

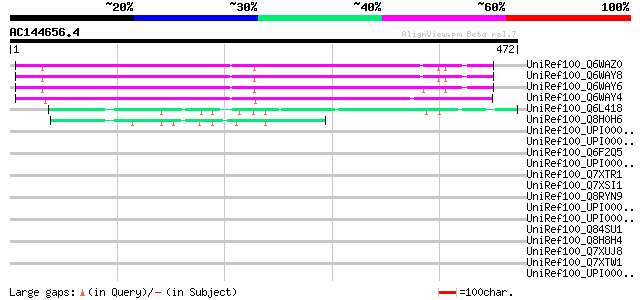
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.4 + phase: 0
(472 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum] 274 3e-72
UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum] 273 8e-72
UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum] 271 2e-71
UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum] 260 5e-68
UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demi... 72 4e-11
UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum] 55 3e-06
UniRef100_UPI000046BA58 UPI000046BA58 UniRef100 entry 38 0.55
UniRef100_UPI0000318B8C UPI0000318B8C UniRef100 entry 38 0.55
UniRef100_Q6F2Q5 Hypothetical protein OSJNBa0088M06.4 [Oryza sat... 38 0.72
UniRef100_UPI0000499F35 UPI0000499F35 UniRef100 entry 37 0.94
UniRef100_Q7XTR1 OSJNBb0085C12.10 protein [Oryza sativa] 37 0.94
UniRef100_Q7XSI1 OSJNBa0073L13.6 protein [Oryza sativa] 37 0.94
UniRef100_Q8RYN9 Putative mutator-like transposase [Oryza sativa] 37 0.94
UniRef100_UPI0000437335 UPI0000437335 UniRef100 entry 37 1.2
UniRef100_UPI000043732F UPI000043732F UniRef100 entry 37 1.2
UniRef100_Q84SU1 Putative transposase [Oryza sativa] 37 1.2
UniRef100_Q8H8H4 Putative mutator-like transposase [Oryza sativa] 37 1.2
UniRef100_Q7XUJ8 OSJNBa0067K08.4 protein [Oryza sativa] 37 1.2
UniRef100_Q7XTW1 OSJNBa0010D21.4 protein [Oryza sativa] 37 1.2
UniRef100_UPI0000437336 UPI0000437336 UniRef100 entry 37 1.6
>UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum]
Length = 562
Score = 274 bits (701), Expect = 3e-72
Identities = 180/478 (37%), Positives = 255/478 (52%), Gaps = 40/478 (8%)
Query: 6 RRTHRYEFKKPDLESLRNLGRMVK--NPEHFRNRYGYLLNILRTNVDEGLINTLVQFYDP 63
R+T Y F + L SL L ++ N + F ++YG +L +L+ VD + TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 64 LYHCFTFSDYQLVPTLEEYSHWVGLPVLDQVPFHGLEPGPKIPAIANALHLEIADIKNKF 123
HCFTF DYQL PTLEEYS + +PV QVPF + +A ALHL I ++ + +
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKEVSDNW 144
Query: 124 ITRAGLQCLPYNFLYQKATICFERSETDAFEAILALLIYGIVLFPNVDKFVDMNAIQIFL 183
+ + LP FL + A E+ +AF A LA++IYGIVLFP++ FVD A+ IF+
Sbjct: 145 KSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 184 TQNPVPTLLADTYVSIHERTDKERGTIVCCAPLLHIWITSHLP--RPKVKPEYL-PWSQK 240
NPVPTLLADTY +IH R K G I CC PLL W S LP P V + W+Q+
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGK-GGAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 241 LMTLTPNDIVWFNPTCDPELIIDSCGDFNNVPLLGTRGGISYSPVLARRQFGFYMEMKPV 300
+M+LT DI W + D +I SCG F NVPL+GTRG I+Y+PVL+ RQ GF M+ +P+
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 301 YLILDRDFFLYKKDDANQRAQFKKAWYSIIRKDRNQLGNRSVIAHEAYVKWVIDRANKWK 360
+ + K+ D + Q +AW SI KD + LG + V+A Y WV +R
Sbjct: 324 EAEIAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETLL 383
Query: 361 MPYPRQRLVTSTVSAIPLPLPPESLEGYQKQLDIERR-----ENSMWEV----------- 404
+PY R + I P E Y++ L RR +++ E+
Sbjct: 384 LPYDRMEPLQEQPPLILAESVP--AEHYKQALMENRRLREKEQDTQMELYKAKAGRLNLA 441
Query: 405 ------------KYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKASIQRKEDFLDK 450
+ R KK+ Y+ ++++LD A +E R++AS Q+K L+K
Sbjct: 442 HQLRGVREEDASRLRSKKRSYEEMESMLD----AEHRECLRLQRVEASYQKKIRDLEK 495
>UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum]
Length = 562
Score = 273 bits (698), Expect = 8e-72
Identities = 180/478 (37%), Positives = 253/478 (52%), Gaps = 40/478 (8%)
Query: 6 RRTHRYEFKKPDLESLRNLGRMVK--NPEHFRNRYGYLLNILRTNVDEGLINTLVQFYDP 63
R+T Y F + L SL L ++ N + F ++YG +L +L+ VD + TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 64 LYHCFTFSDYQLVPTLEEYSHWVGLPVLDQVPFHGLEPGPKIPAIANALHLEIADIKNKF 123
HCFTF DYQL PTLEEYS + +PV QVPF + +A ALHL I ++ + +
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKEVSDSW 144
Query: 124 ITRAGLQCLPYNFLYQKATICFERSETDAFEAILALLIYGIVLFPNVDKFVDMNAIQIFL 183
+ + LP FL + A E+ +AF A LA++IYGIVLFP++ FVD A+ IF+
Sbjct: 145 KSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 184 TQNPVPTLLADTYVSIHERTDKERGTIVCCAPLLHIWITSHLP--RPKVKPEYL-PWSQK 240
NPVPTLLADTY +IH R K G I CC PLL W S LP P V + W+Q+
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGK-GGAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 241 LMTLTPNDIVWFNPTCDPELIIDSCGDFNNVPLLGTRGGISYSPVLARRQFGFYMEMKPV 300
+M+LT DI W + D +I SCG F NVPL+GTRG I+Y+PVL+ RQ GF M+ +P+
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLIGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 301 YLILDRDFFLYKKDDANQRAQFKKAWYSIIRKDRNQLGNRSVIAHEAYVKWVIDRANKWK 360
+ K+ D + Q +AW SI KD + LG + +A Y WV +R
Sbjct: 324 EAEVAESVCFEKRSDPARLEQIGRAWKSIGMKDGSVLGKKFAVAMPDYTDWVKERVETLL 383
Query: 361 MPYPRQRLVTSTVSAIPLPLPPESLEGYQKQLDIERR-----ENSMWEV----------- 404
+PY R + I P E Y++ L RR +++ E+
Sbjct: 384 LPYDRMEPLQEQPPLILAESVP--AEHYKQALMENRRLREKEQDTQMELYKAKADSLNLA 441
Query: 405 ------------KYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKASIQRKEDFLDK 450
+ R KK+ Y+ V+++LD A +E R++AS Q+K L+K
Sbjct: 442 HQLRGVREEDASRLRSKKRSYEEVESMLD----AEHRECLRLQRVEASYQKKIRDLEK 495
>UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum]
Length = 562
Score = 271 bits (694), Expect = 2e-71
Identities = 180/476 (37%), Positives = 254/476 (52%), Gaps = 36/476 (7%)
Query: 6 RRTHRYEFKKPDLESLRNLGRMVK--NPEHFRNRYGYLLNILRTNVDEGLINTLVQFYDP 63
R+T Y F + L SL L ++ N + F +++G +L +L+ VD + TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQHGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 64 LYHCFTFSDYQLVPTLEEYSHWVGLPVLDQVPFHGLEPGPKIPAIANALHLEIADIKNKF 123
HCFTF DYQL PTLEEYS + +PV QVPF + +A ALHL I ++ + +
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFGVVARALHLGIKEVSDSW 144
Query: 124 ITRAGLQCLPYNFLYQKATICFERSETDAFEAILALLIYGIVLFPNVDKFVDMNAIQIFL 183
+ LP FL + A E+ +AF A LA++IYGIVLFP++ FVD A+ IF+
Sbjct: 145 KYSGDVVGLPLKFLLRVAREEAEKGSWEAFRAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 184 TQNPVPTLLADTYVSIHERTDKERGTIVCCAPLLHIWITSHLP--RPKVKPEYL-PWSQK 240
NPVPTLLADTY +IH R K G I CC PLL W S LP P V + W+Q+
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGK-GGAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 241 LMTLTPNDIVWFNPTCDPELIIDSCGDFNNVPLLGTRGGISYSPVLARRQFGFYMEMKPV 300
+M+LT DI W + D +I SCG F NVPL+GTRG I+Y+PVL+ RQ GF M+ +P+
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 301 YLILDRDFFLYKKDDANQRAQFKKAWYSIIRKDRNQLGNRSVIAHEAYVKWVIDRANKWK 360
+ + K+ D + Q +AW SI KD + LG + V+A Y WV +R
Sbjct: 324 EAEVAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETLL 383
Query: 361 MPYPR-QRLVTSTVSAIPLPLPPE-------------------SLEGYQKQLDIERRENS 400
+PY R + L S + +P E LE Y+ + D +
Sbjct: 384 LPYDRMEPLQEQPPSILAESVPAEHYKQALMENRRLRGKEQDTQLELYKAKADKLNLAHQ 443
Query: 401 MWEV------KYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKASIQRKEDFLDK 450
+ V + R KK+ Y+ ++++LD A +E R +AS Q+K L+K
Sbjct: 444 LRGVREEDASRLRSKKRSYEEMESMLD----AEHRECLRLQRAEASYQKKIRDLEK 495
>UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum]
Length = 546
Score = 260 bits (665), Expect = 5e-68
Identities = 169/450 (37%), Positives = 237/450 (52%), Gaps = 10/450 (2%)
Query: 6 RRTHRYEFKKPDLESLRNLGRMVKNPE--HFRNRYGYLLNILRTNVDEGLINTLVQFYDP 63
R+T Y F + L SL LG ++ + + F ++G +L +L+T VD + TL+QFYDP
Sbjct: 9 RKTCSYSFYREPLTSLIELGSLMPSDQLKGFVGQFGDILTLLKTVVDPVPLQTLLQFYDP 68
Query: 64 LYHCFTFSDYQLVPTLEEYSHWVGLPVLDQVPFHGLEPGPKIPAIANALHLEIADIKNKF 123
CFTF DYQL PTLEEYS + +P+ QVPF + IA AL + + ++ + +
Sbjct: 69 ELRCFTFQDYQLAPTLEEYSILMSIPIQHQVPFLDVPKEVDFRVIARALRMSVKEVCDNW 128
Query: 124 ITRAGLQCLPYNFLYQKATICFERSETDAFEAILALLIYGIVLFPNVDKFVDMNAIQIFL 183
+ +P FL + A E+ + F A LA +IYGIVLFP++ F+D AI IF+
Sbjct: 129 KPSGEVVGMPLKFLLRVAREEAEKGNWEVFHAQLAAMIYGIVLFPSMPNFIDHAAISIFI 188
Query: 184 TQNPVPTLLADTYVSIHERTDKERGTIVCCAPLLHIWITSHLPR--PKVKPE-YLPWSQK 240
NPVPTLLADTY +IH R K G I CC PLL W S LP P + + L W+Q+
Sbjct: 189 GGNPVPTLLADTYYAIHSRHGK-GGAIRCCLPLLLRWFMSLLPASGPFMDTQSTLKWTQR 247
Query: 241 LMTLTPNDIVWFNPTCDPELIIDSCGDFNNVPLLGTRGGISYSPVLARRQFGFYMEMKPV 300
+M+LT DI W + D + ++ SCG+F NVPL+GT+G I+Y+PVL+ RQ GF M +P+
Sbjct: 248 VMSLTSYDIRWQSYRMDVKDVVMSCGEFRNVPLVGTKGCINYNPVLSLRQLGFIMSRRPL 307
Query: 301 YLILDRDFFLYKKDDANQRAQFKKAWYSIIRKDRNQLGNRSVIAHEAYVKWVIDRANKWK 360
+ + K+ D + Q +AW SI KD + LG + IA Y WV R
Sbjct: 308 EAEIVESVYFEKRTDPVRLEQIGRAWKSIGVKDGSVLGKKFAIAMPDYTDWVKKRVETLL 367
Query: 361 MPYPRQRLVTSTVSAIPLPLPPESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLL 420
+PY R + PL L + KQ +E R E R + + K L
Sbjct: 368 LPYDRMEPLQEQP---PLILADSVPAEHYKQALMENRRLREKEQDTRMELYKAKADKLNL 424
Query: 421 DQQIQANCKEKNENAR-LKASIQRKEDFLD 449
Q++ E AR K S + E LD
Sbjct: 425 AHQLREMQGEDASRARSKKRSYKEMESMLD 454
>UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demissum]
Length = 607
Score = 71.6 bits (174), Expect = 4e-11
Identities = 117/511 (22%), Positives = 202/511 (38%), Gaps = 105/511 (20%)
Query: 37 RYGYLLNILRTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPVLDQVPF 96
R G+L ++L LI LV F+DP+ + F FSD++L PTLEE ++G
Sbjct: 47 RLGFLRSLLYVEPRRDLIEALVHFWDPIKNVFHFSDFELTPTLEEIGAFIGRG------- 99
Query: 97 HGLEPG-PKIPAIANALH-LEIADI-KNKFITRAGLQCLPYNFLYQK------------- 140
L G P IP N LE+ I +N+ +P FLY++
Sbjct: 100 KNLHEGEPMIPKHINGRRFLELLHINENEIGGCLDNGWVPLEFLYKRYGRKDGFELYGKK 159
Query: 141 --ATICFERSETDAFEAILALLIYGIVLFPNVDKFVDMN--AIQIFLTQNP----VPTLL 192
C ET +++AI + GI++F +++N A+ + P VP +L
Sbjct: 160 LHNNGCRLTWETHSYDAITVAFL-GIMVFLKRGGKININLAAVITAFEKKPNITLVPMIL 218
Query: 193 ADTYVSIHERTDKERGTIVC---------CAPLLHIWITSHL------------------ 225
AD Y R +C C LL +W+ H+
Sbjct: 219 ADIY----------RALTICKNGGDYFEGCNMLLQLWMIEHIRHHPYVVDFKVECNDYIG 268
Query: 226 -PRPKVKPEYLP-----WSQKLMTLTPNDIVWFNPTCDPELIIDSCGDFNN-VPLLGTRG 278
++K P W + L LT + IVW N P + F + + L+G RG
Sbjct: 269 GHEERIKDHSFPKGIEAWKKYLNNLTADKIVW-NYHWFPSAEVIYMSTFRSFIVLMGLRG 327
Query: 279 GISYSPVLARRQFGFYMEMKPVYLILDRDF-FLYKKDDANQRAQFKKAWYS-IIRKDRNQ 336
Y P+ RQ G + P I R+F + + + ++++ K W ++ ++
Sbjct: 328 VQPYMPLRVMRQLGRRQFLPPNEDI--REFMYEFHPEIPLRKSEIFKIWGGCMLSSPHDR 385
Query: 337 LGNRSV-IAHEAYVKWVIDRANKWKMPYPRQRLVTSTVSAIPLPLPPESLE---GYQKQL 392
+ +R+ +AY++WV D+ + +P + + I + + LE Y+ L
Sbjct: 386 VEDRTKGEVDQAYLEWVHDQPSPKVLPEGSVKGPVDREAEIEVRIKQARLEVERSYRSTL 445
Query: 393 DIERRE-----------NSMWEVKYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKASI 441
D + ++++EV+ RK + T++ D I + E+ E K +
Sbjct: 446 DCLSNDLKNAKEELAQRDAIFEVRVRKHRSTILTLQE--DLGIVISAMEQQEEEYTKEKV 503
Query: 442 QRKEDFLDKICPGRKKRRMDLFNGPHLDSEE 472
Q + C + +R+ + HL EE
Sbjct: 504 QAE-------CEVERGQRIAERDALHLQIEE 527
>UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum]
Length = 464
Score = 55.5 bits (132), Expect = 3e-06
Identities = 76/307 (24%), Positives = 116/307 (37%), Gaps = 63/307 (20%)
Query: 39 GYLLNILRTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPVLDQVPFHG 98
G L +I++ + LI LV F+DP+++ F FSD++L PTLEE + G
Sbjct: 39 GALTDIMKIKPRDDLIEALVTFWDPVHNVFCFSDFELTPTLEEIDGYSGFG-------RD 91
Query: 99 LEPGPKIPAIANALH--LEIADIKNKF-ITRAGLQCLPYNFLYQK--ATICFERSETD-- 151
L I A ++H ++ +I + T C + FLY + FE E
Sbjct: 92 LRNQELIFPRALSVHRFFDLLNISKQIRKTNVVKGCCSFYFLYSRFGQPNGFEMHEKGLN 151
Query: 152 ------------AFEAILALLIYGIVLFPNVDKFVD---MNAIQIFLTQNP---VPTLLA 193
F I+A L GI++FPN ++ +D +Q+ T+ P +L+
Sbjct: 152 NKQNKDTWHIHRCFAFIMAFL--GIMVFPNRERTIDTRIARVVQVLTTKEHHTLAPIILS 209
Query: 194 DTYVSIHERTDKERGT--IVCCAPLLHIWITSHLPRPKVKPEYLP--------------- 236
D Y ++ T + G C LL +W+ HL Y P
Sbjct: 210 DIYRAL---TLCKSGAKFFEGCNILLQMWLIEHLRHHPKFMSYGPSKDNFIDSYEERVKE 266
Query: 237 ---------WSQKLMTLTPNDIVWFNPTCDPELIIDSCGDFNNVPLLGTRGGISYSPVLA 287
W L +L I W +I +++ LLG R Y P
Sbjct: 267 YNSPEGVETWISHLRSLNACQIEWTLGWLPLREVIHMSALKSHLLLLGLRSVQPYMPHRV 326
Query: 288 RRQFGFY 294
RQ G Y
Sbjct: 327 LRQLGRY 333
>UniRef100_UPI000046BA58 UPI000046BA58 UniRef100 entry
Length = 211
Score = 38.1 bits (87), Expect = 0.55
Identities = 18/58 (31%), Positives = 33/58 (56%)
Query: 383 ESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKAS 440
+SL Y+ + +R++ ++ K +KKK E D L++ + +N KE N+N +L S
Sbjct: 129 KSLNDYKSCFEKQRKKRKWFQFKKKKKKYELDEGVKYLNESVCSNYKEANKNVKLNNS 186
>UniRef100_UPI0000318B8C UPI0000318B8C UniRef100 entry
Length = 964
Score = 38.1 bits (87), Expect = 0.55
Identities = 28/86 (32%), Positives = 42/86 (48%), Gaps = 11/86 (12%)
Query: 360 KMPYPRQRLVTSTVSAIPLPLPPESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNL 419
K+ P+ R T T S +PL PP+ L +++R S + KKK + K
Sbjct: 81 KLYKPKSRFETQTKSFLPLETPPQFLA-------LDKRNTSKKTTEKEKKKSNLELFKEE 133
Query: 420 LDQQIQANCKEKNENARLKASIQRKE 445
L +QIQ +E++E RLK + R E
Sbjct: 134 L-KQIQ---EERDERHRLKGRVSRFE 155
>UniRef100_Q6F2Q5 Hypothetical protein OSJNBa0088M06.4 [Oryza sativa]
Length = 1635
Score = 37.7 bits (86), Expect = 0.72
Identities = 17/49 (34%), Positives = 30/49 (60%)
Query: 46 RTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPVLDQV 94
R ++D L++ LV + P H F + ++VPTL++ S+ +GLP+ V
Sbjct: 922 RIHIDAALLSALVDRWRPETHTFHLTVGEMVPTLQDVSYLLGLPIAGPV 970
>UniRef100_UPI0000499F35 UPI0000499F35 UniRef100 entry
Length = 186
Score = 37.4 bits (85), Expect = 0.94
Identities = 24/122 (19%), Positives = 60/122 (48%), Gaps = 13/122 (10%)
Query: 330 IRKDRNQLGNRSVIAHEAYVKWVIDRANKWKMPYPRQRLVTSTVSAIPLPLPPESLEGYQ 389
++K + ++ R + A + Y WVI++ K M Y + + + + E +
Sbjct: 69 LKKRKKEIRRRILQARKNYNSWVIEQQQKMLMAYKQGK----------EKIYKQDEEIRK 118
Query: 390 KQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQQIQANCKEKNENARLKASIQRKEDFLD 449
+L + +W+ ++ +KK E++ ++++++ KEKNE A++ + I + + L+
Sbjct: 119 SKLKEMNNQFKLWKKQFEEKKIEFE---KRFEREVESIKKEKNERAKIISQIMVEREQLN 175
Query: 450 KI 451
+
Sbjct: 176 DL 177
>UniRef100_Q7XTR1 OSJNBb0085C12.10 protein [Oryza sativa]
Length = 960
Score = 37.4 bits (85), Expect = 0.94
Identities = 16/45 (35%), Positives = 29/45 (63%)
Query: 46 RTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPV 90
R ++D L++ LV + P H F + ++VPTL++ S+ +GLP+
Sbjct: 606 RIHIDAALLSALVDRWRPETHTFHLTVGEMVPTLQDVSYLLGLPI 650
>UniRef100_Q7XSI1 OSJNBa0073L13.6 protein [Oryza sativa]
Length = 552
Score = 37.4 bits (85), Expect = 0.94
Identities = 16/45 (35%), Positives = 29/45 (63%)
Query: 46 RTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPV 90
R ++D L++ LV + P H F + ++VPTL++ S+ +GLP+
Sbjct: 72 RIHIDAALLSALVDRWRPETHTFHLTVGEMVPTLQDVSYLLGLPI 116
>UniRef100_Q8RYN9 Putative mutator-like transposase [Oryza sativa]
Length = 1701
Score = 37.4 bits (85), Expect = 0.94
Identities = 16/45 (35%), Positives = 29/45 (63%)
Query: 46 RTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPV 90
R ++D L++ LV + P H F + ++VPTL++ S+ +GLP+
Sbjct: 1010 RIHIDAALLSALVDRWRPETHTFHLTVGEMVPTLQDVSYLLGLPI 1054
>UniRef100_UPI0000437335 UPI0000437335 UniRef100 entry
Length = 1880
Score = 37.0 bits (84), Expect = 1.2
Identities = 23/87 (26%), Positives = 45/87 (51%), Gaps = 7/87 (8%)
Query: 383 ESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQ-------QIQANCKEKNENA 435
E+++ +KQLD ++ E + + K+K ++D + LD+ Q Q +EKN+
Sbjct: 844 ETMKNERKQLDKDKEEMEEQKQEMEKEKHDFDQSRKSLDKDLKMMKLQKQVFEEEKNKLE 903
Query: 436 RLKASIQRKEDFLDKICPGRKKRRMDL 462
++K ++R+ D + KI + R L
Sbjct: 904 QMKIELEREADEIRKIKEETQNERQSL 930
>UniRef100_UPI000043732F UPI000043732F UniRef100 entry
Length = 1970
Score = 37.0 bits (84), Expect = 1.2
Identities = 23/87 (26%), Positives = 45/87 (51%), Gaps = 7/87 (8%)
Query: 383 ESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQ-------QIQANCKEKNENA 435
E+++ +KQLD ++ E + + K+K ++D + LD+ Q Q +EKN+
Sbjct: 1061 ETMKNERKQLDKDKEEMEEQKQEMEKEKHDFDQSRKSLDKDLKMMKLQKQVFEEEKNKLE 1120
Query: 436 RLKASIQRKEDFLDKICPGRKKRRMDL 462
++K ++R+ D + KI + R L
Sbjct: 1121 QMKIELEREADEIRKIKEETQNERQSL 1147
Score = 35.4 bits (80), Expect = 3.6
Identities = 23/87 (26%), Positives = 45/87 (51%), Gaps = 7/87 (8%)
Query: 383 ESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQ-------QIQANCKEKNENA 435
E+++ +KQLD ++ E + + K+K ++D +N LD+ Q Q +EKN+
Sbjct: 148 ETMKNERKQLDKDKEEMEEQKREIEKEKHDFDQGRNSLDEDLKMMKLQKQVFEEEKNKLE 207
Query: 436 RLKASIQRKEDFLDKICPGRKKRRMDL 462
++K ++R+ + KI + R L
Sbjct: 208 QMKIELEREAGEIRKIKEEIQNERQSL 234
>UniRef100_Q84SU1 Putative transposase [Oryza sativa]
Length = 1511
Score = 37.0 bits (84), Expect = 1.2
Identities = 16/45 (35%), Positives = 29/45 (63%)
Query: 46 RTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPV 90
R ++D L++ LV + P H F + ++VPTL++ S+ +GLP+
Sbjct: 890 RIHIDAALMSALVDRWRPETHTFHLTVGEMVPTLQDVSYLLGLPI 934
>UniRef100_Q8H8H4 Putative mutator-like transposase [Oryza sativa]
Length = 1436
Score = 37.0 bits (84), Expect = 1.2
Identities = 16/45 (35%), Positives = 29/45 (63%)
Query: 46 RTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPV 90
R ++D L++ LV + P H F + ++VPTL++ S+ +GLP+
Sbjct: 944 RIHIDAALMSALVDRWRPETHTFHLTVGEMVPTLQDVSYLLGLPI 988
>UniRef100_Q7XUJ8 OSJNBa0067K08.4 protein [Oryza sativa]
Length = 1555
Score = 37.0 bits (84), Expect = 1.2
Identities = 16/45 (35%), Positives = 29/45 (63%)
Query: 46 RTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPV 90
R ++D L++ LV + P H F + ++VPTL++ S+ +GLP+
Sbjct: 901 RIHIDAALMSALVDRWRPETHTFHLTVGEMVPTLQDVSYLLGLPI 945
>UniRef100_Q7XTW1 OSJNBa0010D21.4 protein [Oryza sativa]
Length = 1609
Score = 37.0 bits (84), Expect = 1.2
Identities = 16/45 (35%), Positives = 29/45 (63%)
Query: 46 RTNVDEGLINTLVQFYDPLYHCFTFSDYQLVPTLEEYSHWVGLPV 90
R ++D L++ LV + P H F + ++VPTL++ S+ +GLP+
Sbjct: 935 RIHIDAALMSALVDRWRPETHTFHLTVGEMVPTLQDVSYLLGLPI 979
>UniRef100_UPI0000437336 UPI0000437336 UniRef100 entry
Length = 1949
Score = 36.6 bits (83), Expect = 1.6
Identities = 26/89 (29%), Positives = 47/89 (52%), Gaps = 8/89 (8%)
Query: 383 ESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQ-------QIQANCKEKNENA 435
E+++ +KQLD ++ E + + K+K ++D + LD+ Q Q +EKN++
Sbjct: 488 ETMKNERKQLDKDKEEMEEQKQEMEKEKHDFDQSRKSLDKDLKMMKLQKQVFEEEKNKDI 547
Query: 436 RLK-ASIQRKEDFLDKICPGRKKRRMDLF 463
K SIQ +D L+K +K R +L+
Sbjct: 548 EKKIKSIQSDKDMLEKEKHDLEKTRSELY 576
Score = 34.3 bits (77), Expect = 7.9
Identities = 22/87 (25%), Positives = 44/87 (50%), Gaps = 7/87 (8%)
Query: 383 ESLEGYQKQLDIERRENSMWEVKYRKKKQEYDTVKNLLDQ-------QIQANCKEKNENA 435
E+++ +KQLD ++ E + + K++ D + LD+ Q Q +EKN+
Sbjct: 134 ETMKNERKQLDKDKEEMEEQKQEMEKERDNMDQSRKSLDEDQKKMKLQKQMFEEEKNKLE 193
Query: 436 RLKASIQRKEDFLDKICPGRKKRRMDL 462
++K ++R+ D + KI + +R L
Sbjct: 194 QMKIELEREADEISKIKEETQNKRQRL 220
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.140 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 844,020,401
Number of Sequences: 2790947
Number of extensions: 37865409
Number of successful extensions: 97299
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 97082
Number of HSP's gapped (non-prelim): 211
length of query: 472
length of database: 848,049,833
effective HSP length: 131
effective length of query: 341
effective length of database: 482,435,776
effective search space: 164510599616
effective search space used: 164510599616
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC144656.4


