSFGVLLVEMMTGRHPVEPKKKI 337
Query: 378 EERVTLRWAFRKYNEGSVVALLDP-LMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTV 436
+ERVT+RWA + G V +DP L + V K+F LAF C AP RP MK
Sbjct: 338 DERVTIRWAMKMLKNGDAVFAMDPRLRRSPASIKVVKKVFKLAFQCLAPSIHSRPAMKNC 397
Query: 437 GEQLWAIRADY 447
E LW IR D+
Sbjct: 398 AEVLWGIRKDF 408
>UniRef100_Q8VZJ9 Hypothetical protein At4g00330 [Arabidopsis thaliana]
Length = 411
Score = 330 bits (847), Expect = 4e-89
Identities = 161/300 (53%), Positives = 219/300 (72%), Gaps = 5/300 (1%)
Query: 160 QVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLR---TEFSSEVEL 216
++ AT+NFS + +IG+GGFGTVYK L DG AVKRAK+ + + EF SE++
Sbjct: 111 EIYDATKNFSPSFRIGQGGFGTVYKVKLRDGKTFAVKRAKKSMHDDRQGADAEFMSEIQT 170
Query: 217 LAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAH 276
LA++ H +LVK G++ +E+IL+ E+VANGTLR+HLD GK LD RL+IA DVAH
Sbjct: 171 LAQVTHLSLVKYYGFVVHNDEKILVVEYVANGTLRDHLDCKEGKTLDMATRLDIATDVAH 230
Query: 277 GLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGP-VNNDHTHISTKVKGTV 335
+TYLH+Y + IIHRD+KSSNILLTE+ RAKVADFGFA+L P ++ TH+ST+VKGT
Sbjct: 231 AITYLHMYTQPPIIHRDIKSSNILLTENYRAKVADFGFARLAPDTDSGATHVSTQVKGTA 290
Query: 336 GYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSV 395
GYLDPEY+ TY LT KSDVYSFG+LL+E+LTGRRP+EL + +ER+T+RWA +K+ G
Sbjct: 291 GYLDPEYLTTYQLTEKSDVYSFGVLLVELLTGRRPIELSRGQKERITIRWAIKKFTSGDT 350
Query: 396 VALLDPLMQEAVKTDVAV-KMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLKSSSTT 454
+++LDP +++ ++A+ K+ ++AF C AP R RP MK E LW IR DY + +T+
Sbjct: 351 ISVLDPKLEQNSANNLALEKVLEMAFQCLAPHRRSRPSMKKCSEILWGIRKDYRELLNTS 410
>UniRef100_Q8H085 Hypothetical protein OSJNBb0050N02.6 [Oryza sativa]
Length = 430
Score = 329 bits (844), Expect = 9e-89
Identities = 161/305 (52%), Positives = 223/305 (72%), Gaps = 4/305 (1%)
Query: 153 SLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFES-LRTEFS 211
S +L Q+ KAT+NFS L+IG+GG GTVYK L+DG ++AVKRAK+ ++ + EF
Sbjct: 125 STQFSLPQIQKATKNFSPNLKIGQGGSGTVYKGQLNDGTLIAVKRAKKNVYDKHMGREFR 184
Query: 212 SEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIA 271
+E+E L I+H NLV+ GY++ G E+++I E+V NG LREHLD + GKIL+F+ RL+I+
Sbjct: 185 NEIETLQCIEHLNLVRFHGYLEFGGEQLIIVEYVPNGNLREHLDCVNGKILEFSLRLDIS 244
Query: 272 IDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKV 331
IDVAH +TYLH Y++ +IHRD+KSSNILLT + RAKVADFGFAKL P D +H+ST+V
Sbjct: 245 IDVAHAVTYLHTYSDHPVIHRDIKSSNILLTNNCRAKVADFGFAKLAP--TDASHVSTQV 302
Query: 332 KGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYN 391
KGT GYLDPEY++TY L KSDVYSFG+LL+E++TGRRP+E +++ ERVT +WA K+
Sbjct: 303 KGTAGYLDPEYLRTYQLNEKSDVYSFGVLLVELITGRRPIEPRRAIVERVTAKWAMEKFV 362
Query: 392 EGSVVALLDPLMQEAVKTDVAV-KMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLKS 450
EG+ + LDP ++ ++AV K ++LA C A + +RP M+ E LW+IR D+ +
Sbjct: 363 EGNAIQTLDPNLEATDAINLAVEKTYELALQCLATTKRNRPSMRRCAEILWSIRKDFREL 422
Query: 451 SSTTA 455
TA
Sbjct: 423 DIPTA 427
>UniRef100_Q6K703 Receptor protein kinase-like [Oryza sativa]
Length = 472
Score = 326 bits (835), Expect = 1e-87
Identities = 163/305 (53%), Positives = 216/305 (70%), Gaps = 6/305 (1%)
Query: 149 SRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAK-REHFESLR 207
++ QS ++ ++ +AT NFS L++G+GGFG VY+ L DG +VAVKRAK R+ +
Sbjct: 129 TQFQSSVFSMEEILRATNNFSPALKVGQGGFGAVYRGVLPDGTLVAVKRAKLRDQNPHVD 188
Query: 208 TEFSSEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQR 267
EF SEV+ +A+I+H++LV+ GY++ G ER+++ EFV NGTLREHLD G+ LD R
Sbjct: 189 VEFRSEVKAMARIEHQSLVRFYGYLECGQERVIVVEFVPNGTLREHLDRCNGRFLDMGAR 248
Query: 268 LEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLG----PVNND 323
LEIAIDVAH +TYLH+YA+ IIHRD+KSSN+LLT S+RAKV DFGFA+LG +
Sbjct: 249 LEIAIDVAHAVTYLHMYADHPIIHRDIKSSNVLLTPSLRAKVGDFGFARLGVGEAGAADG 308
Query: 324 HTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTL 383
TH++T+VKGT GYLDPEY+KT LT +SDVYSFG+LLLEI +GRRP+E ++ ER+T
Sbjct: 309 VTHVTTQVKGTAGYLDPEYLKTCQLTDRSDVYSFGVLLLEIASGRRPIEARREMRERLTA 368
Query: 384 RWAFRKYNEGSVVALLDPLMQEAVKTDVAVKM-FDLAFNCAAPVRSDRPDMKTVGEQLWA 442
RWA RK EG+ +LDP + T A +M +LAF C APVR +RP M LWA
Sbjct: 369 RWAMRKLAEGAAADVLDPHLPRTPATARAAEMVMELAFRCLAPVRQERPSMGECCRALWA 428
Query: 443 IRADY 447
+R Y
Sbjct: 429 VRKTY 433
>UniRef100_Q8W0H6 P0529E05.2 protein [Oryza sativa]
Length = 333
Score = 320 bits (821), Expect = 4e-86
Identities = 157/296 (53%), Positives = 213/296 (71%), Gaps = 2/296 (0%)
Query: 154 LHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFES-LRTEFSS 212
+ L + ++ AT NFSE +IG G FGTVYK L DG ++AVKRA + ++ L EF S
Sbjct: 4 IFLLVQEICMATSNFSEQNRIGLGNFGTVYKGKLRDGSIIAVKRATKNMYDRHLSEEFRS 63
Query: 213 EVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAI 272
E++ L+K++H NLVK LGY++ +ER+++ E+V NG+LREHLDGLRG+ L+F+QRL IAI
Sbjct: 64 EIQTLSKVEHLNLVKFLGYLEHEDERLILVEYVNNGSLREHLDGLRGEPLEFSQRLNIAI 123
Query: 273 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVK 332
D+ H ++YLH Y + IIHRD+KSSNILLT+ +RAKVADFGFA+L P N + TH+ST VK
Sbjct: 124 DIVHAVSYLHGYTDHPIIHRDIKSSNILLTDQLRAKVADFGFARLAPDNTEATHVSTMVK 183
Query: 333 GTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNE 392
GT GY+DPEYM+T LT +SDVYSFG+LL+E+LTGRRP+E + +R+T +WA RK +
Sbjct: 184 GTAGYVDPEYMRTNQLTDRSDVYSFGVLLVELLTGRRPIERGRGRHQRLTTQWALRKCRD 243
Query: 393 GSVVALLDPLMQEAVKTDVAV-KMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADY 447
G V +D M+ A+ K+ LA C AP R+ RP M+ E LW+IR D+
Sbjct: 244 GDAVVAMDARMRRTSAVVAAMEKVMALAAECTAPDRAARPAMRRCAEVLWSIRRDF 299
>UniRef100_O23069 A_IG005I10.8 protein [Arabidopsis thaliana]
Length = 341
Score = 318 bits (816), Expect = 2e-85
Identities = 162/316 (51%), Positives = 219/316 (69%), Gaps = 21/316 (6%)
Query: 160 QVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLR---TEFSSEVEL 216
++ AT+NFS + +IG+GGFGTVYK L DG AVKRAK+ + + EF SE++
Sbjct: 25 EIYDATKNFSPSFRIGQGGFGTVYKVKLRDGKTFAVKRAKKSMHDDRQGADAEFMSEIQT 84
Query: 217 LAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDG---------LRGKILDFNQR 267
LA++ H +LVK G++ +E+IL+ E+VANGTLR+HLD GK LD R
Sbjct: 85 LAQVTHLSLVKYYGFVVHNDEKILVVEYVANGTLRDHLDCEYTSYYYLLFEGKTLDMATR 144
Query: 268 LEIAIDVAHGLTYLHLYAEKQI-------IHRDVKSSNILLTESMRAKVADFGFAKLGP- 319
L+IA DVAH +TYLH+Y K I IHRD+KSSNILLTE+ RAKVADFGFA+L P
Sbjct: 145 LDIATDVAHAITYLHMYTRKSIVYIEPPIIHRDIKSSNILLTENYRAKVADFGFARLAPD 204
Query: 320 VNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEE 379
++ TH+ST+VKGT GYLDPEY+ TY LT KSDVYSFG+LL+E+LTGRRP+EL + +E
Sbjct: 205 TDSGATHVSTQVKGTAGYLDPEYLTTYQLTEKSDVYSFGVLLVELLTGRRPIELSRGQKE 264
Query: 380 RVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAV-KMFDLAFNCAAPVRSDRPDMKTVGE 438
R+T+RWA +K+ G +++LDP +++ ++A+ K+ ++AF C AP R RP MK E
Sbjct: 265 RITIRWAIKKFTSGDTISVLDPKLEQNSANNLALEKVLEMAFQCLAPHRRSRPSMKKCSE 324
Query: 439 QLWAIRADYLKSSSTT 454
LW IR DY + +T+
Sbjct: 325 ILWGIRKDYRELLNTS 340
>UniRef100_Q94GE5 Putative receptor kinase [Oryza sativa]
Length = 444
Score = 266 bits (679), Expect = 1e-69
Identities = 128/305 (41%), Positives = 202/305 (65%), Gaps = 2/305 (0%)
Query: 152 QSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFS 211
+S L ++++ KAT NFS+ I +G + ++Y+ L DG +A+K A++ + + E
Sbjct: 120 ESTELTVAEIFKATSNFSDKNIIKQGSYSSIYRGKLRDGSEIAIKCARKLNSQYASAELR 179
Query: 212 SEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIA 271
E+E+L KIDH+NLV+ LG+ ++ +E + + E+V+NG+LREHLD G L+ QRL IA
Sbjct: 180 RELEILQKIDHKNLVRFLGFFEREDESLTVVEYVSNGSLREHLDESCGNGLELAQRLNIA 239
Query: 272 IDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKV 331
IDVAH +TYLH + E++IIHR+V+SSN+LLT+++ AK+A G A++ + + T+
Sbjct: 240 IDVAHAITYLHEFKEQRIIHRNVRSSNVLLTDTLTAKLAGVGLARMAGGESSESE-DTQG 298
Query: 332 KGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYN 391
K GY+DPEY+ TY LT KSDVYSFG+LL+E++TGR P+E ++ + R T +WA +++
Sbjct: 299 KSAAGYVDPEYLSTYELTDKSDVYSFGVLLVELVTGRPPIERRRDLDPRPTTKWALQRFR 358
Query: 392 EGSVVALLDPLMQEAVKTDVAV-KMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLKS 450
G VV +DP ++ + + V K+ +LA C AP R +RP M+ E LW++R +Y +
Sbjct: 359 GGEVVVAMDPRIRRSPASVATVEKVMELAEQCVAPARKERPSMRRCTEALWSVRREYHRR 418
Query: 451 SSTTA 455
A
Sbjct: 419 QDAPA 423
>UniRef100_Q8VZH5 Receptor protein kinase-like protein [Capsicum annuum]
Length = 648
Score = 244 bits (622), Expect = 5e-63
Identities = 129/287 (44%), Positives = 178/287 (61%), Gaps = 5/287 (1%)
Query: 155 HLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEV 214
H + +++ AT NF E L +G GGFG VY+ +D G VA+KR + + EF +E+
Sbjct: 279 HFSFAEIKAATNNFDEALLLGVGGFGKVYQGEIDGGTKVAIKRGNPLSEQGVH-EFQTEI 337
Query: 215 ELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDV 274
E+L+K+ HR+LV L+GY ++ E IL+ +++A+GTLREHL + L + QRLEI I
Sbjct: 338 EMLSKLRHRHLVSLIGYCEENCEMILVYDYMAHGTLREHLYKTQKPPLPWKQRLEICIGA 397
Query: 275 AHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGT 334
A GL YLH A+ IIHRDVK++NILL E AKV+DFG +K GP DHTH+ST VKG+
Sbjct: 398 ARGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGP-TLDHTHVSTVVKGS 456
Query: 335 VGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGS 394
GYLDPEY + LT KSDVYSFG++L EIL R + E+ WAF Y +G+
Sbjct: 457 FGYLDPEYFRRQQLTDKSDVYSFGVVLFEILCARPALNPTLPKEQVSLAEWAFHCYKKGT 516
Query: 395 VVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
++DP + + + K + A C + V +DRP M G+ LW
Sbjct: 517 FDQIIDPYLNGKLAPECLKKFTETAVKCVSDVGADRPSM---GDVLW 560
>UniRef100_Q6ZF70 Putative PTH-2, resistance gene (PTO kinase) homologs [Oryza
sativa]
Length = 849
Score = 241 bits (616), Expect = 2e-62
Identities = 149/395 (37%), Positives = 221/395 (55%), Gaps = 30/395 (7%)
Query: 61 STSQGDPKQVSSSQVGIFAGGALLVCCAVV---CPCFYGKRRKATSHAVLEKDPNSMELG 117
+++ G PK+ S V I A L++ ++V CFY + +K TS + +P
Sbjct: 415 NSAMGKPKR-SPKWVLIGAAAGLVIFVSIVGVIFVCFYLRWKKKTSANKTKDNP------ 467
Query: 118 SSFEPSVSDKIPASPLRVPPSPSRFSMSPKL-------SRLQSLHLNLSQVSKATRNFSE 170
+ P V L +P+ S SP L S ++++ +AT NF +
Sbjct: 468 PGWRPLV--------LHGATTPAANSRSPTLRAAGTFGSNRMGRQFTVAEIREATMNFDD 519
Query: 171 TLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLG 230
+L IG GGFG VYK ++DG +VA+KR E + ++ EF +E+E+L+++ HR+LV L+G
Sbjct: 520 SLVIGVGGFGKVYKGEMEDGKLVAIKRGHPESQQGVK-EFETEIEILSRLRHRHLVSLIG 578
Query: 231 YIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQII 290
Y D+ NE IL+ E +ANGTLR HL G L + QRLEI I A GL YLH ++ II
Sbjct: 579 YCDEQNEMILVYEHMANGTLRSHLYGTDLPALTWKQRLEICIGAARGLHYLHTGLDRGII 638
Query: 291 HRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTP 350
HRDVK++NILL ++ AK+ADFG +K GP DHTH+ST VKG+ GYLDPEY + LT
Sbjct: 639 HRDVKTTNILLDDNFVAKMADFGISKDGP-PLDHTHVSTAVKGSFGYLDPEYYRRQQLTQ 697
Query: 351 KSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTD 410
SDVYSFG++L E+L R + ++ WA + + + ++DP ++ +
Sbjct: 698 SSDVYSFGVVLFEVLCARPVINPALPRDQINLAEWALKWQKQKLLETIIDPRLEGNYTLE 757
Query: 411 VAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRA 445
K ++A C A RP ++GE LW + +
Sbjct: 758 SIRKFSEIAEKCLADEGRSRP---SIGEVLWHLES 789
>UniRef100_Q5QMM2 Protein kinase-like [Oryza sativa]
Length = 478
Score = 241 bits (615), Expect = 3e-62
Identities = 128/290 (44%), Positives = 184/290 (63%), Gaps = 10/290 (3%)
Query: 160 QVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAK 219
+++ AT +F+++ Q+G+GG+G VYK +L DG VA+KRA + + EF +E+ELL++
Sbjct: 134 EMAAATNDFTDSAQVGQGGYGKVYKGNLTDGTAVAIKRAHEGSLQGSK-EFCTEIELLSR 192
Query: 220 IDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLT 279
+ HRNLV L+GY D+ +E++L+ EF+ NGTLR+HL + L+F+QR+ IA+ A G+
Sbjct: 193 LHHRNLVSLVGYCDEEDEQMLVYEFMPNGTLRDHLSAKSRRPLNFSQRIHIALGAAKGIL 252
Query: 280 YLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNN-DHT---HISTKVKGTV 335
YLH A+ I HRDVK+SNILL AKVADFG ++L PV + D T HIST VKGT
Sbjct: 253 YLHTEADPPIFHRDVKASNILLDSKFVAKVADFGLSRLAPVPDVDGTMPAHISTVVKGTP 312
Query: 336 GYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSV 395
GYLDPEY T+ LT KSDVYS G++LLE+LTG +P++ K+ V Y G +
Sbjct: 313 GYLDPEYFLTHKLTDKSDVYSLGVVLLELLTGMKPIQHGKNIVREVN-----TAYQSGEI 367
Query: 396 VALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRA 445
++D + + + ++ LA C RP M V +L AIR+
Sbjct: 368 AGVIDERISSSSSPECVARLASLAVKCCKDETDARPSMADVVRELDAIRS 417
>UniRef100_Q9SNA7 Receptor-like protein kinase homolog [Arabidopsis thaliana]
Length = 512
Score = 240 bits (613), Expect = 5e-62
Identities = 164/443 (37%), Positives = 228/443 (51%), Gaps = 39/443 (8%)
Query: 14 LSTIFGSEVVLQT-KGCGDNLVANSYSS-HVNLPSTAGRKLLQKDLNNN----STSQGDP 67
L+ + + V QT KG V+ S+ H + P+ L +NN+ ST P
Sbjct: 17 LAGAYSMDFVTQTPKGSNKVRVSIGPSTVHTDYPNAIVNGLEIMKMNNSKGQLSTGTFVP 76
Query: 68 KQVSSSQ--VGIFAGGALLVCCAVV----CPCFYGKRRKAT---SHAVLEKDPNSMELGS 118
SSS+ +G+ G A+ AVV C Y KR++ S + N +GS
Sbjct: 77 GSSSSSKSNLGLIVGSAIGSLLAVVFLGSCFVLYKKRKRGQDGHSKTWMPFSINGTSMGS 136
Query: 119 SFEPSVSDKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGG 178
+ S + ++ + + + V AT NF E+ IG GG
Sbjct: 137 KY-------------------SNGTTLTSITTNANYRIPFAAVKDATNNFDESRNIGVGG 177
Query: 179 FGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNER 238
FG VYK L+DG VAVKR + + L EF +E+E+L++ HR+LV L+GY D+ NE
Sbjct: 178 FGKVYKGELNDGTKVAVKRGNPKSQQGL-AEFRTEIEMLSQFRHRHLVSLIGYCDENNEM 236
Query: 239 ILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSN 298
ILI E++ NGT++ HL G L + QRLEI I A GL YLH K +IHRDVKS+N
Sbjct: 237 ILIYEYMENGTVKSHLYGSGLPSLTWKQRLEICIGAARGLHYLHTGDSKPVIHRDVKSAN 296
Query: 299 ILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFG 358
ILL E+ AKVADFG +K GP D TH+ST VKG+ GYLDPEY + LT KSDVYSFG
Sbjct: 297 ILLDENFMAKVADFGLSKTGP-ELDQTHVSTAVKGSFGYLDPEYFRRQQLTDKSDVYSFG 355
Query: 359 ILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMFDL 418
++L E+L R ++ E WA + +G + ++D ++ ++ D K +
Sbjct: 356 VVLFEVLCARPVIDPTLPREMVNLAEWAMKWQKKGQLDQIIDQSLRGNIRPDSLRKFAET 415
Query: 419 AFNCAAPVRSDRPDMKTVGEQLW 441
C A DRP M G+ LW
Sbjct: 416 GEKCLADYGVDRPSM---GDVLW 435
>UniRef100_Q9LX66 Receptor protein kinase-like [Arabidopsis thaliana]
Length = 830
Score = 240 bits (613), Expect = 5e-62
Identities = 164/443 (37%), Positives = 228/443 (51%), Gaps = 39/443 (8%)
Query: 14 LSTIFGSEVVLQT-KGCGDNLVANSYSS-HVNLPSTAGRKLLQKDLNNN----STSQGDP 67
L+ + + V QT KG V+ S+ H + P+ L +NN+ ST P
Sbjct: 335 LAGAYSMDFVTQTPKGSNKVRVSIGPSTVHTDYPNAIVNGLEIMKMNNSKGQLSTGTFVP 394
Query: 68 KQVSSSQ--VGIFAGGALLVCCAVV----CPCFYGKRRKAT---SHAVLEKDPNSMELGS 118
SSS+ +G+ G A+ AVV C Y KR++ S + N +GS
Sbjct: 395 GSSSSSKSNLGLIVGSAIGSLLAVVFLGSCFVLYKKRKRGQDGHSKTWMPFSINGTSMGS 454
Query: 119 SFEPSVSDKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGG 178
+ S + ++ + + + V AT NF E+ IG GG
Sbjct: 455 KY-------------------SNGTTLTSITTNANYRIPFAAVKDATNNFDESRNIGVGG 495
Query: 179 FGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNER 238
FG VYK L+DG VAVKR + + L EF +E+E+L++ HR+LV L+GY D+ NE
Sbjct: 496 FGKVYKGELNDGTKVAVKRGNPKSQQGL-AEFRTEIEMLSQFRHRHLVSLIGYCDENNEM 554
Query: 239 ILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSN 298
ILI E++ NGT++ HL G L + QRLEI I A GL YLH K +IHRDVKS+N
Sbjct: 555 ILIYEYMENGTVKSHLYGSGLPSLTWKQRLEICIGAARGLHYLHTGDSKPVIHRDVKSAN 614
Query: 299 ILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFG 358
ILL E+ AKVADFG +K GP D TH+ST VKG+ GYLDPEY + LT KSDVYSFG
Sbjct: 615 ILLDENFMAKVADFGLSKTGP-ELDQTHVSTAVKGSFGYLDPEYFRRQQLTDKSDVYSFG 673
Query: 359 ILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMFDL 418
++L E+L R ++ E WA + +G + ++D ++ ++ D K +
Sbjct: 674 VVLFEVLCARPVIDPTLPREMVNLAEWAMKWQKKGQLDQIIDQSLRGNIRPDSLRKFAET 733
Query: 419 AFNCAAPVRSDRPDMKTVGEQLW 441
C A DRP M G+ LW
Sbjct: 734 GEKCLADYGVDRPSM---GDVLW 753
>UniRef100_Q96387 Receptor-like protein kinase precursor [Catharanthus roseus]
Length = 803
Score = 240 bits (613), Expect = 5e-62
Identities = 138/285 (48%), Positives = 181/285 (63%), Gaps = 6/285 (2%)
Query: 158 LSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRA-KREHFESLRTEFSSEVEL 216
L+ V +AT NFSE IG GGFG VYK DG VAVKR + +EF +EVEL
Sbjct: 461 LAVVQEATDNFSENRVIGIGGFGKVYKGVFKDGTKVAVKRGISCSSSKQGLSEFRTEVEL 520
Query: 217 LAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAH 276
L++ HR+LV L+GY D+ NE I+I EF+ NGTLR+HL G L++ +R+EI I A
Sbjct: 521 LSQFRHRHLVSLIGYCDEKNEMIIIYEFMENGTLRDHLYGSDKPKLNWRKRVEICIGSAK 580
Query: 277 GLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVG 336
GL YLH K+IIHRDVKS+NILL E++ AKVADFG +K GP + D TH+ST VKG+ G
Sbjct: 581 GLHYLHTGTMKRIIHRDVKSANILLDENLMAKVADFGVSKTGPDHFDQTHVSTAVKGSFG 640
Query: 337 YLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVV 396
YLDPEY+ LT KSDVYSFG+++LEILTGR ++ K E + WA + +G
Sbjct: 641 YLDPEYLTMQKLTEKSDVYSFGVVMLEILTGRPVIDPSKPREMVNLVEWAMKCSRKGE-- 698
Query: 397 ALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
++D + V+ + +K + A C A DRP T+G+ LW
Sbjct: 699 EIVDSDIVNEVRPESLIKFQETAEKCLAERGVDRP---TMGDVLW 740
>UniRef100_Q94EF4 Hypothetical protein P0665A11.8 [Oryza sativa]
Length = 896
Score = 240 bits (612), Expect = 7e-62
Identities = 150/385 (38%), Positives = 208/385 (53%), Gaps = 25/385 (6%)
Query: 66 DPKQVSSSQVGIFAGG--------ALLVCCAVVCPCFYGKRRKATSHAVLEKDPNSMELG 117
D V S VG GG AL CC ++C KRR D +S L
Sbjct: 444 DGGAVKKSSVGPIVGGVIGGLVVLALGYCCFMIC-----KRRSRVGKDTGMSDGHSGWLP 498
Query: 118 SSFEPSVSDKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEG 177
S + A S + S + L H + +++ AT NF E+L +G G
Sbjct: 499 LSLYGNSHSSGSAK------SHTTGSYASSLPSNLCRHFSFAEIKAATNNFDESLLLGVG 552
Query: 178 GFGTVYKAHLDDGLV-VAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGN 236
GFG VY+ +D G+ VA+KR + + EF +E+E+L+K+ HR+LV L+GY ++ N
Sbjct: 553 GFGKVYRGEIDGGVTKVAIKRGNPLSEQGVH-EFQTEIEMLSKLRHRHLVSLIGYCEEKN 611
Query: 237 ERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKS 296
E IL+ +++A+GTLREHL + L + QRLEI I A GL YLH A+ IIHRDVK+
Sbjct: 612 EMILVYDYMAHGTLREHLYKTKNAPLTWRQRLEICIGAARGLHYLHTGAKHTIIHRDVKT 671
Query: 297 SNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYS 356
+NILL E AKV+DFG +K GP + DHTH+ST VKG+ GYLDPEY + LT KSDVYS
Sbjct: 672 TNILLDEKWVAKVSDFGLSKTGP-SMDHTHVSTVVKGSFGYLDPEYFRRQQLTEKSDVYS 730
Query: 357 FGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMF 416
FG++L E+L R + + EE WA +G + ++DP ++ + K
Sbjct: 731 FGVVLFEVLCARPALNPTLAKEEVSLAEWALHCQKKGILDQIVDPHLKGKIAPQCFKKFA 790
Query: 417 DLAFNCAAPVRSDRPDMKTVGEQLW 441
+ A C + DRP M G+ LW
Sbjct: 791 ETAEKCVSDEGIDRPSM---GDVLW 812
>UniRef100_Q9SC72 L1332.5 protein [Oryza sativa]
Length = 844
Score = 240 bits (612), Expect = 7e-62
Identities = 147/389 (37%), Positives = 223/389 (56%), Gaps = 22/389 (5%)
Query: 58 NNNSTSQGDPKQVSSSQVGIFAGGALLVCCAVVCP-CFYGKRRKATSHAVLEKDP----N 112
N S+ +++ +VGI + + + V+ C+ ++RKA EK+P +
Sbjct: 414 NQRGISKDRNRKILWEEVGIGSASFVTLTSVVLFAWCYIRRKRKAD-----EKEPPPGWH 468
Query: 113 SMELGSSFEPSVSDKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETL 172
+ L + + S +D A + P + + S+ ++ R ++S++ AT+NF E L
Sbjct: 469 PLVLHEAMK-STTDARAAG--KSPLTRNSSSIGHRMGR----RFSISEIRAATKNFDEAL 521
Query: 173 QIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYI 232
IG GGFG VYK +D+G VA+KRA + L+ EF +E+E+L+K+ HR+LV ++GY
Sbjct: 522 LIGTGGFGKVYKGEVDEGTTVAIKRANPLCGQGLK-EFETEIEMLSKLRHRHLVAMIGYC 580
Query: 233 DKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHR 292
++ E IL+ E++A GTLR HL G L + QR++ I A GL YLH A++ IIHR
Sbjct: 581 EEQKEMILVYEYMAKGTLRSHLYGSDLPPLTWKQRVDACIGAARGLHYLHTGADRGIIHR 640
Query: 293 DVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKS 352
DVK++NILL E+ AK+ADFG +K GP D TH+ST VKG+ GYLDPEY + LT KS
Sbjct: 641 DVKTTNILLDENFVAKIADFGLSKTGP-TLDQTHVSTAVKGSFGYLDPEYFRRQQLTQKS 699
Query: 353 DVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVA 412
DVYSFG++L E+ GR ++ ++ WA R + S+ A++DP + ++
Sbjct: 700 DVYSFGVVLFEVACGRPVIDPTLPKDQINLAEWAMRWQRQRSLDAIVDPRLDGDFSSESL 759
Query: 413 VKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
K ++A C A RP M GE LW
Sbjct: 760 KKFGEIAEKCLADDGRSRPSM---GEVLW 785
>UniRef100_Q9FN92 Receptor-like protein kinase [Arabidopsis thaliana]
Length = 829
Score = 239 bits (611), Expect = 9e-62
Identities = 132/289 (45%), Positives = 176/289 (60%), Gaps = 5/289 (1%)
Query: 153 SLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSS 212
S + L V +AT +F E IG GGFG VYK L DG VAVKRA + + L EF +
Sbjct: 467 SYRIPLVAVKEATNSFDENRAIGVGGFGKVYKGELHDGTKVAVKRANPKSQQGL-AEFRT 525
Query: 213 EVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAI 272
E+E+L++ HR+LV L+GY D+ NE IL+ E++ NGTL+ HL G L + QRLEI I
Sbjct: 526 EIEMLSQFRHRHLVSLIGYCDENNEMILVYEYMENGTLKSHLYGSGLLSLSWKQRLEICI 585
Query: 273 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVK 332
A GL YLH K +IHRDVKS+NILL E++ AKVADFG +K GP D TH+ST VK
Sbjct: 586 GSARGLHYLHTGDAKPVIHRDVKSANILLDENLMAKVADFGLSKTGP-EIDQTHVSTAVK 644
Query: 333 GTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNE 392
G+ GYLDPEY + LT KSDVYSFG+++ E+L R ++ + E WA + +
Sbjct: 645 GSFGYLDPEYFRRQQLTEKSDVYSFGVVMFEVLCARPVIDPTLTREMVNLAEWAMKWQKK 704
Query: 393 GSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
G + ++DP ++ ++ D K + C A DRP M G+ LW
Sbjct: 705 GQLEHIIDPSLRGKIRPDSLRKFGETGEKCLADYGVDRPSM---GDVLW 750
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 716,663,785
Number of Sequences: 2790947
Number of extensions: 29262656
Number of successful extensions: 129088
Number of sequences better than 10.0: 18749
Number of HSP's better than 10.0 without gapping: 9608
Number of HSP's successfully gapped in prelim test: 9143
Number of HSP's that attempted gapping in prelim test: 89503
Number of HSP's gapped (non-prelim): 21681
length of query: 458
length of database: 848,049,833
effective HSP length: 131
effective length of query: 327
effective length of database: 482,435,776
effective search space: 157756498752
effective search space used: 157756498752
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC144657.5


