

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.4 + phase: 0
(776 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
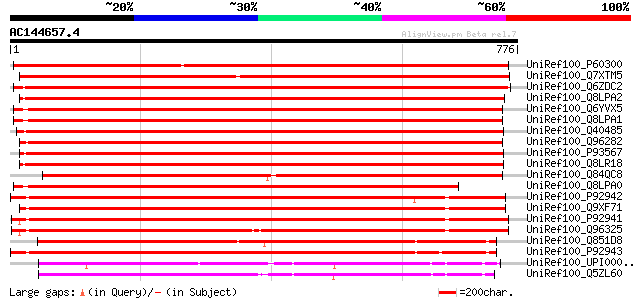
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P60300 Putative chloride channel-like protein CLC-G [A... 1118 0.0
UniRef100_Q7XTM5 OSJNBa0033G05.20 protein [Oryza sativa] 1017 0.0
UniRef100_Q6ZDC2 Putative chloride channel protein [Oryza sativa] 965 0.0
UniRef100_Q8LPA2 Chloride channel [Oryza sativa] 945 0.0
UniRef100_Q6YVX5 Chloride channel [Oryza sativa] 937 0.0
UniRef100_Q8LPA1 Chloride channel [Oryza sativa] 932 0.0
UniRef100_Q40485 C1C-Nt1 protein [Nicotiana tabacum] 926 0.0
UniRef100_Q96282 Chloride channel protein CLC-c [Arabidopsis tha... 924 0.0
UniRef100_P93567 Chloride channel Stclc1 [Solanum tuberosum] 923 0.0
UniRef100_Q8LR18 Putative chloride channel protein [Oryza sativa] 898 0.0
UniRef100_Q84QC8 Chloride channel [Zea mays] 859 0.0
UniRef100_Q8LPA0 Chloride channel [Oryza sativa] 828 0.0
UniRef100_P92942 Chloride channel protein CLC-b [Arabidopsis tha... 823 0.0
UniRef100_Q9XF71 CLC-Nt2 protein [Nicotiana tabacum] 812 0.0
UniRef100_P92941 Chloride channel protein CLC-a [Arabidopsis tha... 811 0.0
UniRef100_Q96325 Voltage-gated chloride channel [Arabidopsis tha... 778 0.0
UniRef100_Q851D8 Putative CLC-d chloride channel protein [Oryza ... 618 e-175
UniRef100_P92943 Chloride channel protein CLC-d [Arabidopsis tha... 614 e-174
UniRef100_UPI00003619FE UPI00003619FE UniRef100 entry 400 e-110
UniRef100_Q5ZL60 Hypothetical protein [Gallus gallus] 392 e-107
>UniRef100_P60300 Putative chloride channel-like protein CLC-G [Arabidopsis thaliana]
Length = 763
Score = 1118 bits (2891), Expect = 0.0
Identities = 549/758 (72%), Positives = 642/758 (84%), Gaps = 2/758 (0%)
Query: 6 LSNGESEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGV 65
+ N +E + PLL S R NSTSQVAIVGANVCPIESLDYEI EN+FFKQDWR R
Sbjct: 1 MPNSTTEDSVAVPLLPSLRRATNSTSQVAIVGANVCPIESLDYEIAENDFFKQDWRGRSK 60
Query: 66 VQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFF 125
V+I QY+ MKWLLCF IG+IV IGF NNLAVENLAG+KFV TSNMM+ RF F +F
Sbjct: 61 VEIFQYVFMKWLLCFCIGIIVSLIGFANNLAVENLAGVKFVVTSNMMIAGRFAMGFVVFS 120
Query: 126 ASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSG 185
+NL LTLFAS+ITAF+AP AAGSGI EVKAYLNGVDAP IF++RTL +KIIG+I+AVS
Sbjct: 121 VTNLILTLFASVITAFVAPAAAGSGIPEVKAYLNGVDAPEIFSLRTLIIKIIGNISAVSA 180
Query: 186 SLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAA 245
SL+IGKAGPMVHTGACVA++LGQGGSKRY +TWRWLRFFKNDRDRRDL+ CG+AAGIAA+
Sbjct: 181 SLLIGKAGPMVHTGACVASILGQGGSKRYRLTWRWLRFFKNDRDRRDLVTCGAAAGIAAS 240
Query: 246 FRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIM 305
FRAPVGGVLFALEEM+SW +ALLWR FF+TA VAI LRA+IDVCLS KCGLFGKGGLIM
Sbjct: 241 FRAPVGGVLFALEEMSSW--SALLWRIFFSTAVVAIVLRALIDVCLSGKCGLFGKGGLIM 298
Query: 306 FDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACL 365
FD YS + SYHL DV PV +L VVGG+LGSL+NF+ +KVLR YN I EKG ++ LAC
Sbjct: 299 FDVYSENASYHLGDVLPVLLLGVVGGILGSLYNFLLDKVLRAYNYIYEKGVTWKILLACA 358
Query: 366 ISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTND 425
ISIFTSCLLFGLP+LA C+PCP DA+E CPTIGRSG +KK+QCPP HYN LASLIFNTND
Sbjct: 359 ISIFTSCLLFGLPFLASCQPCPVDALEECPTIGRSGNFKKYQCPPGHYNDLASLIFNTND 418
Query: 426 DAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGI 485
DAI+NLFS +TD EF S+LVFF+ C FLSIFS GIVAPAGLFVP+IVTGASYGR VG+
Sbjct: 419 DAIKNLFSKNTDFEFHYFSVLVFFVTCFFLSIFSYGIVAPAGLFVPVIVTGASYGRFVGM 478
Query: 486 LVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVA 545
L+G +NL++GL+AVLGAAS LGG+MR TVS CVI+LELTNNLLLLP++M+VL++SK+VA
Sbjct: 479 LLGSNSNLNHGLFAVLGAASFLGGTMRMTVSTCVILLELTNNLLLLPMMMVVLLISKTVA 538
Query: 546 NVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTT 605
+ FNAN+Y+LIMK KG PYL +HAEPYMRQL VGDVVTGPLQ+FNGIEKV IV +L+TT
Sbjct: 539 DGFNANIYNLIMKLKGFPYLYSHAEPYMRQLLVGDVVTGPLQVFNGIEKVETIVHVLKTT 598
Query: 606 AHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKK 665
HNGFPV+D PP + AP+L G+ILR H+ TLLKK+ F+PSPVA + + +F +++FAKK
Sbjct: 599 NHNGFPVVDGPPLAAAPVLHGLILRAHILTLLKKRVFMPSPVACDSNTLSQFKAEEFAKK 658
Query: 666 YSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIP 725
S KIED++L+EEE+ M++DLHPF+NASPYTVVETMSLAKALILFREVG+RHLLVIP
Sbjct: 659 GSGRSDKIEDVELSEEELNMYLDLHPFSNASPYTVVETMSLAKALILFREVGIRHLLVIP 718
Query: 726 KIPGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLR 763
K R PVVGILTRHDF PEHILG+HP + +S+WKRLR
Sbjct: 719 KTSNRPPVVGILTRHDFMPEHILGLHPSVSRSKWKRLR 756
>UniRef100_Q7XTM5 OSJNBa0033G05.20 protein [Oryza sativa]
Length = 802
Score = 1017 bits (2630), Expect = 0.0
Identities = 489/749 (65%), Positives = 606/749 (80%), Gaps = 6/749 (0%)
Query: 16 RRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQYICMK 75
RR L + RS N+TSQVA+VG VCPIESLDYE+ ENE FKQDWR+RG IL+Y+ +K
Sbjct: 50 RRRFLLAGRSQSNTTSQVALVGVGVCPIESLDYELIENEVFKQDWRARGRGHILRYVALK 109
Query: 76 WLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLFA 135
W LCF++G++ GF NL VEN+AG KFV TSN+ML R+ AF +F SN +LT+ A
Sbjct: 110 WALCFLVGVLSAAAGFVANLGVENVAGAKFVVTSNLMLAGRYGTAFAVFLVSNFALTMLA 169
Query: 136 SIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPM 195
+++T ++AP AAGSGI EVKAYLNGVDAP IF+++TL VKI+G I AVS SL +GKAGP+
Sbjct: 170 TVLTVYVAPAAAGSGIPEVKAYLNGVDAPDIFSLKTLVVKIVGCIAAVSSSLHVGKAGPL 229
Query: 196 VHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLF 255
VHTGAC+A++LGQGGS +Y +T +WLR+FKNDRDRRDL+ CG+ AGIAAAFRAPVGGVLF
Sbjct: 230 VHTGACIASILGQGGSSKYHLTCKWLRYFKNDRDRRDLVTCGAGAGIAAAFRAPVGGVLF 289
Query: 256 ALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASISY 315
ALE ++SWWR+ALLWRAFFTTA VA+ LRA+ID C SDKCGLFGKGGLIMFD S I+Y
Sbjct: 290 ALEAVSSWWRSALLWRAFFTTAMVAVVLRALIDFCKSDKCGLFGKGGLIMFDVTSDYITY 349
Query: 316 HLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLF 375
HLVD+PPV L V+GG+LGSL NF +K+ KG +L LA +++I TSC LF
Sbjct: 350 HLVDLPPVITLGVLGGVLGSLHNFFLDKLTF------RKGQKYKLLLAAVVTICTSCCLF 403
Query: 376 GLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSMH 435
GLPW+A C+PCP D E CP+IGRSG +KK+QC N YN LASL FNTNDD IRNL+S
Sbjct: 404 GLPWIASCKPCPSDTEEACPSIGRSGNFKKYQCAMNEYNDLASLFFNTNDDTIRNLYSAG 463
Query: 436 TDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLSN 495
TD+EF +SS+LVFF FL IFS G+ P+GLFVP+I+TGA+YGRLVG+L+G ++ L +
Sbjct: 464 TDDEFHISSILVFFFTSYFLGIFSYGLALPSGLFVPVILTGATYGRLVGMLIGSQSTLDH 523
Query: 496 GLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDL 555
GL+AVLG+A+LLGGSMR TVS+CV++LELTNNLL+LPL+M+VL++SK+VA+ FNAN+YDL
Sbjct: 524 GLFAVLGSAALLGGSMRMTVSVCVVILELTNNLLMLPLVMLVLLISKTVADAFNANIYDL 583
Query: 556 IMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPVIDE 615
++K KG PYLE H EPYMRQL+V DVVTGPLQ FNGIEKV +IV +LRTT HNGFPV+DE
Sbjct: 584 LVKLKGFPYLEGHVEPYMRQLSVSDVVTGPLQAFNGIEKVGHIVHVLRTTGHNGFPVVDE 643
Query: 616 PPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIED 675
PP S++P+LFG++LR HL LL+KK F+P+ A++ D ++F DFAK S + +IE+
Sbjct: 644 PPFSDSPVLFGLVLRAHLLVLLRKKDFIPNCSASALDASKQFLPHDFAKPGSGKHDRIEE 703
Query: 676 IQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVG 735
I+ + EE+ MFVDLHPFTN SPYTVVETMSLAKA +LFREVGLRHLLV+PK R+PVVG
Sbjct: 704 IEFSAEELEMFVDLHPFTNTSPYTVVETMSLAKAHVLFREVGLRHLLVLPKSSKRAPVVG 763
Query: 736 ILTRHDFTPEHILGMHPFLVKSRWKRLRF 764
ILTRHDF PEHILG+HPFL K+RWK++RF
Sbjct: 764 ILTRHDFMPEHILGLHPFLFKTRWKKVRF 792
>UniRef100_Q6ZDC2 Putative chloride channel protein [Oryza sativa]
Length = 796
Score = 965 bits (2494), Expect = 0.0
Identities = 476/768 (61%), Positives = 596/768 (76%), Gaps = 11/768 (1%)
Query: 6 LSNGESEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGV 65
+ E P L R LL RS N+TSQVA+VG+N CPIESLDYEI EN+ F Q+WRSRG
Sbjct: 13 MEEDEHRPPLNRALL--HRSATNNTSQVAMVGSNPCPIESLDYEIIENDLFDQNWRSRGK 70
Query: 66 VQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFF 125
++Y+ +KW CF IG+I G GF NLAVEN+AG+K S +M + AF++F
Sbjct: 71 ADQVRYVVLKWTFCFAIGIITGIAGFVINLAVENVAGLKHTAVSALMESSSYWTAFWLFA 130
Query: 126 ASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSG 185
+NL+L LFAS ITAF++P A GSGI EVKAYLNGVDAP IF++RTL VKIIG+I AVS
Sbjct: 131 GTNLALLLFASSITAFVSPAAGGSGIPEVKAYLNGVDAPNIFSLRTLAVKIIGNIAAVSS 190
Query: 186 SLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAA 245
SL +GKAGPMVHTGAC+AA+ GQGGS++YG+T RWLR+FKNDRDRRDL+ G+ AG+ AA
Sbjct: 191 SLHVGKAGPMVHTGACIAAIFGQGGSRKYGLTCRWLRYFKNDRDRRDLVTIGAGAGVTAA 250
Query: 246 FRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIM 305
FRAPVGGVLFALE ++SWWR+AL+WR+FFTTA VA+ LR I++C S KCGLFGKGGLIM
Sbjct: 251 FRAPVGGVLFALESLSSWWRSALIWRSFFTTAVVAVVLRMFIELCASGKCGLFGKGGLIM 310
Query: 306 FDA---YSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFL 362
+D + ++YHL D+P V ++ V+G +LG+L+NF+ KVLR+Y+VINE+G +L L
Sbjct: 311 YDVSTKFDDLMTYHLKDIPIVVLIGVIGAILGALYNFLMMKVLRVYSVINERGNAHKLLL 370
Query: 363 ACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRS---GIYKKFQCPPNHYNGLASL 419
A ++SI TSC +FGLPWLAPCRPC P A P P G +++F CP HYN LASL
Sbjct: 371 AAVVSILTSCCVFGLPWLAPCRPC-PTAGAPSPPNGTCHSLNRFRRFHCPAGHYNDLASL 429
Query: 420 IFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASY 479
N NDDAIRNL+S T++ + SML FF+ L + S G+VAP+GLFVPII+TGA+Y
Sbjct: 430 FLNINDDAIRNLYSTGTNDVYHPGSMLAFFVASYALGVLSYGVVAPSGLFVPIILTGATY 489
Query: 480 GRLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLV 539
GRLV +L+G R+ L +GL A+LG+AS LGG++R TVS+CVI+LELTNNLLLLPL+M+VL+
Sbjct: 490 GRLVAMLLGGRSGLDHGLVAILGSASFLGGTLRMTVSVCVIILELTNNLLLLPLVMLVLL 549
Query: 540 VSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIV 599
+SK+VA+ FN+++YDLI+ KGLP+L+ HAEPYMRQLTVGDVV GPL+ FNG+EKV +IV
Sbjct: 550 ISKTVADSFNSSIYDLILNLKGLPHLDGHAEPYMRQLTVGDVVAGPLRSFNGVEKVGHIV 609
Query: 600 FILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYD-VVRKFS 658
LRTT H+ FPV+DEPP S AP+L+G++LR HL LLKK+ FL +PV D + +F
Sbjct: 610 HTLRTTGHHAFPVVDEPPFSPAPVLYGLVLRAHLLVLLKKREFLTAPVRCPKDYMAGRFE 669
Query: 659 SDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGL 718
+ DF K+ S ++ I D++L+ EEM M+VDLHPFTN SPYTVVETMSLAKAL+LFREVGL
Sbjct: 670 AQDFDKRGSGKQDTIADVELSPEEMEMYVDLHPFTNTSPYTVVETMSLAKALVLFREVGL 729
Query: 719 RHLLVIPKIPGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLRFWQ 766
RHLLV+PK RSPVVGILTRHDF PEHILG+HP LV SRWKRLR WQ
Sbjct: 730 RHLLVVPKSCDRSPVVGILTRHDFMPEHILGLHPVLVGSRWKRLR-WQ 776
>UniRef100_Q8LPA2 Chloride channel [Oryza sativa]
Length = 801
Score = 945 bits (2442), Expect = 0.0
Identities = 464/745 (62%), Positives = 573/745 (76%), Gaps = 5/745 (0%)
Query: 16 RRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQYICMK 75
R+PLL ++ +N+TSQ+AIVGANVCPIESLDYEI EN+ FKQDWRSR QI QYI +K
Sbjct: 58 RQPLL--RKRTMNTTSQIAIVGANVCPIESLDYEIVENDLFKQDWRSRKKKQIFQYIVLK 115
Query: 76 WLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLER--RFMFAFFIFFASNLSLTL 133
W L +IG++ G +GF NNLAVEN+AG+K + TS++ML++ R+ AF + NL L
Sbjct: 116 WALVLLIGMLTGIVGFFNNLAVENIAGLKLLLTSDLMLKQKCRYFTAFLAYGGCNLVLAT 175
Query: 134 FASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAG 193
A+ I A+IAP AAGSGI EVKAYLNGVDA I TL VKI GSI VS V+GK G
Sbjct: 176 TAAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAPSTLFVKIFGSILGVSAGFVLGKEG 235
Query: 194 PMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGV 253
PMVHTGAC+A LLGQGGS++Y +TW WLR+FKNDRDRRDLI CGSAAG+AAAFRAPVGGV
Sbjct: 236 PMVHTGACIANLLGQGGSRKYHLTWNWLRYFKNDRDRRDLITCGSAAGVAAAFRAPVGGV 295
Query: 254 LFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASI 313
LFALEE ASWWR+ALLWR FFTTA VA+ LR +I+ C S KCGLFG+GGLIMFD S
Sbjct: 296 LFALEEAASWWRSALLWRTFFTTAVVAVVLRGLIEFCRSGKCGLFGQGGLIMFDLSSTIP 355
Query: 314 SYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCL 373
+Y DV + +L ++GG+ G LFNF+ +++LR Y++INE+G ++ L +ISI TS
Sbjct: 356 TYTAQDVVAIIVLGIIGGVFGGLFNFLLDRILRAYSIINERGPPFKILLTMIISIITSAC 415
Query: 374 LFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFS 433
+GLPWLAPC PCP DA E CPTIGRSG +K FQCPP HYNGLASL FNTNDDAIRNLFS
Sbjct: 416 SYGLPWLAPCTPCPADAAEECPTIGRSGNFKNFQCPPGHYNGLASLFFNTNDDAIRNLFS 475
Query: 434 MHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNL 493
T+ EF +S++ VFF L + + GI P+GLF+P+I+ GA+YGR+VG L+G ++L
Sbjct: 476 SGTEKEFHMSTLFVFFTAIYCLGLVTYGIAVPSGLFIPVILAGATYGRIVGTLLGPISDL 535
Query: 494 SNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVY 553
GL+A+LGAAS LGG+MR TVS+CVI+LELTN+L +LPL+M+VL++SK++A+ FN VY
Sbjct: 536 DPGLFALLGAASFLGGTMRMTVSVCVILLELTNDLHMLPLVMLVLLISKTIADSFNKGVY 595
Query: 554 DLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPVI 613
D I+ KGLP++E HAEP+MR L GDVV+GPL F+G+EKV NIV LR T HNGFPV+
Sbjct: 596 DQIVVMKGLPFMEAHAEPFMRNLVAGDVVSGPLITFSGVEKVGNIVHALRITGHNGFPVV 655
Query: 614 DEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYD-VVRKFSSDDFAKKYSVERVK 672
DEPP SEAP L G++LR HL LLK ++F+ V S V+R+F + DFAK S + +K
Sbjct: 656 DEPPVSEAPELVGLVLRSHLLVLLKGRSFMKEKVKTSGSFVLRRFGAFDFAKPGSGKGLK 715
Query: 673 IEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSP 732
IED+ LT+EE+ M+VDLHP TN SPYTVVETMSLAKA +LFR +GLRHLLV+PK PGR P
Sbjct: 716 IEDLDLTDEELDMYVDLHPITNTSPYTVVETMSLAKAAVLFRALGLRHLLVVPKTPGRPP 775
Query: 733 VVGILTRHDFTPEHILGMHPFLVKS 757
+VGILTRHDF EHI G+ P L KS
Sbjct: 776 IVGILTRHDFMHEHIHGLFPNLGKS 800
>UniRef100_Q6YVX5 Chloride channel [Oryza sativa]
Length = 804
Score = 937 bits (2423), Expect = 0.0
Identities = 462/749 (61%), Positives = 566/749 (74%), Gaps = 8/749 (1%)
Query: 7 SNGESEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVV 66
S EPLLR+ + N+TSQ+AIVGANVCPIESLDYE+ EN+ FKQDWRSR
Sbjct: 51 SGSAGEPLLRKRTM-------NTTSQIAIVGANVCPIESLDYEVVENDLFKQDWRSRKKK 103
Query: 67 QILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFA 126
QI QYI +KW L +IGL+ G +GF NNLAVEN+AG K + T N+ML+ R++ AFF +
Sbjct: 104 QIFQYIVLKWTLVLLIGLLTGLVGFFNNLAVENIAGFKLLLTGNLMLKERYLTAFFAYGG 163
Query: 127 SNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGS 186
NL L A+ I A+IAP AAGSGI EVKAYLNGVDA I TL VKI GSI VS
Sbjct: 164 CNLVLAAAAAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAPSTLFVKIFGSILGVSAG 223
Query: 187 LVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAF 246
V+GK GPMVHTGAC+A LLGQGGS++Y +T WLR+FKNDRDRRDLI CGSAAG+AAAF
Sbjct: 224 FVLGKEGPMVHTGACIANLLGQGGSRKYRLTCNWLRYFKNDRDRRDLITCGSAAGVAAAF 283
Query: 247 RAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMF 306
RAPVGGVLFALEE ASWWR+ALLWRAFFTTA VA+ LR++I+ C S KCGLFG+GGLIMF
Sbjct: 284 RAPVGGVLFALEEAASWWRSALLWRAFFTTAVVAVVLRSLIEFCRSGKCGLFGQGGLIMF 343
Query: 307 DAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLI 366
D S +Y D+ + IL ++GG+ G LFNF+ +KVLR+Y++INE+G ++ L I
Sbjct: 344 DLSSTVATYSTPDLIAIIILGIIGGIFGGLFNFLLDKVLRVYSIINERGAPFKILLTITI 403
Query: 367 SIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDD 426
SI TS +GLPWLA C PCP DAVE CPTIGRSG +K FQCPP HYN LASL FNTNDD
Sbjct: 404 SIITSMCSYGLPWLAACTPCPVDAVEQCPTIGRSGNFKNFQCPPGHYNDLASLFFNTNDD 463
Query: 427 AIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGIL 486
AIRNLFS T++EF +S++ +FF L I + G+ P+GLF+P+I+ GA+YGR+VG L
Sbjct: 464 AIRNLFSNGTESEFHMSTLFIFFTAVYCLGILTYGVAVPSGLFIPVILAGATYGRIVGTL 523
Query: 487 VGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVAN 546
+G ++L GL+A+LGAAS LGG+MR TVS+CVI+LELTN+L +LPL+M+VL++SK++A+
Sbjct: 524 LGSISDLDPGLFALLGAASFLGGTMRMTVSVCVILLELTNDLAMLPLVMLVLLISKTIAD 583
Query: 547 VFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTA 606
FN VYD I+ KGLPY+E HAEPYMR L GDVV+GPL F+G+EKV NIV LR T
Sbjct: 584 NFNKGVYDQIVVMKGLPYMEAHAEPYMRHLVAGDVVSGPLITFSGVEKVGNIVHALRFTG 643
Query: 607 HNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYD-VVRKFSSDDFAKK 665
HNGFPV+DEPP +EAP L G++ R HL LL K F+ + S V+++F + DFAK
Sbjct: 644 HNGFPVVDEPPLTEAPELVGLVTRSHLLVLLNGKMFMKDQLKTSGSFVLQRFGAFDFAKP 703
Query: 666 YSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIP 725
S + +KI+D+ T+EEM M+VDLHP TN SPYTVVETMSLAKA ILFR +GLRHLLV+P
Sbjct: 704 GSGKGLKIQDLDFTDEEMEMYVDLHPVTNTSPYTVVETMSLAKAAILFRALGLRHLLVVP 763
Query: 726 KIPGRSPVVGILTRHDFTPEHILGMHPFL 754
K P R P+VGILTRHDF EHI G+ P L
Sbjct: 764 KTPDRPPIVGILTRHDFVEEHIHGLFPNL 792
>UniRef100_Q8LPA1 Chloride channel [Oryza sativa]
Length = 756
Score = 932 bits (2408), Expect = 0.0
Identities = 462/751 (61%), Positives = 566/751 (74%), Gaps = 10/751 (1%)
Query: 7 SNGESEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVV 66
S EPLLR+ + N+TSQ+AIVGANVCPIESLDYE+ EN+ FKQDWRSR
Sbjct: 1 SGSAGEPLLRKRTM-------NTTSQIAIVGANVCPIESLDYEVVENDLFKQDWRSRKKK 53
Query: 67 QILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLER--RFMFAFFIF 124
QI QYI +KW L +IGL+ G +GF NNLAVEN+AG K + T N+ML+ R++ AFF +
Sbjct: 54 QIFQYIVLKWTLVLLIGLLTGLVGFFNNLAVENIAGFKLLLTGNLMLKGKCRYLTAFFAY 113
Query: 125 FASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVS 184
NL L A+ I A+IAP AAGSGI EVKAYLNGVDA I TL VKI GSI VS
Sbjct: 114 GGCNLVLAAAAAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAPSTLFVKIFGSILGVS 173
Query: 185 GSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAA 244
V+GK GPMVHTGAC+A LLGQGGS++Y +T WLR+FKNDRDRRDLI CGSAAG+AA
Sbjct: 174 AGFVLGKEGPMVHTGACIANLLGQGGSRKYRLTCNWLRYFKNDRDRRDLITCGSAAGVAA 233
Query: 245 AFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLI 304
AFRAPVGGVLFALEE ASWWR+ALLWRAFFTTA VA+ LR++I+ C S KCGLFG+GGLI
Sbjct: 234 AFRAPVGGVLFALEEAASWWRSALLWRAFFTTAVVAVVLRSLIEFCRSGKCGLFGQGGLI 293
Query: 305 MFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLAC 364
MFD S +Y D+ + IL ++GG+ G LFNF+ +KVLR+Y++INE+G ++ L
Sbjct: 294 MFDLSSTVATYSTPDLIAIIILGIIGGIFGGLFNFLLDKVLRVYSIINERGAPFKILLTI 353
Query: 365 LISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTN 424
ISI TS +GLPWLA C PCP DAVE CPTIGRSG +K FQCPP HYN LASL FNTN
Sbjct: 354 TISIITSMCSYGLPWLAACTPCPVDAVEQCPTIGRSGNFKNFQCPPGHYNDLASLFFNTN 413
Query: 425 DDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVG 484
DDAIRNLFS T++EF +S++ +FF L I + G+ P+GLF+P+I+ GA+YGR+VG
Sbjct: 414 DDAIRNLFSNGTESEFHMSTLFIFFTAVYCLGILTYGVAVPSGLFIPVILAGATYGRIVG 473
Query: 485 ILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSV 544
L+G ++L GL+A+LGAAS LGG+MR TVS+CVI+LELTN+L +LPL+M+VL++SK++
Sbjct: 474 TLLGSISDLDPGLFALLGAASFLGGTMRMTVSVCVILLELTNDLAMLPLVMLVLLISKTI 533
Query: 545 ANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRT 604
A+ FN VYD I+ KGLPY+E HAEPYMR L GDVV+GPL F+G+EKV NIV LR
Sbjct: 534 ADNFNKGVYDQIVVMKGLPYMEAHAEPYMRHLVAGDVVSGPLITFSGVEKVGNIVHALRF 593
Query: 605 TAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYD-VVRKFSSDDFA 663
T HNGFPV+DEPP +EAP L G++ R HL LL K F+ + S V+++F + DFA
Sbjct: 594 TGHNGFPVVDEPPLTEAPELVGLVTRSHLLVLLNGKMFMKDQLKTSGSFVLQRFGAFDFA 653
Query: 664 KKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLV 723
K S + +KI+D+ T+EEM M+VDLHP TN SPYTVVETMSLAKA ILFR +GLRHLLV
Sbjct: 654 KPGSGKGLKIQDLDFTDEEMEMYVDLHPVTNTSPYTVVETMSLAKAAILFRALGLRHLLV 713
Query: 724 IPKIPGRSPVVGILTRHDFTPEHILGMHPFL 754
+PK P R P+VGILTRHDF EHI G+ P L
Sbjct: 714 VPKTPDRPPIVGILTRHDFVEEHIHGLFPNL 744
>UniRef100_Q40485 C1C-Nt1 protein [Nicotiana tabacum]
Length = 780
Score = 926 bits (2394), Expect = 0.0
Identities = 458/746 (61%), Positives = 565/746 (75%), Gaps = 2/746 (0%)
Query: 11 SEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQ 70
SE +R+PLLSS +S +N+TSQ+AI+GANVCPIESLDYEI EN+ FKQDWRSR VQI Q
Sbjct: 33 SESGVRQPLLSS-KSRVNNTSQIAIIGANVCPIESLDYEIIENDLFKQDWRSRKKVQIFQ 91
Query: 71 YICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLS 130
YI +KW L +IGL VG +GF N+AVEN+AG K + S++ML+ ++ F + NL
Sbjct: 92 YIFLKWTLVLLIGLSVGLVGFFLNIAVENIAGFKLLLISDLMLQDKYFRGFAAYACCNLV 151
Query: 131 LTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIG 190
L A I+ AFIAP AAGSGI EVKAYLNG+DA I TL VKI GS VS V+G
Sbjct: 152 LATCAGILCAFIAPAAAGSGIPEVKAYLNGIDAHSILAPSTLFVKIFGSALGVSAGFVVG 211
Query: 191 KAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPV 250
K GPMVHTGAC+A LLGQGGS++Y +TW+WL++FKNDRDRRDLI CG+AAG+AAAFRAPV
Sbjct: 212 KEGPMVHTGACIANLLGQGGSRKYHLTWKWLKYFKNDRDRRDLITCGAAAGVAAAFRAPV 271
Query: 251 GGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYS 310
GGVLFALEE+ASWWR+ALLWR FF+TA VA+ LR+ I C S KCGLFG+GGLIM+D S
Sbjct: 272 GGVLFALEEVASWWRSALLWRTFFSTAVVAMVLRSFIVFCRSGKCGLFGQGGLIMYDVNS 331
Query: 311 ASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFT 370
+ +Y+ +DV V ++ V+GGLLGSL+N++ +KVLR Y++INE+G ++ L ISI +
Sbjct: 332 GAPNYNTIDVLAVLLIGVLGGLLGSLYNYLVDKVLRTYSIINERGPAFKVLLVMTISILS 391
Query: 371 SCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRN 430
S +GLPW A C PCP + CPTIGRSG YK FQCP HYN LASL NTNDDAIRN
Sbjct: 392 SLCSYGLPWFATCTPCPVGLEDKCPTIGRSGNYKNFQCPAGHYNDLASLFMNTNDDAIRN 451
Query: 431 LFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGER 490
LFS +EF LSS+ VFF L + + GI P+GLF+P+I+ GASYGR VG ++G
Sbjct: 452 LFSSDNSSEFHLSSLFVFFAGVYCLGVVTYGIAIPSGLFIPVILAGASYGRFVGTVLGSI 511
Query: 491 TNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNA 550
+NL+NGL+A+LGAAS LGG+MR TVS+CVI+LELT++LL+LPL+M+VL++SK+VA+ FN
Sbjct: 512 SNLNNGLFALLGAASFLGGTMRMTVSICVILLELTDDLLMLPLVMLVLLISKTVADCFNH 571
Query: 551 NVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGF 610
VYD I+K KGLPYLE HAEPYMRQL GDV +GPL F+G+EKV NI+ L+ T HNGF
Sbjct: 572 GVYDQIVKMKGLPYLEAHAEPYMRQLVAGDVCSGPLITFSGVEKVGNIIHALKFTRHNGF 631
Query: 611 PVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVER 670
PVID PP S+AP G+ LR HL LLK K F V + ++R F + DFAK S +
Sbjct: 632 PVIDAPPFSDAPEFCGLALRSHLLVLLKAKKFTKLSVLSGSSILRSFHAFDFAKPGSGKG 691
Query: 671 VKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIP-KIPG 729
K+ED+ T+EEM M+VDLHP TN SPYTVVETMSLAKA ILFR++GLRHL V+P K G
Sbjct: 692 PKLEDLSFTDEEMEMYVDLHPVTNTSPYTVVETMSLAKAAILFRQLGLRHLCVVPKKTTG 751
Query: 730 RSPVVGILTRHDFTPEHILGMHPFLV 755
R P+VGILTRHDF PEHI G++P LV
Sbjct: 752 RDPIVGILTRHDFMPEHIKGLYPHLV 777
>UniRef100_Q96282 Chloride channel protein CLC-c [Arabidopsis thaliana]
Length = 779
Score = 924 bits (2387), Expect = 0.0
Identities = 452/740 (61%), Positives = 557/740 (75%), Gaps = 3/740 (0%)
Query: 16 RRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQYICMK 75
R+PLL+ R N+TSQ+AIVGAN CPIESLDYEIFEN+FFKQDWRSR ++ILQY +K
Sbjct: 38 RQPLLARNRK--NTTSQIAIVGANTCPIESLDYEIFENDFFKQDWRSRKKIEILQYTFLK 95
Query: 76 WLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLFA 135
W L F+IGL G +GF NNL VEN+AG K + N+ML+ ++ AFF F NL L A
Sbjct: 96 WALAFLIGLATGLVGFLNNLGVENIAGFKLLLIGNLMLKEKYFQAFFAFAGCNLILATAA 155
Query: 136 SIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPM 195
+ + AFIAP AAGSGI EVKAYLNG+DA I TL VKI GSI V+ V+GK GPM
Sbjct: 156 ASLCAFIAPAAAGSGIPEVKAYLNGIDAYSILAPSTLFVKIFGSIFGVAAGFVVGKEGPM 215
Query: 196 VHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLF 255
VHTGAC+A LLGQGGSK+Y +TW+WLRFFKNDRDRRDLI CG+AAG+AAAFRAPVGGVLF
Sbjct: 216 VHTGACIANLLGQGGSKKYRLTWKWLRFFKNDRDRRDLITCGAAAGVAAAFRAPVGGVLF 275
Query: 256 ALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASISY 315
ALEE ASWWR ALLWR FFTTA VA+ LR++I+ C S +CGLFGKGGLIMFD S + Y
Sbjct: 276 ALEEAASWWRNALLWRTFFTTAVVAVVLRSLIEFCRSGRCGLFGKGGLIMFDVNSGPVLY 335
Query: 316 HLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLF 375
D+ + L V+GG+LGSL+N++ +KVLR Y++INEKG ++ L +SI +SC F
Sbjct: 336 STPDLLAIVFLGVIGGVLGSLYNYLVDKVLRTYSIINEKGPRFKIMLVMAVSILSSCCAF 395
Query: 376 GLPWLAPCRPCPPDAVE-PCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSM 434
GLPWL+ C PCP E CP++GRS IYK FQCPPNHYN L+SL+ NTNDDAIRNLF+
Sbjct: 396 GLPWLSQCTPCPIGIEEGKCPSVGRSSIYKSFQCPPNHYNDLSSLLLNTNDDAIRNLFTS 455
Query: 435 HTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLS 494
++NEF +S++ +FF+ L I + GI P+GLF+P+I+ GASYGRLVG L+G + L
Sbjct: 456 RSENEFHISTLAIFFVAVYCLGIITYGIAIPSGLFIPVILAGASYGRLVGRLLGPVSQLD 515
Query: 495 NGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYD 554
GL+++LGAAS LGG+MR TVSLCVI+LELTNNLL+LPL+M+VL++SK+VA+ FN VYD
Sbjct: 516 VGLFSLLGAASFLGGTMRMTVSLCVILLELTNNLLMLPLVMLVLLISKTVADCFNRGVYD 575
Query: 555 LIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPVID 614
I+ KGLPY+E HAEPYMR L DVV+G L F+ +EKV I L+ T HNGFPVID
Sbjct: 576 QIVTMKGLPYMEDHAEPYMRNLVAKDVVSGALISFSRVEKVGVIWQALKMTRHNGFPVID 635
Query: 615 EPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIE 674
EPP +EA L GI LR HL LL+ K F ++R + DF K + +KIE
Sbjct: 636 EPPFTEASELCGIALRSHLLVLLQGKKFSKQRTTFGSQILRSCKARDFGKAGLGKGLKIE 695
Query: 675 DIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVV 734
D+ L+EEEM M+VDLHP TN SPYTV+ET+SLAKA ILFR++GLRHL V+PK PGR P+V
Sbjct: 696 DLDLSEEEMEMYVDLHPITNTSPYTVLETLSLAKAAILFRQLGLRHLCVVPKTPGRPPIV 755
Query: 735 GILTRHDFTPEHILGMHPFL 754
GILTRHDF PEH+LG++P +
Sbjct: 756 GILTRHDFMPEHVLGLYPHI 775
>UniRef100_P93567 Chloride channel Stclc1 [Solanum tuberosum]
Length = 764
Score = 923 bits (2385), Expect = 0.0
Identities = 451/741 (60%), Positives = 561/741 (74%), Gaps = 1/741 (0%)
Query: 15 LRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQYICM 74
+R PLL S +S +N+TSQ+AIVGANV PIESLDY+I EN+ FKQDWRSR V+I QYI +
Sbjct: 22 IRVPLLKS-KSRVNNTSQIAIVGANVYPIESLDYDIVENDLFKQDWRSRKKVEIFQYIFL 80
Query: 75 KWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLF 134
KW L +IGL G +GF NN+ VEN+AG K + TSN+ML+ ++ AF F N+
Sbjct: 81 KWTLVLLIGLSTGLVGFFNNIGVENIAGFKLLLTSNLMLDGKYFQAFAAFAGCNVFFATC 140
Query: 135 ASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGP 194
A+ + AFIAP AAGSGI EVKAYLNG+DA I TL VKI GSI VS V+GK GP
Sbjct: 141 AAALCAFIAPAAAGSGIPEVKAYLNGIDAHSILAPSTLLVKIFGSILGVSAGFVVGKEGP 200
Query: 195 MVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVL 254
MVHTGAC+A LLGQGGS++Y +TW+WL++FKNDRDRRDLI CG+AAG+AAAFRAPVGGVL
Sbjct: 201 MVHTGACIANLLGQGGSRKYHLTWKWLKYFKNDRDRRDLITCGAAAGVAAAFRAPVGGVL 260
Query: 255 FALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASIS 314
FALEE+ASWWR+ALLWR FFTTA VA+ LR++I C CGLFG+GGLIMFD S +
Sbjct: 261 FALEEIASWWRSALLWRTFFTTAIVAMVLRSLIQFCRGGNCGLFGQGGLIMFDVNSGVSN 320
Query: 315 YHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLL 374
Y+ +DV + + V+GGLLGSL+N++ +KVLR Y VINE+G ++ L +SI TSC
Sbjct: 321 YNTIDVLALIFIGVLGGLLGSLYNYLVDKVLRTYAVINERGPAFKILLVMSVSILTSCCS 380
Query: 375 FGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSM 434
+GLPW A C PCP E CPTIGRSG YK FQCP HYN LASL NTNDDAIRNLFS
Sbjct: 381 YGLPWFAGCIPCPVGLEEKCPTIGRSGNYKNFQCPAGHYNDLASLFLNTNDDAIRNLFSS 440
Query: 435 HTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLS 494
+ NEF +S++L+FF L I + GI P+GLF+P+I+ GASYGR+ G +G +NL+
Sbjct: 441 NNSNEFHISTLLIFFAGVYCLGIITYGIAIPSGLFIPVILAGASYGRIFGRALGSLSNLN 500
Query: 495 NGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYD 554
GL+++LGAAS LGG+MR TVS+CVI+LELTNNLL+LPL+M+VL++SK+VA++FN VYD
Sbjct: 501 VGLFSLLGAASFLGGTMRMTVSICVILLELTNNLLMLPLVMLVLLISKTVADIFNKGVYD 560
Query: 555 LIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPVID 614
I+K KGLP+LE HAEP+MR L GDV +GPL F+G+EKV NIV L+ T HNGFPVID
Sbjct: 561 QIVKMKGLPFLEAHAEPFMRNLVAGDVCSGPLLSFSGVEKVGNIVHALKYTRHNGFPVID 620
Query: 615 EPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIE 674
EPP SE P L G++LR HL LL K F V ++ +++ +F + DFAK S + +K E
Sbjct: 621 EPPFSETPELCGLVLRSHLLVLLNGKKFTKQRVLSASNILSRFHAFDFAKPGSGKGLKFE 680
Query: 675 DIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVV 734
D+ +TEEEM M++DLHP TN SPYTVVETMSLAKA ILFR++GLRHL V+PK GR+P+V
Sbjct: 681 DLVITEEEMEMYIDLHPITNTSPYTVVETMSLAKAAILFRQLGLRHLCVVPKKTGRAPIV 740
Query: 735 GILTRHDFTPEHILGMHPFLV 755
GILTRHDF EHI ++P LV
Sbjct: 741 GILTRHDFMHEHISNLYPHLV 761
>UniRef100_Q8LR18 Putative chloride channel protein [Oryza sativa]
Length = 773
Score = 898 bits (2321), Expect = 0.0
Identities = 443/745 (59%), Positives = 554/745 (73%), Gaps = 5/745 (0%)
Query: 15 LRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQYICM 74
L RPLL +R N+TSQ+AIVGANVCPIESLDYE+ ENE +KQDWRSRG +QI Y +
Sbjct: 30 LERPLL--RRRGTNTTSQMAIVGANVCPIESLDYELVENEVYKQDWRSRGKLQIFHYQIL 87
Query: 75 KWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLF 134
KW+L ++GLIVG IGF NN+AVEN+AG K + T+N+ML+ R+ AF F + N L
Sbjct: 88 KWVLALLVGLIVGLIGFFNNIAVENIAGFKLLLTTNLMLQNRYKAAFLWFISCNAMLAAA 147
Query: 135 ASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGP 194
A+ + A+ P AAGSGI EVKAYLNGVDAP I TL VKI+GSI VS V+GK GP
Sbjct: 148 AAALCAYFGPAAAGSGIPEVKAYLNGVDAPSILAPSTLFVKIVGSIFGVSAGFVLGKEGP 207
Query: 195 MVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVL 254
MVHTGACVA+ LGQGGS++YG TW WLR+FKND DRRDLI CG+AAG+ AAFRAPVGGVL
Sbjct: 208 MVHTGACVASFLGQGGSRKYGFTWNWLRYFKNDLDRRDLITCGAAAGVTAAFRAPVGGVL 267
Query: 255 FALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASIS 314
FALEE SWWR+ALLWR F TTA A+ LR++I+ C S CGLFGKGGLIMFD S S
Sbjct: 268 FALEEATSWWRSALLWRTFSTTAVAAMVLRSLIEYCRSGNCGLFGKGGLIMFDVSSQVTS 327
Query: 315 YHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLL 374
Y +D+ V +LA+VGGLLG+LFNF+ N++LR+Y+ INEKG ++ L +IS+ TSC
Sbjct: 328 YTTMDLAAVVLLAIVGGLLGALFNFLLNRILRVYSYINEKGAPYKIILTVVISLVTSCCS 387
Query: 375 FGLPWLAPCRPCPPD--AVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLF 432
FGLPWL C PCPP+ A CPTIGRSG +K F+CPP YN +ASL NTNDDAIRNLF
Sbjct: 388 FGLPWLTACTPCPPELAASGHCPTIGRSGNFKNFRCPPGQYNAMASLFLNTNDDAIRNLF 447
Query: 433 SMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTN 492
S T++EF + +L FF L + + G+ P+GLF+P+I++GAS+GRL+G L+G T
Sbjct: 448 SGGTESEFGVPMLLAFFTAVYSLGLVTYGVAVPSGLFIPVILSGASFGRLLGKLLGVLTG 507
Query: 493 LSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANV 552
L GL+A+LGAAS LGG+MR TVS+CVI+LELTN+LLLLPLIM+VL+VSK+VA+ FN V
Sbjct: 508 LDTGLFALLGAASFLGGTMRMTVSVCVILLELTNDLLLLPLIMLVLLVSKTVADCFNKGV 567
Query: 553 YDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPV 612
Y+ +++ KGLPYLE HAEP MR L GDVV+ PL F+ +E V +V LR T HNGFPV
Sbjct: 568 YEQMVRMKGLPYLEAHAEPCMRSLVAGDVVSAPLIAFSSVESVGTVVDTLRRTGHNGFPV 627
Query: 613 IDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPV-ANSYDVVRKFSSDDFAKKYSVERV 671
I++ P + P L G++LR HL LL+ K F V + +V RK + DFAK S + +
Sbjct: 628 IEDAPFAPEPELCGLVLRSHLLVLLRAKTFTADRVKTGAAEVFRKLAPFDFAKPGSGKGL 687
Query: 672 KIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRS 731
++D+ LTEEEM M+VDLHP N SPYTVVE MSLAKA +LFR++GLRH+ V+P+ PGR
Sbjct: 688 TVDDLDLTEEEMAMYVDLHPIANRSPYTVVENMSLAKAAVLFRQLGLRHMCVVPRTPGRP 747
Query: 732 PVVGILTRHDFTPEHILGMHPFLVK 756
PVVGILTRHDF P +I G+ P +++
Sbjct: 748 PVVGILTRHDFMPGYIRGLFPNVLR 772
>UniRef100_Q84QC8 Chloride channel [Zea mays]
Length = 719
Score = 859 bits (2219), Expect = 0.0
Identities = 429/712 (60%), Positives = 528/712 (73%), Gaps = 12/712 (1%)
Query: 50 IFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTS 109
+ EN+ FKQDWRSR QI QYI +KW L +IGL+ GF+GF NNLAVEN+AG K + TS
Sbjct: 1 VVENDLFKQDWRSRKKKQIFQYIVLKWSLVLLIGLLTGFVGFFNNLAVENIAGFKLLLTS 60
Query: 110 NMMLERRFMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTV 169
++ML+ R++ AFF++ NL L A+ I A+IAP AAGSGI EVKAYLNGVDA I
Sbjct: 61 DLMLKGRYIRAFFVYGGCNLVLAAAAAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAP 120
Query: 170 RTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRD 229
TL VKI GSI VS V+GK GPMVHTGA +A LLGQGGS++Y +T WLR+FKNDRD
Sbjct: 121 STLFVKIFGSILGVSAGFVLGKEGPMVHTGAFIANLLGQGGSRKYHLTCNWLRYFKNDRD 180
Query: 230 RRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDV 289
RRDLI CGSAAG+AAAFRAPVGGVLFALEE ASWWR+ALLWR FFTTA VA+ LR +I+
Sbjct: 181 RRDLITCGSAAGVAAAFRAPVGGVLFALEEAASWWRSALLWRTFFTTAVVAVVLRGLIEF 240
Query: 290 CLSDKCGLFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYN 349
C S KCGLFG+GGLIMFD S Y D+ + +L ++GG+ G LFN++ +K+LR+Y+
Sbjct: 241 CRSGKCGLFGQGGLIMFDLSSTVAVYSTPDLIAIIVLGIIGGIFGGLFNYLLDKILRVYS 300
Query: 350 VINEKGTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVE------PCPTIGRSGIY 403
+INE+G ++ L ISI TS +GLPWLA C PCP DAV P P I R+
Sbjct: 301 IINERGAPFKILLTIPISIITSMGSYGLPWLAACTPCPVDAVGAVSHCWPLPEIIRTS-- 358
Query: 404 KKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIV 463
+CPP HYNGLASL FNTNDDAIRNLFS T EF++SS+ +FF L + + GI
Sbjct: 359 ---KCPPGHYNGLASLFFNTNDDAIRNLFSNGTSTEFQMSSLFIFFTAIYCLGLVTYGIA 415
Query: 464 APAGLFVPIIVTGASYGRLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLE 523
P+GLF+P+I+ GA+YGR+VG L+G ++L GL+A+LGAAS LGG+MR TVS+CVI+LE
Sbjct: 416 VPSGLFIPVILAGATYGRIVGTLLGSISDLDPGLFALLGAASFLGGTMRMTVSVCVILLE 475
Query: 524 LTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVT 583
LTN+L +LPL+M+VL++SK++A+ FN VYD I+ KGLPY+E HAEPYMR L GDVV+
Sbjct: 476 LTNDLPMLPLVMLVLLISKTIADSFNKGVYDQIVVMKGLPYMEAHAEPYMRHLVAGDVVS 535
Query: 584 GPLQMFNGIEKVRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFL 643
GPL F+G+EKV NIV LR T HNGFPV+DEPP +E P L G++ R HL LL K F+
Sbjct: 536 GPLITFSGVEKVGNIVHALRLTGHNGFPVLDEPPITETPELVGLVTRSHLLVLLNSKNFM 595
Query: 644 PSPVANSYD-VVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVE 702
V S V+R+F + DFAK S + +KIED+ T+EEM M+VDLHP TN SPYTVVE
Sbjct: 596 KGRVKTSGSFVLRRFGAFDFAKPGSGKGLKIEDLDFTDEEMDMYVDLHPITNTSPYTVVE 655
Query: 703 TMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHDFTPEHILGMHPFL 754
TMSLAKA ILFRE+GLRHLLV+PK P R P+VGILTRHDF PEHI + P L
Sbjct: 656 TMSLAKAAILFRELGLRHLLVVPKTPDRPPIVGILTRHDFMPEHIHSLFPNL 707
>UniRef100_Q8LPA0 Chloride channel [Oryza sativa]
Length = 726
Score = 828 bits (2139), Expect = 0.0
Identities = 410/683 (60%), Positives = 510/683 (74%), Gaps = 10/683 (1%)
Query: 7 SNGESEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVV 66
S EPLLR+ + N+TSQ+AIVGANVCPIESLDYE+ EN+ FKQDWRSR
Sbjct: 51 SGSAGEPLLRKRTM-------NTTSQIAIVGANVCPIESLDYEVVENDLFKQDWRSRKKK 103
Query: 67 QILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLER--RFMFAFFIF 124
QI QYI +KW L +IGL+ G +GF NNLAVEN+AG K + T N+ML+ R++ AFF +
Sbjct: 104 QIFQYIVLKWTLVLLIGLLTGLVGFFNNLAVENIAGFKLLLTGNLMLKGKCRYLTAFFAY 163
Query: 125 FASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVS 184
NL L A+ I A+IAP AAGSGI EVKAYLNGVDA I TL VKI GSI VS
Sbjct: 164 GGCNLVLAAAAAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAPSTLFVKIFGSILGVS 223
Query: 185 GSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAA 244
V+GK GPMVHTGAC+A LLGQGGS++Y +T WLR+FKNDRDRRDLI CGSAAG+AA
Sbjct: 224 AGFVLGKEGPMVHTGACIANLLGQGGSRKYRLTCNWLRYFKNDRDRRDLITCGSAAGVAA 283
Query: 245 AFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLI 304
AFRAPVGGVLFALEE ASWWR+ALLWRAFFTTA VA+ LR++I+ C S KCGLFG+GGLI
Sbjct: 284 AFRAPVGGVLFALEEAASWWRSALLWRAFFTTAVVAVVLRSLIEFCRSGKCGLFGQGGLI 343
Query: 305 MFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLAC 364
MFD S +Y D+ + IL ++GG+ G LFNF+ +KVLR+Y++INE+G ++ L
Sbjct: 344 MFDLSSTVATYSTPDLIAIIILGIIGGIFGGLFNFLLDKVLRVYSIINERGAPFKILLTI 403
Query: 365 LISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTN 424
ISI TS +GLPWLA C PCP DAVE CPTIGRSG +K FQCPP HYN LASL FNTN
Sbjct: 404 TISIITSMCSYGLPWLAACTPCPVDAVEQCPTIGRSGNFKNFQCPPGHYNDLASLFFNTN 463
Query: 425 DDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVG 484
DDAIRNLFS T++EF +S++ +FF L I + G+ P+GLF+P+I+ GA+YGR+VG
Sbjct: 464 DDAIRNLFSNGTESEFHMSTLFIFFTAVYCLGILTYGVAVPSGLFIPVILAGATYGRIVG 523
Query: 485 ILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSV 544
L+G ++L GL+A+LGAAS LGG+MR TVS+CVI+LELTN+L +LPL+M+VL++SK++
Sbjct: 524 TLLGSISDLDPGLFALLGAASFLGGTMRMTVSVCVILLELTNDLAMLPLVMLVLLISKTI 583
Query: 545 ANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRT 604
A+ FN VYD I+ KGLPY+E HAEPYMR L GDVV+GPL F+G+EKV NIV LR
Sbjct: 584 ADNFNKGVYDQIVVMKGLPYMEAHAEPYMRHLVAGDVVSGPLITFSGVEKVGNIVHALRF 643
Query: 605 TAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYD-VVRKFSSDDFA 663
T HNGFPV+DEPP +EAP L G++ R HL LL K F+ + S V+++F + DFA
Sbjct: 644 TGHNGFPVVDEPPLTEAPELVGLVTRSHLLVLLNGKMFMKDQLKTSGSFVLQRFGAFDFA 703
Query: 664 KKYSVERVKIEDIQLTEEEMGMF 686
K S + +KI+D+ T+EEM M+
Sbjct: 704 KPGSGKGLKIQDLDFTDEEMEMY 726
>UniRef100_P92942 Chloride channel protein CLC-b [Arabidopsis thaliana]
Length = 780
Score = 823 bits (2126), Expect = 0.0
Identities = 408/769 (53%), Positives = 547/769 (71%), Gaps = 18/769 (2%)
Query: 2 SNNHLSNGESEP-LLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDW 60
SN + G+ E L +PL+ + R++ S++ +A+VGA V IESLDYEI EN+ FK DW
Sbjct: 13 SNYNGEGGDPESNTLNQPLVKANRTL--SSTPLALVGAKVSHIESLDYEINENDLFKHDW 70
Query: 61 RSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFA 120
R R Q+LQY+ +KW L ++GL G I NLAVEN+AG K + + + + R++
Sbjct: 71 RKRSKAQVLQYVFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGHFLTQERYVTG 130
Query: 121 FFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSI 180
+ +NL LTL AS++ APTAAG GI E+KAYLNGVD P +F T+ VKI+GSI
Sbjct: 131 LMVLVGANLGLTLVASVLCVCFAPTAAGPGIPEIKAYLNGVDTPNMFGATTMIVKIVGSI 190
Query: 181 TAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAA 240
AV+ L +GK GP+VH G+C+A+LLGQGG+ + I WRWLR+F NDRDRRDLI CGSAA
Sbjct: 191 GAVAAGLDLGKEGPLVHIGSCIASLLGQGGTDNHRIKWRWLRYFNNDRDRRDLITCGSAA 250
Query: 241 GIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGK 300
G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA V + LR I++C S KCGLFGK
Sbjct: 251 GVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLREFIEICNSGKCGLFGK 310
Query: 301 GGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRL 360
GGLIMFD + +YH+ D+ PV ++ V+GG+LGSL+N + +KVLR+YN+INEKG I ++
Sbjct: 311 GGLIMFDVSHVTYTYHVTDIIPVMLIGVIGGILGSLYNHLLHKVLRLYNLINEKGKIHKV 370
Query: 361 FLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLI 420
L+ +S+FTS L+GLP+LA C+PC P E CPT GRSG +K+F CP +YN LA+L+
Sbjct: 371 LLSLTVSLFTSVCLYGLPFLAKCKPCDPSIDEICPTNGRSGNFKQFHCPKGYYNDLATLL 430
Query: 421 FNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYG 480
TNDDA+RNLFS +T NEF + S+ +FF++ L +F+ GI P+GLF+PII+ GA+YG
Sbjct: 431 LTTNDDAVRNLFSSNTPNEFGMGSLWIFFVLYCILGLFTFGIATPSGLFLPIILMGAAYG 490
Query: 481 RLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVV 540
R++G +G T++ GLYAVLGAA+L+ GSMR TVSLCVI LELTNNLLLLP+ M+VL++
Sbjct: 491 RMLGAAMGSYTSIDQGLYAVLGAAALMAGSMRMTVSLCVIFLELTNNLLLLPITMIVLLI 550
Query: 541 SKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDV--VTGPLQMFNGIEKVRNI 598
+K+V + FN ++YD+I+ KGLP+LE + EP+MR LTVG++ P+ G+EKV NI
Sbjct: 551 AKTVGDSFNPSIYDIILHLKGLPFLEANPEPWMRNLTVGELGDAKPPVVTLQGVEKVSNI 610
Query: 599 VFILRTTAHNGFPVIDEPP------GSEAPILFGIILRHHLTTLLKKKAFLPSP-VANSY 651
V +L+ T HN FPV+DE + A L G+ILR HL +LKK+ FL +
Sbjct: 611 VDVLKNTTHNAFPVLDEAEVPQVGLATGATELHGLILRAHLVKVLKKRWFLTEKRRTEEW 670
Query: 652 DVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALI 711
+V KF D+ A++ +D+ +T EM M+VDLHP TN +PYTV+E MS+AKAL+
Sbjct: 671 EVREKFPWDELAERED----NFDDVAITSAEMEMYVDLHPLTNTTPYTVMENMSVAKALV 726
Query: 712 LFREVGLRHLLVIPKI--PGRSPVVGILTRHDFTPEHILGMHPFLVKSR 758
LFR+VGLRHLL++PKI G PVVGILTR D +IL P L KS+
Sbjct: 727 LFRQVGLRHLLIVPKIQASGMCPVVGILTRQDLRAYNILQAFPLLEKSK 775
>UniRef100_Q9XF71 CLC-Nt2 protein [Nicotiana tabacum]
Length = 786
Score = 812 bits (2098), Expect = 0.0
Identities = 403/755 (53%), Positives = 544/755 (71%), Gaps = 17/755 (2%)
Query: 15 LRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQYICM 74
L +PLL R++ S+S A+VGA V IESLDYEI EN+ FK DWR R VQ+LQY+ +
Sbjct: 32 LHQPLLKRNRTL--SSSPFALVGAKVSHIESLDYEINENDLFKHDWRRRSRVQVLQYVFL 89
Query: 75 KWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLF 134
KW L F++GL+ G NLA+EN+AG K N + +RR++ F F +N LTL
Sbjct: 90 KWTLAFLVGLLTGVTATLINLAIENMAGYKLRAVVNYIEDRRYLMGFAYFAGANFVLTLI 149
Query: 135 ASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGP 194
A+++ APTAAG GI E+KAYLNGVD P ++ TL VKIIGSI AVS SL +GK GP
Sbjct: 150 AALLCVCFAPTAAGPGIPEIKAYLNGVDTPNMYGATTLFVKIIGSIAAVSASLDLGKEGP 209
Query: 195 MVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVL 254
+VH GAC A+LLGQGG Y + WRWLR+F NDRDRRDLI CGS++G+ AAFR+PVGGVL
Sbjct: 210 LVHIGACFASLLGQGGPDNYRLRWRWLRYFNNDRDRRDLITCGSSSGVCAAFRSPVGGVL 269
Query: 255 FALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASIS 314
FALEE+A+WWR+ALLWR FF+TA V + LRA I+ C S CGLFG+GGLIMFD S+S
Sbjct: 270 FALEEVATWWRSALLWRTFFSTAVVVVILRAFIEYCKSGNCGLFGRGGLIMFDVSGVSVS 329
Query: 315 YHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLL 374
YH+VD+ PV ++ ++GGLLGSL+N + +K+LR+YN+INEKG + ++ LA +S+FTS +
Sbjct: 330 YHVVDIIPVVVIGIIGGLLGSLYNHVLHKILRLYNLINEKGKLHKVLLALSVSLFTSICM 389
Query: 375 FGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSM 434
+GLP+LA C+PC P CP G +G +K+F CP +YN LA+L+ TNDDA+RN+FS+
Sbjct: 390 YGLPFLAKCKPCDPSLPGSCPGTGGTGNFKQFNCPDGYYNDLATLLLTTNDDAVRNIFSI 449
Query: 435 HTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLS 494
+T EF++ S++ +F++ L + + GI P+GLF+PII+ G++YGRL+ I +G T +
Sbjct: 450 NTPGEFQVMSLISYFVLYCILGLITFGIAVPSGLFLPIILMGSAYGRLLAIAMGSYTKID 509
Query: 495 NGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYD 554
GLYAVLGAASL+ GSMR TVSLCVI LELTNNLLLLP+ M+VL+++KSV + FN ++Y+
Sbjct: 510 PGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMLVLLIAKSVGDCFNLSIYE 569
Query: 555 LIMKAKGLPYLETHAEPYMRQLTVGDV--VTGPLQMFNGIEKVRNIVFILRTTAHNGFPV 612
+I++ KGLP+L+ + EP+MR +T G++ V P+ G+EKV IV L+ T +NGFPV
Sbjct: 570 IILELKGLPFLDANPEPWMRNITAGELADVKPPVVTLCGVEKVGRIVEALKNTTYNGFPV 629
Query: 613 IDE---PPGS---EAPILFGIILRHHLTTLLKKKAFL-PSPVANSYDVVRKFSSDDFAKK 665
+DE PP A L G++LR HL +LKKK FL ++V KF+ D A++
Sbjct: 630 VDEGVVPPVGLPVGATELHGLVLRTHLLLVLKKKWFLHERRRTEEWEVREKFTWIDLAER 689
Query: 666 YSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIP 725
KIED+ +T++EM M+VDLHP TN +PYTVVE++S+AKA++LFR+VGLRH+L++P
Sbjct: 690 GG----KIEDVLVTKDEMEMYVDLHPLTNTTPYTVVESLSVAKAMVLFRQVGLRHMLIVP 745
Query: 726 K--IPGRSPVVGILTRHDFTPEHILGMHPFLVKSR 758
K G SPVVGILTR D +IL + P L KS+
Sbjct: 746 KYQAAGVSPVVGILTRQDLRAHNILSVFPHLEKSK 780
>UniRef100_P92941 Chloride channel protein CLC-a [Arabidopsis thaliana]
Length = 775
Score = 811 bits (2096), Expect = 0.0
Identities = 406/772 (52%), Positives = 540/772 (69%), Gaps = 17/772 (2%)
Query: 3 NNHLSNGESEP------LLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFF 56
+N NGE E L +PLL R++ S++ +A+VGA V IESLDYEI EN+ F
Sbjct: 10 SNSNYNGEEEGEDPENNTLNQPLLKRHRTL--SSTPLALVGAKVSHIESLDYEINENDLF 67
Query: 57 KQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERR 116
K DWRSR Q+ QYI +KW L ++GL G I NLAVEN+AG K + + + R
Sbjct: 68 KHDWRSRSKAQVFQYIFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGYYIAQDR 127
Query: 117 FMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKI 176
F +F +NL LTL A+++ + APTAAG GI E+KAYLNG+D P +F T+ VKI
Sbjct: 128 FWTGLMVFTGANLGLTLVATVLVVYFAPTAAGPGIPEIKAYLNGIDTPNMFGFTTMMVKI 187
Query: 177 IGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIIC 236
+GSI AV+ L +GK GP+VH G+C+A+LLGQGG + I WRWLR+F NDRDRRDLI C
Sbjct: 188 VGSIGAVAAGLDLGKEGPLVHIGSCIASLLGQGGPDNHRIKWRWLRYFNNDRDRRDLITC 247
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCG 296
GSA+G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA V + LRA I++C S KCG
Sbjct: 248 GSASGVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLRAFIEICNSGKCG 307
Query: 297 LFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGT 356
LFG GGLIMFD + YH D+ PV ++ V GG+LGSL+N + +KVLR+YN+IN+KG
Sbjct: 308 LFGSGGLIMFDVSHVEVRYHAADIIPVTLIGVFGGILGSLYNHLLHKVLRLYNLINQKGK 367
Query: 357 ICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGL 416
I ++ L+ +S+FTS LFGLP+LA C+PC P E CPT GRSG +K+F CP +YN L
Sbjct: 368 IHKVLLSLGVSLFTSVCLFGLPFLAECKPCDPSIDEICPTNGRSGNFKQFNCPNGYYNDL 427
Query: 417 ASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTG 476
++L+ TNDDA+RN+FS +T NEF + S+ +FF + L + + GI P+GLF+PII+ G
Sbjct: 428 STLLLTTNDDAVRNIFSSNTPNEFGMVSLWIFFGLYCILGLITFGIATPSGLFLPIILMG 487
Query: 477 ASYGRLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMM 536
++YGR++G +G TN+ GLYAVLGAASL+ GSMR TVSLCVI LELTNNLLLLP+ M
Sbjct: 488 SAYGRMLGTAMGSYTNIDQGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMF 547
Query: 537 VLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDV--VTGPLQMFNGIEK 594
VL+++K+V + FN ++Y++I+ KGLP+LE + EP+MR LTVG++ P+ NG+EK
Sbjct: 548 VLLIAKTVGDSFNLSIYEIILHLKGLPFLEANPEPWMRNLTVGELNDAKPPVVTLNGVEK 607
Query: 595 VRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFL-PSPVANSYDV 653
V NIV +LR T HN FPV+D + L G+ILR HL +LKK+ FL ++V
Sbjct: 608 VANIVDVLRNTTHNAFPVLDGADQNTGTELHGLILRAHLVKVLKKRWFLNEKRRTEEWEV 667
Query: 654 VRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILF 713
KF+ + A++ +D+ +T EM ++VDLHP TN +PYTVV++MS+AKAL+LF
Sbjct: 668 REKFTPVELAERED----NFDDVAITSSEMQLYVDLHPLTNTTPYTVVQSMSVAKALVLF 723
Query: 714 REVGLRHLLVIPKI--PGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLR 763
R VGLRHLLV+PKI G SPV+GILTR D +IL P L K + + R
Sbjct: 724 RSVGLRHLLVVPKIQASGMSPVIGILTRQDLRAYNILQAFPHLDKHKSGKAR 775
>UniRef100_Q96325 Voltage-gated chloride channel [Arabidopsis thaliana]
Length = 773
Score = 778 bits (2008), Expect = 0.0
Identities = 398/772 (51%), Positives = 529/772 (67%), Gaps = 19/772 (2%)
Query: 3 NNHLSNGESEP------LLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFF 56
+N NGE E L +PLL R++ S++ +A+VGA V IESLDYEI EN+ F
Sbjct: 10 SNSNYNGEEEGEDPENNTLNQPLLKRHRTL--SSTPLALVGAKVSHIESLDYEINENDLF 67
Query: 57 KQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERR 116
K DWRSR Q+ QYI +KW L ++GL G I NLAVEN+AG K + + + R
Sbjct: 68 KHDWRSRSKAQVFQYIFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGYYIAQDR 127
Query: 117 FMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKI 176
F +F +NL LTL A+++ + APTAAG GI E+KAYLNG+D P +F T+ VKI
Sbjct: 128 FWTGLMVFTGANLGLTLVATVLVVYFAPTAAGPGIPEIKAYLNGIDTPNMFGFTTMMVKI 187
Query: 177 IGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIIC 236
+GSI AV+ L +GK GP+VH G+C+A+LLGQGG + I WRWLR+F NDRDRRDLI C
Sbjct: 188 VGSIGAVAAGLDLGKEGPLVHIGSCIASLLGQGGPDNHRIKWRWLRYFNNDRDRRDLITC 247
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCG 296
GSA+G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA V + LRA I++C S KCG
Sbjct: 248 GSASGVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLRAFIEICNSGKCG 307
Query: 297 LFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGT 356
LFG GGLIMFD + YH D+ PV ++ V GG+LGSL+N + +KVLR+YN+IN+KG
Sbjct: 308 LFGSGGLIMFDVSHVEVRYHAADIIPVTLIGVFGGILGSLYNHLLHKVLRLYNLINQKGK 367
Query: 357 ICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGL 416
I ++ L+ +S+ LLFGLP+LA C P E CPT GRSG +K+F CP +YN L
Sbjct: 368 IHKVLLSLGVSLLHQ-LLFGLPFLAN-EACDPTIDEICPTNGRSGNFKQFNCPNGYYNDL 425
Query: 417 ASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTG 476
++L+ TNDDA+R F EF + S+ +FF + L + + GI P+GLF+PII+ G
Sbjct: 426 STLLLTTNDDAVRKHFLFKHSYEFGMVSLWIFFGLYCILGLITFGIATPSGLFLPIILMG 485
Query: 477 ASYGRLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMM 536
++YGR++G +G TN+ GLYAVLGAASL+ GSMR TVSLCVI LELTNNLLLLP+ M
Sbjct: 486 SAYGRMLGTAMGSYTNIDQGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMF 545
Query: 537 VLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDV--VTGPLQMFNGIEK 594
VL+++K+V + FN ++Y++I+ KGLP+LE + EP+MR LTVG++ P+ NG+EK
Sbjct: 546 VLLIAKTVGDSFNLSIYEIILHLKGLPFLEANPEPWMRNLTVGELNDAKPPVVTLNGVEK 605
Query: 595 VRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFL-PSPVANSYDV 653
V NIV +LR T HN FPV+D + L G+ILR HL +LKK+ FL ++V
Sbjct: 606 VANIVDVLRNTTHNAFPVLDGADQNTGTELHGLILRAHLVKVLKKRWFLNEKRRTEEWEV 665
Query: 654 VRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILF 713
KF+ + A++ +D+ +T EM ++VDLHP TN +PYTVV++MS+AKAL+LF
Sbjct: 666 REKFTPVELAERED----NFDDVAITSSEMQLYVDLHPLTNTTPYTVVQSMSVAKALVLF 721
Query: 714 REVGLRHLLVIPKI--PGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLR 763
R VGLRHLLV+PKI G SPV+GILTR D +IL P L K + + R
Sbjct: 722 RSVGLRHLLVVPKIQASGMSPVIGILTRQDLRAYNILQAFPHLDKHKSGKAR 773
>UniRef100_Q851D8 Putative CLC-d chloride channel protein [Oryza sativa]
Length = 782
Score = 618 bits (1593), Expect = e-175
Identities = 328/716 (45%), Positives = 461/716 (63%), Gaps = 20/716 (2%)
Query: 43 IESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAG 102
+ESLDYE+ EN ++++ R + Y+ +KWL +IG+ G NLAVEN +G
Sbjct: 41 VESLDYEVIENYAYREEQAQRSKFWVPYYVMLKWLFSLLIGVGTGLAAIFINLAVENFSG 100
Query: 103 IKFVTTSNMMLERRFMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVD 162
K+ T +++ + FF++ NL+L + I AP AAGSGI E+K YLNGVD
Sbjct: 101 WKYAATF-AIIQHSYFVGFFVYIVFNLALVFSSVYIVTNFAPAAAGSGIPEIKGYLNGVD 159
Query: 163 APGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLR 222
GI RTL KI GSI +V G L +GK GP+VHTGAC+A+LLGQGGS +Y ++ RW+R
Sbjct: 160 THGILLFRTLVGKIFGSIGSVGGGLALGKEGPLVHTGACIASLLGQGGSAKYHLSSRWVR 219
Query: 223 FFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIF 282
F++DRDRRDL+ CG AAG+AAAFRAPVGGVLFALEE+ SWWR+ L+WR FFT+A VA+
Sbjct: 220 IFESDRDRRDLVTCGCAAGVAAAFRAPVGGVLFALEEVTSWWRSHLMWRVFFTSAVVAVV 279
Query: 283 LRAMIDVCLSDKCGLFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMT- 341
+R+ ++ C S KCG FG GG I++D Y ++ P+ I+ V+GGLLG+LFN +T
Sbjct: 280 VRSAMNWCKSGKCGHFGSGGFIIWDISGGQEDYSYQELLPMAIIGVIGGLLGALFNQLTL 339
Query: 342 --NKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGLPWLAPCRPCPP----DAVEPCP 395
K R Y +++KG ++F ACLIS+ TS + F LP + C CP +E
Sbjct: 340 YITKWRRTY--LHKKGKRVKIFEACLISLVTSTISFVLPLMRKCSSCPQLETNSGIECPR 397
Query: 396 TIGRSGIYKKFQC-PPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLF 454
G G + F C N YN LA++ FNT DDAIRNLFS T +E+ S++ F ++
Sbjct: 398 PPGTDGNFVNFYCSKDNEYNDLATIFFNTQDDAIRNLFSAKTFHEYSAQSLITFLVMFYS 457
Query: 455 LSIFSCGIVAPAGLFVPIIVTGASYGRLVGILV---GERTNLSNGLYAVLGAASLLGGSM 511
L++ + G PAG FVP I+ G++YGRLVG+ V ++ N+ G YA+LGAAS LGGSM
Sbjct: 458 LAVVTFGTAVPAGQFVPGIMIGSTYGRLVGMFVVKFYKKLNVEEGTYALLGAASFLGGSM 517
Query: 512 RTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEP 571
R TVSLCVIM+E+TNNL LLPLIM+VL++SK+V + FN +Y++ + +G+P L++ +
Sbjct: 518 RMTVSLCVIMVEITNNLKLLPLIMLVLLISKAVGDFFNEGLYEVQAQLRGIPLLDSRPKQ 577
Query: 572 YMRQLTVGDVVTG-PLQMFNGIEKVRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILR 630
MR ++ D + + ++ +I+ +LR+ HNGFPV+D E+ ++ G+ILR
Sbjct: 578 VMRNMSAKDACKNQKVVSLPRVSRIVDIISVLRSNKHNGFPVVDRGQNGES-LVIGLILR 636
Query: 631 HHLTTLLKKKA-FLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDL 689
HL LL+ K F SP ++ + ++ DF K S + I+DI LTE+E+G+++DL
Sbjct: 637 SHLLVLLQSKVDFQNSPFPCGPGILNRHNTSDFVKPASSKGKSIDDIHLTEDELGLYLDL 696
Query: 690 HPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHDFTPE 745
PF N SPY V E MSLAK LFR++GLRH+ V+P+ P R VVG++TR D E
Sbjct: 697 APFLNPSPYIVPEDMSLAKVYNLFRQLGLRHIFVVPR-PSR--VVGLITRQDLLLE 749
>UniRef100_P92943 Chloride channel protein CLC-d [Arabidopsis thaliana]
Length = 792
Score = 614 bits (1583), Expect = e-174
Identities = 344/763 (45%), Positives = 479/763 (62%), Gaps = 28/763 (3%)
Query: 1 MSNNHLSNG-ESEPLLRRPLLSSQRSIINSTSQVAIVGANVCP---IESLDYEIFENEFF 56
M +NHL NG ES+ LL + S + ST + ++ ++ + SLDYE+ EN +
Sbjct: 1 MLSNHLQNGIESDNLLWSRVPESDDT---STDDITLLNSHRDGDGGVNSLDYEVIENYAY 57
Query: 57 KQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERR 116
+++ RG + + Y+ +KW +IG+ G NL+VEN AG KF T ++++
Sbjct: 58 REEQAHRGKLYVGYYVAVKWFFSLLIGIGTGLAAVFINLSVENFAGWKFALTF-AIIQKS 116
Query: 117 FMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKI 176
+ F ++ NL L ++ I AP AAGSGI E+K YLNG+D PG RTL KI
Sbjct: 117 YFAGFIVYLLINLVLVFSSAYIITQFAPAAAGSGIPEIKGYLNGIDIPGTLLFRTLIGKI 176
Query: 177 IGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIIC 236
GSI +V G L +GK GP+VHTGAC+A+LLGQGGS +Y + RW + FK+DRDRRDL+ C
Sbjct: 177 FGSIGSVGGGLALGKEGPLVHTGACIASLLGQGGSTKYHLNSRWPQLFKSDRDRRDLVTC 236
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCG 296
G AAG+AAAFRAPVGGVLFALEE+ SWWR+ L+WR FFT+A VA+ +R + C S CG
Sbjct: 237 GCAAGVAAAFRAPVGGVLFALEEVTSWWRSQLMWRVFFTSAIVAVVVRTAMGWCKSGICG 296
Query: 297 LFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLR-IYNVINEKG 355
FG GG I++D Y+ ++ P+ ++ V+GGLLG+LFN +T + N +++KG
Sbjct: 297 HFGGGGFIIWDVSDGQDDYYFKELLPMAVIGVIGGLLGALFNQLTLYMTSWRRNSLHKKG 356
Query: 356 TICRLFLACLISIFTSCLLFGLPWLAPCRPCP---PDAVEPCP-TIGRSGIYKKFQC-PP 410
++ AC+IS TS + FGLP L C PCP PD+ CP G G Y F C
Sbjct: 357 NRVKIIEACIISCITSAISFGLPLLRKCSPCPESVPDSGIECPRPPGMYGNYVNFFCKTD 416
Query: 411 NHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFV 470
N YN LA++ FNT DDAIRNLFS T EF S+L F + L++ + G PAG FV
Sbjct: 417 NEYNDLATIFFNTQDDAIRNLFSAKTMREFSAQSLLTFLAMFYTLAVVTFGTAVPAGQFV 476
Query: 471 PIIVTGASYGRLVGILV---GERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNN 527
P I+ G++YGRLVG+ V ++ N+ G YA+LGAAS LGGSMR TVSLCVIM+E+TNN
Sbjct: 477 PGIMIGSTYGRLVGMFVVRFYKKLNIEEGTYALLGAASFLGGSMRMTVSLCVIMVEITNN 536
Query: 528 LLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVV-TGPL 586
L LLPLIM+VL++SK+V + FN +Y++ + KG+P LE+ + +MRQ+ + + +
Sbjct: 537 LKLLPLIMLVLLISKAVGDAFNEGLYEVQARLKGIPLLESRPKYHMRQMIAKEACQSQKV 596
Query: 587 QMFNGIEKVRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKA-FLPS 645
+ +V ++ IL + HNGFPVID E ++ G++LR HL LL+ K F S
Sbjct: 597 ISLPRVIRVADVASILGSNKHNGFPVIDHTRSGET-LVIGLVLRSHLLVLLQSKVDFQHS 655
Query: 646 PV---ANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVE 702
P+ ++ ++ FS +FAK S + + IEDI LT +++ M++DL PF N SPY V E
Sbjct: 656 PLPCDPSARNIRHSFS--EFAKPVSSKGLCIEDIHLTSDDLEMYIDLAPFLNPSPYVVPE 713
Query: 703 TMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHDFTPE 745
MSL K LFR++GLRHL V+P+ P R V+G++TR D E
Sbjct: 714 DMSLTKVYNLFRQLGLRHLFVVPR-PSR--VIGLITRKDLLIE 753
>UniRef100_UPI00003619FE UPI00003619FE UniRef100 entry
Length = 729
Score = 400 bits (1028), Expect = e-110
Identities = 254/722 (35%), Positives = 392/722 (54%), Gaps = 32/722 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGI 103
ESLDY+ EN+ F ++ R + +W++C +IG + G I ++ VE LAGI
Sbjct: 16 ESLDYDNIENQLFLEEERRMSYMSFRCLEISRWVVCGLIGFLTGLIACFIDIVVEELAGI 75
Query: 104 KFVTTSNMMLERR----FMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLN 159
K+ + + + ++ N + SII AF P AAGSGI ++K YLN
Sbjct: 76 KYQVVKENIDKFTEVGGLSISLILWAVLNSVFVMLGSIIVAFFEPIAAGSGIPQIKCYLN 135
Query: 160 GVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWR 219
GV P + ++TL VK+ G I +V G L +GK GPM+H+GA VAA + QG S ++
Sbjct: 136 GVKIPRVVRLKTLLVKVCGVICSVVGGLAVGKEGPMIHSGAVVAAGVSQGRSTSLKRDFK 195
Query: 220 WLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATV 279
+F+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF +
Sbjct: 196 MFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQMLTWRIFFASMIS 255
Query: 280 AIFLRAMIDVCLSDKCGLFGKGGLIMFDAY-SASISYHLVDVPPVFILAVVGGLLGSLFN 338
L + + ++ G GLI F + S S++Y+L ++P + +GGLLG+LFN
Sbjct: 256 TFTLNFFLSI-YNNNPGDLSNPGLINFGRFESESVAYNLYEIPLFIAMGAIGGLLGALFN 314
Query: 339 FMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGLPWLA-PCRPCPPDAVEPCPTI 397
+ N L I+ + ++ A L++ T+ + F + + + C+P + E P
Sbjct: 315 IL-NYWLTIFRIRYVHRPCLQVMEAMLVAAVTATVSFTMIYFSNDCQPLGSEHSEEYPL- 372
Query: 398 GRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSI 457
+ C YN +A+ FNT + ++R+LF + + ++ +F + FL+
Sbjct: 373 -------QLFCADGEYNSMATAFFNTPERSVRSLFH-NQPRTYNPLTLGLFTLTYFFLAC 424
Query: 458 FSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLSN-----GLYAVLGAASLLGGSMR 512
++ G+ AG+F+P ++ GA++GRL GIL+ T+ + G YA++GAA+ LGG +R
Sbjct: 425 WTYGLAVSAGVFIPSLLIGAAWGRLCGILLASITSTGSIWADPGKYALIGAAAQLGGIVR 484
Query: 513 TTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPY 572
T+SL VIM+E T N+ IM+VL+ +K V + F +YD+ +K + +P+L A
Sbjct: 485 MTLSLTVIMVEATGNVTYGLPIMLVLMTAKIVGDYFVEGLYDIHIKLQSVPFLHGEAPGT 544
Query: 573 MRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTA--HNGFPVIDEPPGSEAPI-LFGIIL 629
LT +V++ P+ N IEKV IV L T+ HNGFPV+ + P L G+IL
Sbjct: 545 SHWLTAREVMSSPVTCLNRIEKVGTIVDTLSNTSTNHNGFPVVVQADVLFQPAKLCGLIL 604
Query: 630 RHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDL 689
R L LLK K F+ +A S RK DF Y I+ I ++++E +DL
Sbjct: 605 RSQLIVLLKHKVFV--ELARSRLTQRKLQLKDFRDAYP-RFPPIQSIHVSQDERECMMDL 661
Query: 690 HPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHDFTPEHILG 749
F N +PYTV + SL + LFR +GLRHL+V + + VVG++TR D H LG
Sbjct: 662 TEFMNPTPYTVPQETSLPRVFKLFRALGLRHLVV---VDDENRVVGLVTRKDLARYH-LG 717
Query: 750 MH 751
H
Sbjct: 718 KH 719
>UniRef100_Q5ZL60 Hypothetical protein [Gallus gallus]
Length = 802
Score = 392 bits (1007), Expect = e-107
Identities = 250/711 (35%), Positives = 392/711 (54%), Gaps = 31/711 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGI 103
ESLDY+ EN+ F ++ R +W++C MIG++ G + ++ VENLAG+
Sbjct: 91 ESLDYDNSENQLFLEEERRINHAAFRTVEIKRWVICAMIGILTGLVACFIDIVVENLAGL 150
Query: 104 KF-VTTSNM---MLERRFMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLN 159
K+ V N+ + F+ ++ N S+ + S+I AFI P AAGSGI ++K YLN
Sbjct: 151 KYRVVKDNIDKFTEKGGLSFSLLLWATLNASVVMVGSVIVAFIEPVAAGSGIPQIKCYLN 210
Query: 160 GVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWR 219
GV P + ++TL +K+ G I +V G L +GK GPM+H+GA +AA + QG S ++
Sbjct: 211 GVKIPHVVRLKTLVIKVCGVILSVVGGLAVGKEGPMIHSGAVIAAGISQGRSTSLKRDFK 270
Query: 220 WLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATV 279
+F+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF +
Sbjct: 271 IFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQFLTWRIFFASMIS 330
Query: 280 AIFLRAMIDVCLSDKCGLFGKGGLIMFDAY-SASISYHLVDVPPVFILAVVGGLLGSLFN 338
L +++ V + L GLI F + S + Y + ++P + VVGG+LG+LFN
Sbjct: 331 TFTLNSVLSVYHGNAWDL-SSPGLINFGRFDSEKMGYTIQEIPIFIFMGVVGGILGALFN 389
Query: 339 FMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIG 398
+ N L ++ + ++ A L++ T+ + F + + + R C P G
Sbjct: 390 AL-NYWLTMFRIRYIHRPCLQVIEAMLVAAVTAAVGFVMIYCS--RDCQ-------PIQG 439
Query: 399 RSGIYK-KFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSI 457
S Y + C YN +A+ FNT + ++ NLF + ++ +F ++ FL+
Sbjct: 440 SSVAYPLQLFCADGEYNSMATAFFNTPEKSVVNLFH-DPPGSYNPMTLGMFTLMYFFLAC 498
Query: 458 FSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLS----NGLYAVLGAASLLGGSMRT 513
++ G+ AG+F+P ++ GA++GRL GI + + S G YA++GAA+ LGG +R
Sbjct: 499 WTYGLTVSAGVFIPSLLIGAAWGRLFGISLSYLSKGSIWADPGKYALMGAAAQLGGIVRM 558
Query: 514 TVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYM 573
T+SL VIM+E T N+ IM+VL+ +K V + F +YD+ ++ + +P+L A
Sbjct: 559 TLSLTVIMMEATGNVTYGFPIMLVLMTAKIVGDYFVEGLYDMHIQLQSVPFLHWEAPVTS 618
Query: 574 RQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTA--HNGFPVIDEPPG-SEAPILFGIILR 630
LT +V++ P+ IE+V +V IL T+ HNGFPV++ P ++ L G+ILR
Sbjct: 619 HSLTAREVMSTPVTCLRRIERVGTVVDILSDTSSNHNGFPVVESNPNTTQVAGLRGLILR 678
Query: 631 HHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLH 690
L LLK K F+ AN V R+ DF Y I+ I ++++E +DL
Sbjct: 679 SQLIVLLKHKVFVER--ANLNLVQRRLKLKDFRDAYP-RFPPIQSIHVSQDERECMIDLS 735
Query: 691 PFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHD 741
F N SPYTV SL + LFR +GLRHL+V + + VVG++TR D
Sbjct: 736 EFMNPSPYTVPREASLPRVFKLFRALGLRHLVV---VNNHNEVVGMVTRKD 783
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.328 0.143 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,290,530,305
Number of Sequences: 2790947
Number of extensions: 55419205
Number of successful extensions: 177796
Number of sequences better than 10.0: 627
Number of HSP's better than 10.0 without gapping: 491
Number of HSP's successfully gapped in prelim test: 136
Number of HSP's that attempted gapping in prelim test: 175142
Number of HSP's gapped (non-prelim): 1360
length of query: 776
length of database: 848,049,833
effective HSP length: 135
effective length of query: 641
effective length of database: 471,271,988
effective search space: 302085344308
effective search space used: 302085344308
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC144657.4


