

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.13 + phase: 0 /pseudo/partial
(1235 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
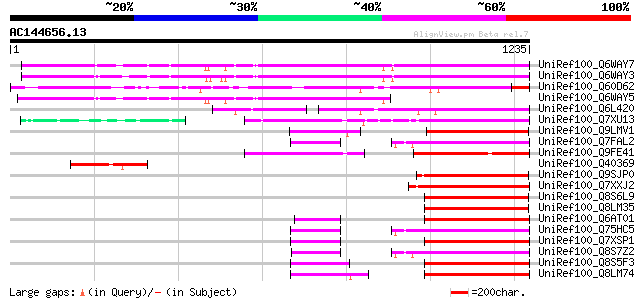
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 947 0.0
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 945 0.0
UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solan... 546 e-153
UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum] 523 e-146
UniRef100_Q6L420 Putative polyprotein [Solanum demissum] 345 5e-93
UniRef100_Q7XU13 OSJNBa0091D06.8 protein [Oryza sativa] 344 1e-92
UniRef100_Q9LMV1 F5M15.26 [Arabidopsis thaliana] 246 3e-63
UniRef100_Q7FAL2 OSJNBa0042N22.17 protein [Oryza sativa] 244 1e-62
UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic ... 239 3e-61
UniRef100_Q40369 RPE15 protein [Medicago sativa] 239 3e-61
UniRef100_Q9SJP0 Putative retroelement pol polyprotein [Arabidop... 237 2e-60
UniRef100_Q7XXJ2 OSJNBa0094O15.10 protein [Oryza sativa] 237 2e-60
UniRef100_Q8S6L9 Putative retroelement [Oryza sativa] 236 4e-60
UniRef100_Q8LM35 Putative retroelement [Oryza sativa] 236 4e-60
UniRef100_Q6AT01 Putative polyprotein [Oryza sativa] 235 7e-60
UniRef100_Q75HC5 Putative reverse transcriptase [Oryza sativa] 234 9e-60
UniRef100_Q7XSP1 OSJNBa0070M12.15 protein [Oryza sativa] 234 1e-59
UniRef100_Q8S7Z2 Putative retroelement [Oryza sativa] 233 4e-59
UniRef100_Q8S5F3 Putative retroelement [Oryza sativa] 233 4e-59
UniRef100_Q8LM74 Putative retroelement [Oryza sativa] 233 4e-59
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 947 bits (2448), Expect = 0.0
Identities = 559/1279 (43%), Positives = 762/1279 (58%), Gaps = 101/1279 (7%)
Query: 29 LVPKVDVPKKFKVPEFDRYNGLTCPHNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAE 88
LV + +P KFK P FD+YNG +CP H+ Y RK+ Y+D++ + ++ FQDSL + +
Sbjct: 159 LVEGLRIPYKFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLD 218
Query: 89 WYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKDESFREYAQRWRGAAA 148
WY L +D + + +L AF Y N + P+R L+SL QK ESF+EYAQRWR AA
Sbjct: 219 WYMELKRDSIRCWKDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWRELAA 278
Query: 149 RITPALDEEEMTQRFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVREGIIVFEKVE 208
R+ P + E E+T F+ TL+ +++RM +FS++V G R E ++ G I +
Sbjct: 279 RVQPPMLERELTDMFIGTLQGVFMDRMGSCPFVSFSDVVICGERTESLIKTGKIQ----D 334
Query: 209 SSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAA*LPPSYPYAPYSQHPFFPY 268
+ ++SK+ G +++E E V Q+ + + A +P
Sbjct: 335 AGSSSSKKPFAGAPRRREGETNAVQYRRDQNRSQRCQVAAVTIPA--------------- 379
Query: 269 PLPSGQPQVPVNAVVQQMQQQPPVQQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRQ 328
P P Q Q V QQ QQQ P Q +Q R F +PM YAELLP LL G R
Sbjct: 380 PQPRQQQQQRVQQPQQQQQQQRPYQPRQRMPDR-RFDSLPMSYAELLPELLRLGMVELRT 438
Query: 329 DKPPPDPLPPRFRSDLKCDFHQGALGHDVKGCYALKYIVKKLIDQGKLTFENNVPHVLDN 388
PP LPP + ++++CDFH GA GH + C AL++ V+ LID + F VP+V++N
Sbjct: 439 -MAPPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFAP-VPNVVNN 496
Query: 389 PLPNHAA--VNMIEVYE-EAPRLDVRNIATPLVPLHIKLCQASLFNHDHTNCPECFRNPL 445
P+P H VN IE E E ++V ++ T L+ + +L +++ +C C +
Sbjct: 497 PMPQHGGHRVNNIEGKEAEDLVVNVDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESEN 556
Query: 446 GCYVVQNDIQSLMNGNYL----------TVSDVCVI-------------VPVFHDPPVK- 481
GC ++ IQ +M+ L TVS + + P + PV
Sbjct: 557 GCDQLRTGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPSEGRGQRVVSAPATNGTPVTI 616
Query: 482 SIPSKENVEPLVIRLPGPVSYTS*KAIPYKY-----------------NATMIENGV--E 522
S+P + P I G + + +A+P+KY N + G+
Sbjct: 617 SVPVTISA-PTTIAASGRRAVENSRAVPWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNR 675
Query: 523 VPLASLAMVSNIAEGTTAALRSGTV-RPPLFQKKAATPTTPPIDKATLTDVSHVTRDASR 581
VP V N+ G RSG + P + A K + + V ++A
Sbjct: 676 VPATVGPAVDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEA-- 732
Query: 582 PSQSIEDSNLDEILRLIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQAYV 641
P S E +++E +++IK+SDYKIVDQL QTPSKIS+LSLLL SEAHRN LLK+L AYV
Sbjct: 733 PEGSFE-KDVEEFMKIIKKSDYKIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNLAYV 791
Query: 642 DHEVTMDRFGSIVGNITACNNLWFSEDELPEAGKYHNLALHISVNCKSDMLSNVLVDTGS 701
E+++++ ++ N++ + + F+ +L G+ HN ALHI++ CK +LS+VLVDTGS
Sbjct: 792 PQEISVNQLEGVMANVSTRHGVGFTNLDLTPEGRNHNKALHITMECKGAVLSHVLVDTGS 851
Query: 702 SLNVMPKSTLDQLSYRETSLRISTFLVKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMD 761
SLNV+PK L ++ L S +V+AFDGS+++V GE+ LP+ IGPE F I F VMD
Sbjct: 852 SLNVLPKQILKKIDVEGFVLTPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMD 911
Query: 762 INASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNGKLVTVHGEEAYLVSQLSSFSCIEA-G 820
I +YSCLLGRPWIH AGAV+STLHQ K+V NG++VTV GEE LVS LSSF +E G
Sbjct: 912 IQPAYSCLLGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDG 971
Query: 821 SAEGT---AFQGLTIEGM---ELKKTGTAMASLKDAQKAVQEGQAAGRGKLIQLRENKHK 874
T AF+ + +E + E +K G ++ S K A++ V G+A G GK++ L + K
Sbjct: 972 EIHETLCQAFETVALEKVAYAEQRKPGASITSYKQAKEVVDSGKAEGWGKMVYLPVKEDK 1031
Query: 875 EGLGFSPTSGVSTG-----TFHSAGFVNAITEEATGFGP-----------RPVFVIPGGI 918
G+G+ P G TF SAG +N ATG RP PGG
Sbjct: 1032 FGVGYEPLQAEQNGQAGPSTFTSAGLMNHGDVSATGSEDCDSDCDLDNWVRPC--APGGS 1089
Query: 919 ARDWDAIDIPSIMHVSELNHNKPVGHSNPTFPPNFEFPMYEAEAEEGDD--IPYEITRLL 976
+W A ++ + ++E + +++ +F P+Y+AE E +D +P E+ RLL
Sbjct: 1090 INNWTAEEVVQVTLLTESDLMAFTMNNSVMAQYDFGNPIYQAEEESEEDCELPAELVRLL 1149
Query: 977 EQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDM 1036
QE++ IQPHQEE+E +N+GTE+ KREIK+GAALE+ VK+++I++LREY +IFAWSY+DM
Sbjct: 1150 RQEERVIQPHQEELEVVNLGTEDAKREIKIGAALEDSVKRRLIEMLREYVEIFAWSYQDM 1209
Query: 1037 PGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVAN 1096
PGLD IV HR+P + CP V+QKLRRT PDMA KIK EVQKQ DAGFL YP WVAN
Sbjct: 1210 PGLDTDIVVHRLPLREGCPSVKQKLRRTSPDMATKIKEEVQKQWDAGFLAVTSYPPWVAN 1269
Query: 1097 IVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIK 1156
IVPVPKKDGKVRMCVD+RDLN+ASPKD+FPL HIDVLVDN AQS VF FMDGFSGYNQIK
Sbjct: 1270 IVPVPKKDGKVRMCVDYRDLNRASPKDDFPLPHIDVLVDNTAQSSVFSFMDGFSGYNQIK 1329
Query: 1157 MSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVK 1216
M+PED EKT+FITPWGTF YKVMP GL NAGATYQR MTTLFHDM+HKE+EVYVDDMI K
Sbjct: 1330 MAPEDMEKTTFITPWGTFYYKVMPFGLKNAGATYQRAMTTLFHDMMHKEIEVYVDDMIAK 1389
Query: 1217 SKDEEQHVEYLTKMFERLR 1235
S+ EE+H+ L K+F+RLR
Sbjct: 1390 SQTEEEHLVNLQKLFDRLR 1408
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 945 bits (2443), Expect = 0.0
Identities = 562/1282 (43%), Positives = 760/1282 (58%), Gaps = 108/1282 (8%)
Query: 29 LVPKVDVPKKFKVPEFDRYNGLTCPHNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAE 88
LV + +P KFK P FD+YNG +CP H+ Y RK+ Y+D++ + ++ FQDSL + +
Sbjct: 158 LVEGLRIPYKFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLD 217
Query: 89 WYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKDESFREYAQRWRGAAA 148
WY L D + + +L AF Y N + P+R L+SL QK +ESF+EYAQRWR AA
Sbjct: 218 WYMELKSDSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSNESFKEYAQRWRELAA 277
Query: 149 RITPALDEEEMTQRFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVREGIIVFEKVE 208
R+ P + E E+T F+ TL+ +++RM +FS++V G R E ++ G I + V
Sbjct: 278 RVQPPMLERELTDMFIGTLQGVFMDRMGSCPFGSFSDVVICGERTESLIKTGKI--QDVG 335
Query: 209 SSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAA*LPPSYPYAPYSQHPFFPY 268
SS +SK+ G +++E E V Q+ A +P P Q P
Sbjct: 336 SS--SSKKPFAGAPRRREGETNAVQHRRDQNRIEYRQAAAVTIPAPQPRQQQQQRVQQP- 392
Query: 269 PLPSGQPQVPVNAVVQQMQQQPPVQQQQHQQARPTFPPIPMLYAELLPTLLLRGH---CT 325
QQ QQQ P Q +Q R F +PM YAELLP LL G CT
Sbjct: 393 ---------------QQQQQQRPYQPRQRMPDR-RFDSLPMSYAELLPELLRLGLVELCT 436
Query: 326 TRQDKPPPDPLPPRFRSDLKCDFHQGALGHDVKGCYALKYIVKKLIDQGKLTFENNVPHV 385
PP LPP + ++++CDFH GA GH + C AL++ V+ LID + F VP+V
Sbjct: 437 MA----PPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDANAINFAP-VPNV 491
Query: 386 LDNPLPNHAA--VNMIEVYEEAPRLD-VRNIATPLVPLHIKLCQASLFNHDHTNCPECFR 442
++NP+P H VN IE E +D V ++ T L+ + +L +++ +C C
Sbjct: 492 VNNPMPQHGGHRVNNIEGKEAEDLVDNVDDVQTSLLVVKSRLLNEGVYSGCDEDCLGCAE 551
Query: 443 NPLGCYVVQNDIQSLMNGNYLTVS-------DVCVIVPVF-----HDPPVKSIPSKENVE 490
+ GC ++ IQ +M+ L S V I F H V S P+ N
Sbjct: 552 SENGCDQLRAGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPSEGHGQRVVSAPAT-NGT 610
Query: 491 PLVIRLPGPVS------------YTS*KAIPYKY-----------------NATMIENGV 521
P+ I +P +S + +A+P+KY N T + G+
Sbjct: 611 PVTIPVPVTISAPTTIVASGRRAVENSRAVPWKYDNAYRSNRRVESQTKPVNQTPVTIGL 670
Query: 522 --EVPLASLAMVSNIAEGTTAALRSGTV-RPPLFQKKAATPTTPPIDKATLTDVSHVTRD 578
VP V N+ G RSG + P + A K + + V ++
Sbjct: 671 ANRVPATVGPAVDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKE 729
Query: 579 ASRPSQSIEDSNLDEILRLIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQ 638
A P S E +++E +++IK+SDYKIVDQL QTPSKIS+LSLLL SEAHRN LLK+L
Sbjct: 730 A--PEGSFE-KDVEEFMKMIKKSDYKIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNL 786
Query: 639 AYVDHEVTMDRFGSIVGNITACNNLWFSEDELPEAGKYHNLALHISVNCKSDMLSNVLVD 698
AYV E+++++ ++ N++ + + F+ +LP G+ HN ALHI++ CK +LS+VLVD
Sbjct: 787 AYVPQEISVNQLEGVMANVSTRHGVGFTNLDLPPEGRNHNKALHITMECKGAVLSHVLVD 846
Query: 699 TGSSLNVMPKSTLDQLSYRETSLRISTFLVKAFDGSRKNVLGEIDLPMTIGPETFLITFQ 758
TGSSLNV+PK L ++ L S +V+AFD S+++V GE+ LP+ IGPE F I F
Sbjct: 847 TGSSLNVLPKQILKKIDVEGFVLTPSDLIVRAFDRSKRSVCGEVTLPVKIGPEVFDIIFY 906
Query: 759 VMDINASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNGKLVTVHGEEAYLVSQLSSFSCIE 818
VMDI +YSCLLGRPWIH AGAV+STLHQ K+V NG++VTV GEE LVS LSSF +E
Sbjct: 907 VMDIQPAYSCLLGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVE 966
Query: 819 A-GSAEGT---AFQGLTIEGM---ELKKTGTAMASLKDAQKAVQEGQAAGRGKLIQLREN 871
G T F+ + +E + E +K G ++ S K A++ V G+A G GK++ L
Sbjct: 967 VDGEIHETLCQVFETVALEKVAYAEQRKPGVSITSYKQAKEVVDSGKAEGWGKMVDLPVK 1026
Query: 872 KHKEGLGFSPTSGVSTG-----TFHSAGFVNAITEEATGFGP-----------RPVFVIP 915
+ K G+G+ P G TF SAG +N ATG RP P
Sbjct: 1027 EDKFGVGYEPLQAEQNGQAGPSTFTSAGLMNHGDVSATGSEDCDSDCDLDNWVRPC--AP 1084
Query: 916 GGIARDWDAIDIPSIMHVSELNHNKPVGHSNPTFPPNFEFPMYEAEAEEGDD--IPYEIT 973
GG +W A ++ + ++E + + + +F+ P+Y+AE E +D +P E+
Sbjct: 1085 GGSINNWTAEEVVQVTLLTESDLMAFTMNGSVIAQYDFDNPIYQAEEESEEDCELPAELV 1144
Query: 974 RLLEQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSY 1033
RLL+QE++ IQPHQEE+E +N+GTE+ REIK+GAALE+ VK+++I++LREY +IFAWSY
Sbjct: 1145 RLLKQEERVIQPHQEELEVVNLGTEDATREIKIGAALEDSVKRRLIEMLREYVEIFAWSY 1204
Query: 1034 EDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEW 1093
+DMPGLD IV HR+P + CP V+QKLRRT PDMA KIK EVQKQ DAGFL YP W
Sbjct: 1205 QDMPGLDTDIVVHRLPLREGCPSVKQKLRRTSPDMATKIKEEVQKQWDAGFLAVTSYPPW 1264
Query: 1094 VANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYN 1153
+ANIVPVPKKDGKVRMCVD+RDLN+ASPKD+FPL HIDVLVDN AQS VF FMDGFSGYN
Sbjct: 1265 MANIVPVPKKDGKVRMCVDYRDLNRASPKDDFPLPHIDVLVDNTAQSSVFSFMDGFSGYN 1324
Query: 1154 QIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDM 1213
QIKM+PED EKT+FITPWGTFCYKVMP GL NAGATYQR MTTLFHDM+HKE+EVYVDDM
Sbjct: 1325 QIKMAPEDMEKTTFITPWGTFCYKVMPFGLKNAGATYQRAMTTLFHDMMHKEIEVYVDDM 1384
Query: 1214 IVKSKDEEQHVEYLTKMFERLR 1235
I KS+ EE+H+ L K+F+RLR
Sbjct: 1385 IAKSQTEEEHLVNLQKLFDRLR 1406
>UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solanum demissum]
Length = 1784
Score = 546 bits (1407), Expect = e-153
Identities = 394/1284 (30%), Positives = 608/1284 (46%), Gaps = 224/1284 (17%)
Query: 2 EELAKELR---REIKANRGNGD--SVKTHDLCLVPKVDVPKKFKVPEFDRYNGLTCPHNH 56
EE+AK+++ + I+ +G G + DLC+ P V +P
Sbjct: 675 EEMAKKMKSLEQSIRDMQGLGGHKGISFSDLCMFPHVHLPT------------------- 715
Query: 57 IIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNT 116
+ L++ F +SL+ A+EW+ + HT+D LA F + +N
Sbjct: 716 ---------GAEGKEELLMAYFGESLVGIASEWFIDQDIANWHTWDNLARCFVQQFQYNI 766
Query: 117 RLKPNREFLRSLSQKKDESFREYAQRWRGAAARITPALDEEEMTQRFLKTLKKDYVERMI 176
+ P+R L ++ +K E+FREYA RWR AAR+ ++ E EM FL+ + DY ++
Sbjct: 767 DIVPDRSSLANMRKKTTENFREYAVRWREQAARVKLSMKESEMIDVFLQAQEPDYFHYLL 826
Query: 177 IAAPNNFSEMVTMGTRLEEAVREGIIVFEKVESSVNASKRYGNGH--HKKKETEVGMVSA 234
A F E++ +G +E ++ G IV + + + + G+G+ KK+ +V V
Sbjct: 827 SAVGKTFVEVIKVGEMVENGIKSGKIVSQAALKATTQALQNGSGNIGGKKRREDVATVGN 886
Query: 235 GAGQSMATVAPINAA*LPPSYPYAPYSQHPFFPYPLPSGQPQVPVNAVVQQMQQQPPVQQ 294
GA P G+ + ++ + P+++
Sbjct: 887 GAKDEFT-----------------------------PIGESYASLFQKLRTLNVLSPIER 917
Query: 295 QQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRQDKPPPDPLPPRFRSDLKCDFHQGALG 354
+ PP + Y++ HC D P G
Sbjct: 918 KMSNP-----PPRNLDYSQ---------HCAYCSDAP----------------------G 941
Query: 355 HDVKGCYALKYIVKKLIDQGKLTFEN-NVPHVLDNPLPNHAAVNMIEVYEEAPRLDVRNI 413
H+++ C+ LK ++ LID ++ E+ N P++ NPLP H NM+E+ + +
Sbjct: 942 HNIERCWYLKKAIQDLIDTRRIIVESPNGPNINQNPLPRHTETNMLEMVN-----NHEEV 996
Query: 414 ATPLVPLHIKLCQASLFNHDHTNCPECFR-NPLGCYVVQN----------DIQSLMNGNY 462
A P P + + TN + + P G I+ + +
Sbjct: 997 AVPYKP----ILKVETGMESSTNVIDLTKIMPSGAESTSEKLTPSSAPILTIKGALEDVW 1052
Query: 463 LTVSDVCVIVPVFHDPPVKSIPSKENVEPLVIRLPGPVSYTS*KAIPYKYNATMIE-NGV 521
+ + ++VP D P+ I + P++IR + T+ KA+P+ Y T++ G
Sbjct: 1053 ASQREARLVVPRGPDKPI-LIVQGAYIPPVIIRPVSQLPMTNPKAVPWNYEPTVVTYEGK 1111
Query: 522 EVPLASLAMVSNIAEGTTAALRSGTVRPPLFQKKAATPTTPPIDKATLTDVSHVTRDASR 581
E ++ + RSG LT++ D +
Sbjct: 1112 E--------INEEVDEIGGMTRSGRC-------------------YALTELRKNKNDQMQ 1144
Query: 582 PSQSIEDSNLDEILRLIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQAYV 641
+ + +E LR +K SDY IV+QL +TP++IS+LSLL+ S+AHR ++K+L +A+V
Sbjct: 1145 VKSPVTEGEAEEFLRKMKLSDYSIVEQLRKTPAQISLLSLLIHSDAHRKAVIKILNEAHV 1204
Query: 642 DHEVTMDRFGSIVGNITACNNLWFSEDELPEAGKYHNLALHISVNCKSDMLSNVLVDTGS 701
+EVT+ + I G I N + FS+DELP GS
Sbjct: 1205 PNEVTVSQLEKIAGRIFEVNRITFSDDELPV--------------------------EGS 1238
Query: 702 SLNVMPKSTLDQLSYRETSLRISTFLVKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMD 761
N+ P STL +L+ +R + V+AFDG++ + +GEI+L + IG F + FQV++
Sbjct: 1239 GANICPLSTLQKLNVNVERVRPNNVCVRAFDGTKTDAIGEIELILKIGHVDFTVNFQVLN 1298
Query: 762 INASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNGKLVTVHGEEAYLVSQLSSFSCIEAGS 821
INASY+ LLGRPW+H AGAV STLHQ KF + + V VH E V + SS I+A +
Sbjct: 1299 INASYNLLLGRPWVHRAGAVPSTLHQMVKFEYDRQEVIVHSEGDLSVYKDSSLPFIKANN 1358
Query: 822 A-EGTAFQGLTI-------EGMELKKTGTAMASLKDAQKAVQEGQAAGRGKLIQLRENKH 873
E +Q + EG + K MAS+ + ++ G G+G I L
Sbjct: 1359 ENEALVYQAFEVVVVEHILEGNLISKPQLPMASVMMVNEMLKHGFEPGKGLGIFL----- 1413
Query: 874 KEGLGFSPTSGVSTGTFHSAGFVNAITEEATGFGPRPVFVIPG-GIARDWDAIDIPSIMH 932
+G + + S GTF G+ PR + +D ++ P
Sbjct: 1414 -QGRAYPVSPRKSLGTF------------GLGYKPRAKDKMKAKKQKKDVWSLTKPIPPI 1460
Query: 933 VSELNHNKPVGHSNPTFP-PNFEFPMYE-AEAEEGDDIPY--EITRLLEQEKKAIQPHQE 988
N + S P P E M E E D+ + +L E + E
Sbjct: 1461 YKSFNKARTTESSESLLPEPVLEANMVELGEGTSDRDVQFIGPDVQLNNWEATPLPTKNE 1520
Query: 989 EIEFINIGTEEN-----KREIKVGAALE---EWVKKKI---------IQLLREYPDIFAW 1031
F ++ ++K+ + L E + ++I IQ L +Y D+FA
Sbjct: 1521 SCSFYADSSDMTCMQNFSPDLKIQSNLRPNPEIISQEIEYDEDEEDIIQALFDYKDVFAS 1580
Query: 1032 SYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYP 1091
SY+DMPGL +V H++P P+ PPV+QKLR+ DM++ IK E+ KQ++A + +YP
Sbjct: 1581 SYDDMPGLSTDMVVHKLPIDPKFPPVKQKLRKLKTDMSVIIKEEITKQLEAKVIRVAQYP 1640
Query: 1092 EWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSG 1151
W+ANIVPVPKKDGKVRMCVD+RDLNKASPKD+F L +I +L+DN A+ ++ F+D ++G
Sbjct: 1641 SWLANIVPVPKKDGKVRMCVDYRDLNKASPKDDFLLPNIHILLDNCAKHEIASFVDCYAG 1700
Query: 1152 YNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVD 1211
Y+QI M ED EKTSFITPWGT+CY+VMP GL NAGATY R MTT+FHDM+HKE+EVYVD
Sbjct: 1701 YHQIIMDDEDAEKTSFITPWGTYCYRVMPFGLKNAGATYMRAMTTMFHDMMHKEIEVYVD 1760
Query: 1212 DMIVKSKDEEQHVEYLTKMFERLR 1235
D+I+KSK + HV+ L + FERLR
Sbjct: 1761 DVIIKSKKQSNHVQDLRRFFERLR 1784
Score = 63.9 bits (154), Expect = 3e-08
Identities = 28/42 (66%), Positives = 35/42 (82%)
Query: 1194 MTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
MTT+FHDM+HKE+EVYVDD+I+KSK HV+ L + FERLR
Sbjct: 1 MTTMFHDMMHKEIEVYVDDVIIKSKKLPNHVQDLRRFFERLR 42
>UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 523 bits (1347), Expect = e-146
Identities = 348/947 (36%), Positives = 506/947 (52%), Gaps = 86/947 (9%)
Query: 18 NGDSVKTHDLCLVPKVDVPKKFKVPEFDRYNGLTCPHNHIIKYVRKMGNYSDNDSLMIHC 77
N S ++ LV + +P KFK P FD+YN +CP H+ Y RK+ Y+D++ + ++
Sbjct: 148 NSLSFDVTNMGLVEGLRIPYKFKAPSFDKYNDTSCPRTHVQAYYRKISAYTDDEKMWMYF 207
Query: 78 FQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKDESFR 137
FQDSL + +WY L +D + + +L AF Y N + P+R L+SL QK ESF+
Sbjct: 208 FQDSLSGASLDWYMELKRDSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFK 267
Query: 138 EYAQRWRGAAARITPALDEEEMTQRFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAV 197
EYAQRWR AAR+ P + E E+T F+ TL+ +++RM +FS++V G R E +
Sbjct: 268 EYAQRWRELAARVQPPMLERELTDMFIGTLQGVFMDRMGSCPFGSFSDVVICGERTESLI 327
Query: 198 REGIIVFEKVESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAA*LPPSYPY 257
+ G I + V SS +SK+ G +++E E +V Q+ A +P
Sbjct: 328 KTGKI--QDVGSS--SSKKPFAGAPRRREGETNVVQHRRDQNRIEYRQAAAVTIPA---- 379
Query: 258 APYSQHPFFPYPLPSGQPQVPVNAVVQQMQQQPPVQQQQHQQARPTFPPIPMLYAELLPT 317
P P Q Q V QQ QQQ P Q +Q R F +PM YAELLP
Sbjct: 380 -----------PQPRQQQQQRVQQPQQQQQQQRPYQPRQRMPDR-RFDSLPMSYAELLPE 427
Query: 318 LLLRGHCTTRQDKPPPDPLPPRFRSDLKCDFHQGALGHDVKGCYALKYIVKKLIDQGKLT 377
LL G R PP LPP + ++++CDFH GA GH + C AL++ V+ LID +
Sbjct: 428 LLRLGMVELRT-MAPPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAIN 486
Query: 378 FENNVPHVLDNPLPNHAA--VNMIEVYE-EAPRLDVRNIATPLVPLHIKLCQASLFNHDH 434
F VP+V++NP+P H VN IE E E ++V ++ T L+ + +L +++
Sbjct: 487 FAP-VPNVVNNPMPQHGGHRVNNIEGKEAEDLVVNVDDVQTSLLVVKGRLLNGGVYSGCD 545
Query: 435 TNCPECFRNPLGCYVVQNDIQSLMNGNYL----------TVSDVCVI------------- 471
+C C + GC ++ IQ +M+ L TVS + +
Sbjct: 546 EDCLGCAESENGCDQLRAGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPTEGRGQRVVS 605
Query: 472 VPVFHDPPVK-SIPSKENVEPLVIRLPGPVSYTS*KAIPYKY-----------------N 513
P +D PV S+P + P I G + + + +P+KY N
Sbjct: 606 APATNDTPVTISVPVTISA-PTTIVASGRRAVENSRVVPWKYDNAYRSNRRAESQTRPVN 664
Query: 514 ATMIENGV--EVPLASLAMVSNIAEGTTAALRSGTV-RPPLFQKKAATPTTPPIDKATLT 570
+ G+ VP V N+ G RSG + P + A K +
Sbjct: 665 QAPVTIGLPNRVPATVGPAVDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVV 723
Query: 571 DVSHVTRDASRPSQSIEDSNLDEILRLIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRN 630
+ V ++A P + E +++E +++IK+SDYKIVDQL QTPSKIS+LSLL+ SEAHRN
Sbjct: 724 EEEPVQKEA--PEGTFE-KDVEEFMKIIKKSDYKIVDQLNQTPSKISILSLLMCSEAHRN 780
Query: 631 TLLKVLEQAYVDHEVTMDRFGSIVGNITACNNLWFSEDELPEAGKYHNLALHISVNCKSD 690
LLK+L AYV E+++++ ++ N++ + + F+ +LP G+ HN ALHI++ CK
Sbjct: 781 ALLKMLNLAYVPQEISVNQLEGVMANVSTRHGVGFTNLDLPPEGRNHNKALHITMECKGA 840
Query: 691 MLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFLVKAFDGSRKNVLGEIDLPMTIGP 750
+LS+VLVDTGSSLNV+P L ++ L S +V+AFDGS+++V GE+ LP+ IGP
Sbjct: 841 VLSHVLVDTGSSLNVLPNQILKKIDVEGFVLTPSDLIVRAFDGSKRSVCGEVTLPVKIGP 900
Query: 751 ETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNGKLVTVHGEEAYLVSQ 810
E F I F VMDI +YSCLLGRPWIH AGAV+STLHQ K+V NG++VTV GEE LVS
Sbjct: 901 EVFDIIFYVMDIQPAYSCLLGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSH 960
Query: 811 LSSFSCIEA-GSAEGT---AFQGLTIEGM---ELKKTGTAMASLKDAQKAVQEGQAAGRG 863
LSSF +E G T AF+ + +E + E +K G ++ S K A++ V G+A G G
Sbjct: 961 LSSFKYVEVDGEIHETLCQAFETVALEKVAYAEQRKPGASITSYKQAKEVVDSGKAEGWG 1020
Query: 864 KLIQLRENKHKEGLGFSPTSGVSTG-----TFHSAGFVNAITEEATG 905
K++ L + K G+G+ P G TF SAG +N ATG
Sbjct: 1021 KMVDLPVKEDKFGVGYEPLRAEQNGQAGPSTFTSAGLMNHGDVSATG 1067
>UniRef100_Q6L420 Putative polyprotein [Solanum demissum]
Length = 1483
Score = 345 bits (885), Expect = 5e-93
Identities = 213/533 (39%), Positives = 308/533 (56%), Gaps = 40/533 (7%)
Query: 736 KNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNG 795
++ +GEI+L + IGP F + FQV++INASY+ LLGRPW+H AGAV STLHQ KF +
Sbjct: 213 EDAIGEIELILKIGPVDFTVNFQVLNINASYNLLLGRPWVHRAGAVPSTLHQMVKFEYDR 272
Query: 796 KLVTVHGEEAYLVSQLSSFSCIEAGSA-EGTAFQGLTI-------EGMELKKTGTAMASL 847
+ V VH E V + SS I+A + E +Q + EG + K MAS+
Sbjct: 273 QEVIVHCERDLSVYKDSSLPFIKANNENEALVYQAFEVVVVEHNLEGNLISKPQLPMASV 332
Query: 848 KDAQKAVQEGQAAGRGKLIQLRENKHKEGLGFSPTSGVSTGTFHSAGFVNAITEEATGFG 907
+ ++ G G+G I L +G + + S GTF G+ + ++
Sbjct: 333 MMVNEMLKHGFKLGKGLGIFL------QGRAYPVSPRKSLGTF-GLGYKPRVEDKMKAKK 385
Query: 908 PR-PVFVIPGGIARDWDAIDIPSIMHVSELNHNKPVGHSNPTFPPNFEFPMYEA---EAE 963
R V+ + I + + + + S+ +PV + F+ EA E E
Sbjct: 386 QRRDVWSLTKPIPPIYKSFNKARTIESSKSLLPEPVLEVHEELINYFQDLFVEADMIELE 445
Query: 964 EGD---DIPY----------EITRL-LEQEKKAIQPHQEEIEFINIGTEENKREIKVGAA 1009
EG D+ + E T L ++ E + ++ I + + K + +
Sbjct: 446 EGTSDRDVQFIGPDVQLNNWEATPLPIKNESCSFYADSSDMTCIQNYSPDLKIQSNLRPN 505
Query: 1010 LE------EWVKKKIIQ-LLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLR 1062
E E+ + ++ + L +Y D+FA SY+DMPGL +V H++P P+ PPV++KLR
Sbjct: 506 PEILSQEIEYDEDEVFEEALLDYKDVFASSYDDMPGLSTDMVVHKLPIDPKFPPVKKKLR 565
Query: 1063 RTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPK 1122
+ DM++ IK E+ KQ++A + +YP W+ANIVPVPKKDGKVRMCVD RDLNKASPK
Sbjct: 566 KLKTDMSVIIKEEITKQLEAKVIRVAQYPSWLANIVPVPKKDGKVRMCVDCRDLNKASPK 625
Query: 1123 DNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIG 1182
D+FPL + +L+DN A+ ++ F+D ++GY+QI M ED EK SFITPWGT+CY+VMP G
Sbjct: 626 DDFPLPNSHILLDNCAEHEIASFVDCYAGYHQIIMDDEDAEKMSFITPWGTYCYRVMPFG 685
Query: 1183 LINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
L NAGATY R MTT+FHDM+HKE+EVYVDD+I+KSK + HV+ L + FERLR
Sbjct: 686 LKNAGATYMRAMTTMFHDMMHKEIEVYVDDVIIKSKKQSNHVQDLRRFFERLR 738
Score = 85.1 bits (209), Expect = 1e-14
Identities = 62/229 (27%), Positives = 107/229 (46%), Gaps = 8/229 (3%)
Query: 483 IPSKENVEPLVIRLPGPVSYTS*KAIPYKYNATMIENGVEVPLASLAMVSNIA-----EG 537
I S NV L +P TS K P ++ +E L M + A E
Sbjct: 21 IESSANVVDLTKMMPSGAESTSEKLTPSSAPILTVKGALEDASVPLPMTNPKAVPWKYEP 80
Query: 538 TTAALRSGTVRPPLFQKKAATPTTPPIDKATLTDVSHVTRDASRPSQSIEDSNLDEILRL 597
T + + + + + T + A L ++ D + + + +E LR
Sbjct: 81 TVVTYKGKEINEKVDEIEGMTRSRRCYAPAELRKNNN---DQMQVKSPVTEREAEEFLRK 137
Query: 598 IKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRNTLLKVLEQAYVDHEVTMDRFGSIVGNI 657
+K SDY IV+QL +TP++IS+LSLL+ S+ HR ++K+L +A+V +E TM + I G I
Sbjct: 138 MKLSDYSIVEQLRKTPAQISLLSLLIHSDEHRKVVMKILNEAHVPNEGTMSQLEKIAGRI 197
Query: 658 TACNNLWFSEDELPEAGKYHNLALHISVNCKSDMLSNVLVDTGSSLNVM 706
N + FS+DELP + L + + ++ +++ +S N++
Sbjct: 198 FEVNCITFSDDELPVEDAIGEIELILKIGPVDFTVNFQVLNINASYNLL 246
>UniRef100_Q7XU13 OSJNBa0091D06.8 protein [Oryza sativa]
Length = 2657
Score = 344 bits (882), Expect = 1e-92
Identities = 231/686 (33%), Positives = 350/686 (50%), Gaps = 35/686 (5%)
Query: 560 TTPPIDKATLTDVSHVTRDASRPSQSIEDSNLDEILRLIKRSDYKIVDQLLQTPSKISVL 619
T P KA + V ++AS P ++ E + I DY ++ L + P+ +SV
Sbjct: 1348 TLPDPHKAKPSKVDKPIKEASPPGEAPETKARSKEKPDI---DYNVLAHLKRIPALLSVY 1404
Query: 620 SLLLSSEAHRNTLLKVLEQAYVDHEVTMDRFGSIVGNITACNNLWFSEDELPEAGKYHNL 679
L+ R L+K L+ V +EV M + + N N + F++++ G HN
Sbjct: 1405 DALILVPDLREALIKALQTPEV-YEVDMAKH-RLFNNPLFVNEITFADEDNIIEGSDHNR 1462
Query: 680 ALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFLVKAFDGSRKNVL 739
L+I N L +L+D GS++N++P +L + + L + ++ FD K+ L
Sbjct: 1463 PLYIEGNIGPAHLRRILIDPGSAVNILPVRSLTRAGFTTKDLEPTDMVICGFDNQGKSTL 1522
Query: 740 GEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNGKLVT 799
G I + + + +F + F V++ N SYS LLGRPWIH V STLHQ KF+ +GK
Sbjct: 1523 GAITVKIQMSTFSFKVHFFVIEANTSYSALLGRPWIHKYRVVPSTLHQCLKFL-DGK--- 1578
Query: 800 VHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGMELKKT----------GTAMASLKD 849
G + +V SS++ E+ A+ + + +L +T GTAM +
Sbjct: 1579 --GAQQRIVGNSSSYTIQESYHADAKYYFPVEENMQQLGRTAPAADVLIQPGTAMTP--E 1634
Query: 850 AQKAVQEGQAAGRGKLIQLRENKHKEGLGFSPTSGVSTGTFHSAGFVNAITEEATGFGPR 909
A++ + R +++ R + +E S + F+ +E +G
Sbjct: 1635 ARRTPNKEATPTRSGVVKKRVAEIEEAKA-GVQSICEDWWAQAEDFIKRKCKEQPKYG-- 1691
Query: 910 PVFVIPGGIARDWDAIDIPSIMHVSELNHNKPVGHSNPTFPPNFEFPMYEAEAEEGDDIP 969
+ I D D H + P P F + AE EE D
Sbjct: 1692 -LGYINTDETDDEDEALDDHTFHCCTASVTTIADAQLPQHP----FQVAAAEVEEELDT- 1745
Query: 970 YEITRLLEQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIF 1029
+ +Q + QP +E+ +N+GTE++ R I V A L E ++ L EY D F
Sbjct: 1746 ---MQAPQQLEDGGQPTIDELVEMNLGTEDDPRPIFVSAMLTEEEREDYRSFLMEYRDCF 1802
Query: 1030 AWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVE 1089
W+Y++MPGLDP++ H++ P+ PV+Q RR P+ ++ EV + I+ GF+ ++
Sbjct: 1803 TWTYKEMPGLDPRVATHKLAIDPQFRPVKQPPRRLRPEFQDQVIAEVDRLINVGFIKEIQ 1862
Query: 1090 YPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGF 1149
YP W+ANI+PV KK+G+VR+CVDFRDLN+A PKD+FPLL+ +++V + FMDG
Sbjct: 1863 YPRWLANIIPVEKKNGQVRVCVDFRDLNRACPKDDFPLLNTELVVASTTGYGALSFMDGS 1922
Query: 1150 SGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVY 1209
SGY QIKM P D T+F TP G F Y VMP GL NAGATYQR M + D+IH VE Y
Sbjct: 1923 SGYKQIKMDPLDASDTAFRTPKGNFYYTVMPFGLKNAGATYQRAMQFILDDLIHHSVECY 1982
Query: 1210 VDDMIVKSKDEEQHVEYLTKMFERLR 1235
VDDM+VK+ D ++H E L +FERLR
Sbjct: 1983 VDDMVVKTHDPKRHQEDLRVVFERLR 2008
Score = 53.1 bits (126), Expect = 5e-05
Identities = 78/402 (19%), Positives = 150/402 (36%), Gaps = 82/402 (20%)
Query: 25 HDLCLVPKVDVPKKFKVPEFDRYNGLTCPHNHIIKYVRKMGNYSDNDSLMIHCFQDSLME 84
HDL P P KF+ +FDR H+ + G+ ++N SL++ F SL
Sbjct: 839 HDLVAFPARWHPPKFR--QFDRTGDA---REHLAYFEAACGDTANNSSLLLRQFSGSLTG 893
Query: 85 DAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREF----LRSLSQKKDESFREYA 140
A WY+ L + ++ + K H+ + ++F L + Q++DE+ +Y
Sbjct: 894 PAFHWYSRLPTGSIRSWASMKEVLKKHF-----VAMKKDFSIVELSQVRQRRDEAIDDYV 948
Query: 141 QRWRGAAARITPALDEEEMTQRFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVREG 200
R+R + R+ + E+ + L + T+LE
Sbjct: 949 IRFRNSFVRLAREMHLEDAIEIALSS--------------------AVAATKLEFENSPQ 988
Query: 201 IIVFEKVESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAA*LPPSYPYAPY 260
I+ K S+ + +K + +G G A N A + + P
Sbjct: 989 IMELYKNASAFDPAKHFS-----------ATKPSGGGSKPKAPAEANVARVFSTAP---- 1033
Query: 261 SQHPFFPYPLPSGQPQVPVNAVVQQM---QQQPPVQQQQHQQARPTFPPIPMLYAELLPT 317
Q QVP+ + +Q+P +Q +Q + EL+
Sbjct: 1034 -------------QGQVPMLGAKNEQARGRQRPSIQDLLKKQY--------IFRRELVKD 1072
Query: 318 LL--LRGHCTTRQDKPPPDPLPPRFRSDLKCDFHQGALGHDVKGCYALKYIVKKLIDQGK 375
+ L+ H +P + L C +H+ +G+ ++ C A K +++ +++ +
Sbjct: 1073 MFNQLKEHRALNLPEPWRPDQVTMVDNPLYCPYHR-YVGYAIEDCVAFKEWLQRAVNEKR 1131
Query: 376 LTFENNVPHVLDNPLPNHAAVNMIEVYEEAPRLDVRNIATPL 417
+ + D P++ AVNM+ V + + +I PL
Sbjct: 1132 INLD------ADAINPDYRAVNMVSVEPGSREQEGADIWVPL 1167
>UniRef100_Q9LMV1 F5M15.26 [Arabidopsis thaliana]
Length = 1838
Score = 246 bits (628), Expect = 3e-63
Identities = 122/244 (50%), Positives = 164/244 (67%)
Query: 991 EFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHRIPT 1050
E +NI + R + VGA + ++ ++I LL+ FAWS EDM G+DP I H +
Sbjct: 759 EMVNIDESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNV 818
Query: 1051 KPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMC 1110
P PV+QK R+ P+ A + EV+K + AG ++ V+YPEW+AN V V KK+GK R+C
Sbjct: 819 DPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVC 878
Query: 1111 VDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITP 1170
VD+ DLNKA PKD++PL HID LV+ + + + FMD FSGYNQI M +D+EKTSF+T
Sbjct: 879 VDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTD 938
Query: 1171 WGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKM 1230
GT+CYKVM GL NAGATYQR + + D I + VEVY+DDM+VKS E HVE+L+K
Sbjct: 939 RGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKC 998
Query: 1231 FERL 1234
F+ L
Sbjct: 999 FDVL 1002
Score = 71.2 bits (173), Expect = 2e-10
Identities = 46/176 (26%), Positives = 82/176 (46%), Gaps = 7/176 (3%)
Query: 665 FSEDELPEAGKYHNLALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRIS 724
F++ +L HN L + + ++ VL+DTGSS++++ K L ++ + ++
Sbjct: 602 FTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPV 661
Query: 725 TFLVKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTST 784
+ + FDG +G I LP+ +G + F V+ A Y+ +LG PWIH A+ ST
Sbjct: 662 SKPLAGFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPST 721
Query: 785 LHQNFKFVKNGKLVTVHG-------EEAYLVSQLSSFSCIEAGSAEGTAFQGLTIE 833
HQ KF + + T+ +Y S+L + ++ T G+ E
Sbjct: 722 YHQCVKFPTHNGIFTLRAPKEAKTPSRSYEESELCRTEMVNIDESDPTRCVGVGAE 777
Score = 45.4 bits (106), Expect = 0.011
Identities = 36/172 (20%), Positives = 71/172 (40%), Gaps = 4/172 (2%)
Query: 43 EFDRYNGLTCPHNHIIKY---VRKMGNYSDN-DSLMIHCFQDSLMEDAAEWYTSLSKDDV 98
+ + YNG P + + + + DN D+ F + L A W++ L + +
Sbjct: 220 KLESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAHNWFSRLKPNSI 279
Query: 99 HTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKDESFREYAQRWRGAAARITPALDEEE 158
+F +L ++F HY + + L S+SQ ES R + R++ IT +
Sbjct: 280 DSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFKLVVTNITVPDEAAI 339
Query: 159 MTQRFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVREGIIVFEKVESS 210
+ R + + + + AP+ + + +R E E +I+ K S+
Sbjct: 340 VALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELEEEKLILARKHNST 391
>UniRef100_Q7FAL2 OSJNBa0042N22.17 protein [Oryza sativa]
Length = 1436
Score = 244 bits (623), Expect = 1e-62
Identities = 143/344 (41%), Positives = 197/344 (56%), Gaps = 19/344 (5%)
Query: 908 PRPVFVIPGG------IARDWDAID--IPSIMHVSELNHNKPVGHSNPTFPPNFEFPM-- 957
P FVI G + RDW ++ IPS MH + G P + M
Sbjct: 962 PTSFFVIDGKGSYSLLLGRDWIHVNCCIPSTMHQCLIQWQ---GDKVEIVPADSRLKMEN 1018
Query: 958 ----YEAEAEEGDDIPYEITRLLEQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEW 1013
+E E + +I L+ ++ +++E I+IG + R + L
Sbjct: 1019 PSYYFEGVVEGSNVYTKDIVDDLDDKQGQGFMSTDDLEEIDIGPGDRPRPTFISKNLSSE 1078
Query: 1014 VKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIK 1073
+ K+I+LL+E+ D FAW Y +MPGL IVEHR+P KP P +Q LRR DM +K
Sbjct: 1079 FRTKLIELLKEFRDCFAWEYHEMPGLSRSIVEHRLPLKPGFKPYQQPLRRCKADMLEPVK 1138
Query: 1074 NEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVL 1133
E+++ DAGF+ Y EWV++IVPV KK+GKVR+C+DFRDLNKA+PKD +P+ D L
Sbjct: 1139 AEIKRLYDAGFIRPCRYAEWVSSIVPVIKKNGKVRVCIDFRDLNKATPKDEYPMPVADQL 1198
Query: 1134 VDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPW--GTFCYKVMPIGLINAGATYQ 1191
VD + K+ FMDG +GYNQI M+ ED KT+F P G F + VM GL +AGATYQ
Sbjct: 1199 VDAASGHKILSFMDGNAGYNQIFMAEEDIHKTAFRCPGAIGLFEWVVMTFGLKSAGATYQ 1258
Query: 1192 RGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
R M ++HD+I VEVY+DD++VKSK+ H+ L K+FER R
Sbjct: 1259 RAMNYIYHDLIGWLVEVYIDDVVVKSKEIGDHIADLRKVFERKR 1302
Score = 71.2 bits (173), Expect = 2e-10
Identities = 40/120 (33%), Positives = 66/120 (54%), Gaps = 1/120 (0%)
Query: 669 ELPEAGKYHNLA-LHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFL 727
E PE + +L L+I+ +S ++VD G+++N+MP +T +L L +
Sbjct: 876 EKPEGTENRDLKPLYINGYVNGKPMSKMMVDGGAAVNLMPYTTFRKLGRNAEDLIRMNMV 935
Query: 728 VKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 787
+K F G+ G +++ +T+G +T +F V+D SYS LLGR WIH + ST+HQ
Sbjct: 936 LKDFGGNSSETKGVLNVELTVGSKTIPTSFFVIDGKGSYSLLLGRDWIHVNCCIPSTMHQ 995
>UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic DNA, chromosome 1, PAC
clone:P0433F09 [Oryza sativa]
Length = 2876
Score = 239 bits (611), Expect = 3e-61
Identities = 125/274 (45%), Positives = 171/274 (61%), Gaps = 6/274 (2%)
Query: 962 AEEGDDIPYEITRLLEQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQL 1021
A G + ++ L+Q QP +E+ +N+GTE++ R I V L E ++
Sbjct: 1750 AAVGVEEELDVAGALKQLDDGGQPTIDELVEMNLGTEDDPRPIFVSGMLTEEEREDYRSF 1809
Query: 1022 LREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQID 1081
L E+ D FAW+Y++MPGLD ++ H++ P+ PV+Q RR P+ ++ EV + I+
Sbjct: 1810 LMEFRDCFAWTYKEMPGLDSRVATHKLAIDPQFRPVKQPPRRLRPEFQDQVIAEVDRLIN 1869
Query: 1082 AGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSK 1141
GF+ ++YP W+ANIVPV KK+G+VR+CVDFRDLN+A PKD+FPL +++VD+
Sbjct: 1870 VGFIKEIQYPRWLANIVPVEKKNGQVRVCVDFRDLNRACPKDDFPLPITEMVVDSTTG-- 1927
Query: 1142 VFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDM 1201
SGYNQIKM D T+F TP G F Y VMP GL NAGATYQR M + D+
Sbjct: 1928 ----YGALSGYNQIKMDLLDAFDTAFRTPKGNFYYTVMPFGLKNAGATYQRAMQFVLDDL 1983
Query: 1202 IHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
IH VE YVDDM+VK+KD E H E L +FERLR
Sbjct: 1984 IHHSVECYVDDMVVKTKDHEHHQEDLRIVFERLR 2017
Score = 112 bits (280), Expect = 7e-23
Identities = 80/284 (28%), Positives = 134/284 (47%), Gaps = 8/284 (2%)
Query: 560 TTPPIDKATLTDVSHVTRDASRPSQSIEDSNLDEILRLIKRSDYKIVDQLLQTPSKISVL 619
T P K+ + +V + AS P ++ E + DYK++ L + P+ +SV
Sbjct: 1263 TLPDPHKSKVPNVDKPAKKASPPGEAPEAPETKTGSKEKPAVDYKVLAHLKRIPALLSVY 1322
Query: 620 SLLLSSEAHRNTLLKVLEQAYVDHEVTMDRFGSIVGNITACNNLWFSEDELPEAGKYHNL 679
L+ R L+K L+ V +EV M + + N N + F++++ G HN
Sbjct: 1323 DALMMVPDLREALIKALQAPEV-YEVDMAKH-RLYDNPLFVNEITFADEDNIIKGGDHNR 1380
Query: 680 ALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFLVKAFDGSRKNVL 739
L+I N S L +L+D GS++N++P +L + + L ++ FD K L
Sbjct: 1381 PLYIEGNIGSAHLRRILIDLGSAVNILPVRSLTRAGFTTKDLEPIDVVICGFDNQGKPTL 1440
Query: 740 GEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNGKLVT 799
G I + + + +F + F V++ N SYS LLGRPWIH V STLHQ KF+
Sbjct: 1441 GAITIKIQMSTFSFKVRFFVIEANTSYSALLGRPWIHKYRVVPSTLHQCLKFLDG----- 1495
Query: 800 VHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGMELKKTGTA 843
+G + + S S ++ E+ A+ + + +L +T A
Sbjct: 1496 -NGVQQRITSNFSPYTIQESYHADAKYYFPVEENKQQLGRTTPA 1538
>UniRef100_Q40369 RPE15 protein [Medicago sativa]
Length = 219
Score = 239 bits (611), Expect = 3e-61
Identities = 126/192 (65%), Positives = 143/192 (73%), Gaps = 16/192 (8%)
Query: 146 AAARITPALDEEEMTQRFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVREGIIVFE 205
AAAR++P LDEEE+TQ FLKTLKK++ ERMI++APNNFS+MVTMGTRLEEAVREGIIV E
Sbjct: 13 AAARVSPRLDEEELTQTFLKTLKKEHKERMIVSAPNNFSDMVTMGTRLEEAVREGIIVLE 72
Query: 206 KVESSVNASKRYGNGHHKKKET-EVGMVSAGAGQSMATVAPINAA*LPPSYPYAPYSQHP 264
K E S +A K+YGNGHHK+KE+ +VGMVS P+NA LP Y Y SQHP
Sbjct: 73 KGECSASAPKKYGNGHHKRKESKKVGMVSGNHD------TPVNATQLPLPYQYMQISQHP 126
Query: 265 FFP-----YPLPSGQPQVPVNAVVQQMQQQPPVQQQQHQQARPT----FPPIPMLYAELL 315
FFP YPLP GQP VPVNA+ QQ+Q QPP QQQQ QQ P FPPIPM YAELL
Sbjct: 127 FFPPFYQQYPLPPGQPPVPVNAMAQQVQHQPPAQQQQQQQQGPRPKTYFPPIPMRYAELL 186
Query: 316 PTLLLRGHCTTR 327
PTLL +GHC TR
Sbjct: 187 PTLLAKGHCVTR 198
>UniRef100_Q9SJP0 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1787
Score = 237 bits (604), Expect = 2e-60
Identities = 118/267 (44%), Positives = 168/267 (62%), Gaps = 1/267 (0%)
Query: 968 IPYEITRLLEQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPD 1027
I Y + + Q+++ P + + +NI + R + + L ++ +++ LR+
Sbjct: 695 IIYNVILVPTQDERP-NPQKGTVVQVNIDESDPSRCVGIRIDLPSELQNELVNFLRQNAA 753
Query: 1028 IFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMT 1087
FAWS EDMPG+D + H + P P++QK R+ PD + EV+K +DAG ++
Sbjct: 754 TFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVE 813
Query: 1088 VEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMD 1147
V YP+W+ N V V KK+GK R+C+DF DLNKA PKD+FPL HID LV+ A +++ FMD
Sbjct: 814 VRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMD 873
Query: 1148 GFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVE 1207
FSGYNQI M DREKT FIT GT+CYKVMP GL NAGATY R + +F D + +E
Sbjct: 874 AFSGYNQILMHQNDREKTVFITDQGTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSME 933
Query: 1208 VYVDDMIVKSKDEEQHVEYLTKMFERL 1234
VY+DDM+VKS E+H+ +L + F+ L
Sbjct: 934 VYIDDMLVKSLRAEEHITHLRQCFQVL 960
>UniRef100_Q7XXJ2 OSJNBa0094O15.10 protein [Oryza sativa]
Length = 1197
Score = 237 bits (604), Expect = 2e-60
Identities = 131/291 (45%), Positives = 181/291 (62%), Gaps = 6/291 (2%)
Query: 950 PPNFEFPMYEAEAE-EGDDIPYEITRLLEQEKKAIQPHQ--EEIEFINIGTEENKREIKV 1006
P E P Y E EG D+ Y + + + K Q +++E I+IG + R +
Sbjct: 291 PRQMENPSYYFEGVVEGSDV-YAKDTIDDLDDKQGQGFMSADDLEEIDIGPGDRPRPTFI 349
Query: 1007 GAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHP 1066
L + K+I+LL+E+ D AW Y +MPGL IVEHR+P KP P +Q RR
Sbjct: 350 SKHLSLEFRAKLIELLKEFRDCLAWEYYEMPGLSRSIVEHRLPIKPRVRPHQQLPRRCKA 409
Query: 1067 DMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFP 1126
DM +K E+++ DAGF+ Y EWV++IVPV KK+GKVR+C+DF+DLNKA+PKD +P
Sbjct: 410 DMLEPVKAEIKRLYDAGFIHPCRYAEWVSSIVPVIKKNGKVRVCIDFQDLNKATPKDEYP 469
Query: 1127 LLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPW--GTFCYKVMPIGLI 1184
+ D LVD + +K+ FMDG +GYNQI M+ ED KT+F P G F + VM GL
Sbjct: 470 MPVADHLVDAASGNKILSFMDGNAGYNQIFMAEEDIHKTAFRCPGAIGLFEWVVMTFGLK 529
Query: 1185 NAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
+AGATYQR M ++HD+I VEVY+DD++VKSK+ + H+ L K+FER R
Sbjct: 530 SAGATYQRAMNYIYHDLIGSLVEVYIDDVVVKSKEVDDHIADLRKVFERTR 580
>UniRef100_Q8S6L9 Putative retroelement [Oryza sativa]
Length = 1445
Score = 236 bits (601), Expect = 4e-60
Identities = 120/248 (48%), Positives = 163/248 (65%), Gaps = 2/248 (0%)
Query: 988 EEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHR 1047
+++E ++IG + R + L + K+I+LL+EY D FAW Y +MPGL IVEHR
Sbjct: 524 DDLEEVDIGPGDRPRPTFIIKNLSSDFRTKLIELLKEYRDCFAWEYYEMPGLSRSIVEHR 583
Query: 1048 IPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV 1107
+P KPE P +Q RR DM +K E++ D GF+ Y EWV++I+PV KK+GK+
Sbjct: 584 LPIKPEVRPHQQPPRRCKADMLEPVKGEIKCLYDTGFIRPCRYAEWVSSIIPVIKKNGKI 643
Query: 1108 RMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSF 1167
R+C+DFRDLNKA+ KD +P+ D LVD + K+ FMDG +GYNQI M+ ED KTSF
Sbjct: 644 RVCIDFRDLNKATSKDKYPMPVADQLVDATSGHKILSFMDGNAGYNQIIMAEEDIHKTSF 703
Query: 1168 ITPW--GTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 1225
P G F + VM GL +AGATYQR M ++HD+I VEVY+DD++VKSK+ E H+
Sbjct: 704 RCPGAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSKEIEDHIA 763
Query: 1226 YLTKMFER 1233
L K+FER
Sbjct: 764 NLRKVFER 771
>UniRef100_Q8LM35 Putative retroelement [Oryza sativa]
Length = 1384
Score = 236 bits (601), Expect = 4e-60
Identities = 120/248 (48%), Positives = 163/248 (65%), Gaps = 2/248 (0%)
Query: 988 EEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHR 1047
+++E ++IG + R + L + K+I+LL+EY D FAW Y +MPGL IVEHR
Sbjct: 463 DDLEEVDIGPGDRPRPTFIIKNLSSDFRTKLIELLKEYRDCFAWEYYEMPGLSRSIVEHR 522
Query: 1048 IPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV 1107
+P KPE P +Q RR DM +K E++ D GF+ Y EWV++I+PV KK+GK+
Sbjct: 523 LPIKPEVRPHQQPPRRCKADMLEPVKGEIKCLYDTGFIRPCRYAEWVSSIIPVIKKNGKI 582
Query: 1108 RMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSF 1167
R+C+DFRDLNKA+ KD +P+ D LVD + K+ FMDG +GYNQI M+ ED KTSF
Sbjct: 583 RVCIDFRDLNKATSKDKYPMPVADQLVDATSGHKILSFMDGNAGYNQIIMAEEDIHKTSF 642
Query: 1168 ITPW--GTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 1225
P G F + VM GL +AGATYQR M ++HD+I VEVY+DD++VKSK+ E H+
Sbjct: 643 RCPGAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSKEIEDHIA 702
Query: 1226 YLTKMFER 1233
L K+FER
Sbjct: 703 NLRKVFER 710
>UniRef100_Q6AT01 Putative polyprotein [Oryza sativa]
Length = 1739
Score = 235 bits (599), Expect = 7e-60
Identities = 122/250 (48%), Positives = 166/250 (65%), Gaps = 2/250 (0%)
Query: 988 EEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHR 1047
+++E I+IG + R V L + K+I+LL+E+ D FAW Y +MPGL IVEHR
Sbjct: 859 DDLEEIDIGPGDRPRPTFVSKNLSPEFRTKLIELLKEFRDCFAWEYYEMPGLSRSIVEHR 918
Query: 1048 IPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV 1107
+P KP P +Q LRR DM +K E+++ DAGF+ Y +WV++IVPV KK+GKV
Sbjct: 919 LPIKPGVRPRQQPLRRCKADMLEPVKAEIKRLYDAGFIRPWRYAKWVSSIVPVIKKNGKV 978
Query: 1108 RMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSF 1167
R+C+DFRDLNKA+PKD +P+ D LVD + +K+ FMDG +GYNQI M+ ED KT+F
Sbjct: 979 RVCIDFRDLNKATPKDEYPMPVADQLVDAASGNKILSFMDGNAGYNQIFMAEEDIHKTAF 1038
Query: 1168 ITPW--GTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 1225
P G F + VM GL +AGATYQR M ++HD+I V VY+DD++VKSK+ E +
Sbjct: 1039 RCPCAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVGVYIDDVVVKSKEIEDQIA 1098
Query: 1226 YLTKMFERLR 1235
L K+FER R
Sbjct: 1099 DLRKVFERTR 1108
Score = 72.8 bits (177), Expect = 6e-11
Identities = 37/111 (33%), Positives = 62/111 (55%)
Query: 677 HNLALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFLVKAFDGSRK 736
H L+I+ ++S ++VD G+++N+MP +T +L L + ++K F G+
Sbjct: 691 HLKPLYINGYVNGKLMSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMVLKDFGGNPS 750
Query: 737 NVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 787
G +++ +T+G +T TF V+D SYS LGR WIH + ST+HQ
Sbjct: 751 ETKGVLNVGLTVGSKTIPTTFLVIDGKGSYSLFLGRDWIHANCCIPSTMHQ 801
Score = 36.2 bits (82), Expect = 6.4
Identities = 22/80 (27%), Positives = 41/80 (50%), Gaps = 1/80 (1%)
Query: 65 GNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREF 124
G S D+L + F SL A W++SL ++++ +L F S++ ++ +
Sbjct: 343 GEASAVDALKVRLFWLSLSGSAFTWFSSLPYGSINSWADLEKQFHSYF-YSGVHEMKLSD 401
Query: 125 LRSLSQKKDESFREYAQRWR 144
L ++ Q+ DE E+ QR+R
Sbjct: 402 LTAIKQRHDEPVHEHIQRFR 421
>UniRef100_Q75HC5 Putative reverse transcriptase [Oryza sativa]
Length = 1149
Score = 234 bits (598), Expect = 9e-60
Identities = 143/344 (41%), Positives = 195/344 (56%), Gaps = 19/344 (5%)
Query: 908 PRPVFVIPGG------IARDWDAID--IPSIMHVSELNHNKPVGHSNPTFPPN----FEF 955
P FVI G + RDW + IPS MH + G P + E
Sbjct: 736 PTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQ---GDKIEIVPADSQLKMEN 792
Query: 956 PMYEAEAEEGDDIPYEITRLLEQEKKAIQPHQ--EEIEFINIGTEENKREIKVGAALEEW 1013
P Y E G Y + + + K Q +++E I+IG + R + L
Sbjct: 793 PSYYFEGVMGGSSVYLKDTVDDLDDKQGQGFMSADDLEEIDIGPGDRPRPTFISKNLSPE 852
Query: 1014 VKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIK 1073
+ K+I+LL+E+ D FAW Y +MPGL IVEHR+P KP P +Q RR DM +K
Sbjct: 853 FRTKLIELLKEFRDCFAWEYYEMPGLSRSIVEHRLPIKPGVRPRQQHPRRCKDDMLEPVK 912
Query: 1074 NEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVL 1133
E ++ DAGF+ Y EWV++IVPV KK+GKVR+C+DFRDLNKA+PKD +P+ D L
Sbjct: 913 AETKRLYDAGFICPCRYAEWVSSIVPVIKKNGKVRVCIDFRDLNKATPKDEYPMPVADQL 972
Query: 1134 VDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPW--GTFCYKVMPIGLINAGATYQ 1191
VD + +K+ FMDG +GYNQI M+ ED KT+F P G F + VM GL +AGATYQ
Sbjct: 973 VDAVSGNKILSFMDGNAGYNQIFMAEEDIHKTTFRCPGAIGLFEWVVMTFGLKSAGATYQ 1032
Query: 1192 RGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
R M +++D+I VEVY+DD++VKSK+ H+ L K+FER +
Sbjct: 1033 RAMNYIYYDVIGWLVEVYIDDVVVKSKEIGDHIADLRKVFERTK 1076
Score = 72.0 bits (175), Expect = 1e-10
Identities = 40/120 (33%), Positives = 67/120 (55%), Gaps = 1/120 (0%)
Query: 669 ELPEAGKYHNLA-LHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFL 727
E PE + +L L+I+ +S ++VD G+++N+MP +T +L L + +
Sbjct: 650 EKPEGTENRHLKPLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMV 709
Query: 728 VKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 787
++ F G+ G +++ +T+G +T TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 710 LEDFGGNPSETKGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQ 769
>UniRef100_Q7XSP1 OSJNBa0070M12.15 protein [Oryza sativa]
Length = 1685
Score = 234 bits (597), Expect = 1e-59
Identities = 119/250 (47%), Positives = 165/250 (65%), Gaps = 2/250 (0%)
Query: 988 EEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHR 1047
++++ ++IG + R + L + K+++LL+E+ D FAW Y +MPGL IVEHR
Sbjct: 904 DDLDEVDIGPGDRPRPTFISKNLSSEFRTKLMELLKEFRDCFAWEYYEMPGLSRSIVEHR 963
Query: 1048 IPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV 1107
+P KP P +Q RR DM +K E+++ DAGF+ Y EWV++IVPV KK+GKV
Sbjct: 964 LPLKPGIRPHQQPPRRCKADMLEPVKAEIKRLYDAGFIHPCRYAEWVSSIVPVIKKNGKV 1023
Query: 1108 RMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSF 1167
R+C+DFRDLNKA+PKD +P+ D LVD + K+ FMDG +GYNQI M+ ED KT F
Sbjct: 1024 RVCIDFRDLNKATPKDEYPMPVADQLVDAASGHKILSFMDGNAGYNQIFMTEEDIHKTVF 1083
Query: 1168 ITPW--GTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 1225
P G F + VM GL +AGATYQR M ++HD+I VEVY+DD++VKS++ E H+
Sbjct: 1084 RCPGAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSREIEDHIA 1143
Query: 1226 YLTKMFERLR 1235
L K+FER R
Sbjct: 1144 DLRKVFERTR 1153
Score = 73.9 bits (180), Expect = 3e-11
Identities = 41/120 (34%), Positives = 67/120 (55%), Gaps = 1/120 (0%)
Query: 669 ELPEAGKYHNLA-LHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFL 727
E PE + +L L+I+ +S ++VD G+++N+MP +T +L L + +
Sbjct: 727 EKPEGTENRHLKPLYINGYVNGKPMSKMMVDGGAAVNLMPYTTFRKLGRTTEDLMKTNMV 786
Query: 728 VKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 787
+K F G+ G +++ +T+G +T TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 787 LKDFGGNPSETKGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQ 846
>UniRef100_Q8S7Z2 Putative retroelement [Oryza sativa]
Length = 2058
Score = 233 bits (593), Expect = 4e-59
Identities = 140/344 (40%), Positives = 193/344 (55%), Gaps = 19/344 (5%)
Query: 908 PRPVFVIPGG------IARDWDAID--IPSIMHVSELNHNKPVGHSNPTFPPNFEFPM-- 957
P FVI G + RDW + IPS MH + G P + + M
Sbjct: 1159 PTTFFVIDGKGSYSLLLGRDWVHANCCIPSTMHQCLIQWQ---GDKIEIVPADSQLKMEN 1215
Query: 958 ----YEAEAEEGDDIPYEITRLLEQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEW 1013
+E E + + L+ ++ +++E I+IG + R + L
Sbjct: 1216 PSYYFEGIVEGSNVYTKDTVDDLDDKQGQGFMSADDLEDIDIGPGDRPRPTFISQNLSSE 1275
Query: 1014 VKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIK 1073
+ K+I+LL+E+ D FAW Y +MPGL IVEHR+PT+P P +Q RR DM +K
Sbjct: 1276 FRTKLIELLKEFRDCFAWQYYEMPGLSRSIVEHRLPTEPGVRPHQQPPRRCKADMLEPVK 1335
Query: 1074 NEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVL 1133
E++ DA F+ Y EWV++IVPV KK+GK R+C+DFRDLNKA+PKD +P+ D L
Sbjct: 1336 AEIKCLYDASFIRRCRYAEWVSSIVPVIKKNGKERVCIDFRDLNKATPKDEYPMPVADQL 1395
Query: 1134 VDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITP--WGTFCYKVMPIGLINAGATYQ 1191
VD + K+ FMDG GYNQI M+ ED KT+F P G F + VM GL +AGATYQ
Sbjct: 1396 VDAASGYKILSFMDGNVGYNQIFMAEEDIHKTAFRCPSAIGLFEWVVMTFGLKSAGATYQ 1455
Query: 1192 RGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
R M ++HD+I VEVY+DD++VKSK+ E H+ L K+FER R
Sbjct: 1456 RAMNYIYHDLISWLVEVYIDDVVVKSKEIEDHIADLRKVFERTR 1499
Score = 69.7 bits (169), Expect = 5e-10
Identities = 39/118 (33%), Positives = 65/118 (55%), Gaps = 1/118 (0%)
Query: 671 PEAGKYHNLA-LHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFLVK 729
PE + +L L+I+ +S ++VD G++LN+M +T +L L + ++K
Sbjct: 1075 PEGAENRHLKPLYINDYVNGKPMSKMMVDGGAALNLMLYATFRKLGRNAEDLIKTNMVLK 1134
Query: 730 AFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 787
F G+ G +++ +T+G +T TF V+D SYS LLGR W+H + ST+HQ
Sbjct: 1135 DFGGNPSETKGVLNVELTVGGKTIPTTFFVIDGKGSYSLLLGRDWVHANCCIPSTMHQ 1192
>UniRef100_Q8S5F3 Putative retroelement [Oryza sativa]
Length = 1627
Score = 233 bits (593), Expect = 4e-59
Identities = 124/250 (49%), Positives = 160/250 (63%), Gaps = 2/250 (0%)
Query: 988 EEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHR 1047
+++E I+IG + R + L + K+I+LL+EY D FAW Y +MPGL IVEHR
Sbjct: 914 DDLEEIDIGPGDRPRPTFISKNLSPEFRTKLIELLKEYRDCFAWEYYEMPGLSRSIVEHR 973
Query: 1048 IPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV 1107
+P K P +Q RR DM +K EV + DA F+ Y EWV+N+VPV KK+GKV
Sbjct: 974 LPIKTGVRPYQQPPRRCKADMLEAVKAEVARLYDADFIRPCRYAEWVSNMVPVIKKNGKV 1033
Query: 1108 RMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSF 1167
R+C+DFRDLNKA+ KD +P+ D LVD + K+ FMDG +GYNQI M ED KT+F
Sbjct: 1034 RVCIDFRDLNKATLKDEYPMSVADQLVDAASGHKILNFMDGNAGYNQIFMVEEDIHKTAF 1093
Query: 1168 ITPW--GTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 1225
P G F + VM GL AGATYQR M +FHD+I VEVYVDD++VKSK+ E H+
Sbjct: 1094 RCPGAIGLFEWVVMTFGLKRAGATYQRAMNYIFHDLIGWLVEVYVDDVVVKSKEIENHIA 1153
Query: 1226 YLTKMFERLR 1235
L K+FER R
Sbjct: 1154 NLRKVFERTR 1163
Score = 71.2 bits (173), Expect = 2e-10
Identities = 43/142 (30%), Positives = 76/142 (53%), Gaps = 1/142 (0%)
Query: 669 ELPEAGKYHNL-ALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFL 727
E PE + +L +L+I+ +S ++VD G+++N+MP +T +L L + +
Sbjct: 714 EKPEGTENRHLKSLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGGNSEDLIKTNMV 773
Query: 728 VKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 787
+K F G+ G +++ +T+ +T TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 774 LKDFGGNPSENKGVLNVELTVDSKTVPTTFFVIDRKGSYSLLLGRDWIHANCCIPSTMHQ 833
Query: 788 NFKFVKNGKLVTVHGEEAYLVS 809
+ + K+ V + + V+
Sbjct: 834 SLIQWQGDKIEIVPADRSVNVA 855
>UniRef100_Q8LM74 Putative retroelement [Oryza sativa]
Length = 1781
Score = 233 bits (593), Expect = 4e-59
Identities = 124/250 (49%), Positives = 160/250 (63%), Gaps = 2/250 (0%)
Query: 988 EEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHR 1047
+++E I+IG + R + L + K+I+LL+EY D FAW Y +MPGL IVEHR
Sbjct: 895 DDLEEIDIGPGDRPRPTFISKNLSPEFRTKLIELLKEYRDCFAWEYYEMPGLSRSIVEHR 954
Query: 1048 IPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKV 1107
+P K P +Q RR DM +K EV + DA F+ Y EWV+N+VPV KK+GKV
Sbjct: 955 LPIKTGVRPYQQPPRRCKADMLEAVKAEVARLYDADFIRPCRYAEWVSNMVPVIKKNGKV 1014
Query: 1108 RMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSF 1167
R+C+DFRDLNKA+ KD +P+ D LVD + K+ FMDG +GYNQI M ED KT+F
Sbjct: 1015 RVCIDFRDLNKATLKDEYPMSVADQLVDAASGHKILNFMDGNAGYNQIFMVEEDIHKTAF 1074
Query: 1168 ITPW--GTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVE 1225
P G F + VM GL AGATYQR M +FHD+I VEVYVDD++VKSK+ E H+
Sbjct: 1075 RCPGAIGLFEWVVMTFGLKRAGATYQRAMNYIFHDLIGWLVEVYVDDVVVKSKEIENHIA 1134
Query: 1226 YLTKMFERLR 1235
L K+FER R
Sbjct: 1135 NLRKVFERTR 1144
Score = 72.4 bits (176), Expect = 8e-11
Identities = 52/194 (26%), Positives = 96/194 (48%), Gaps = 11/194 (5%)
Query: 669 ELPEAGKYHNL-ALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFL 727
E PE + +L +L+I+ +S ++VD G+++N+MP +T +L L + +
Sbjct: 707 EKPEGTENRHLKSLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGGNSEDLIKTNMV 766
Query: 728 VKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 787
+K F G+ G +++ +T+ +T TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 767 LKDFGGNPSENKGVLNVELTVDSKTVPTTFFVIDRKGSYSLLLGRDWIHANCCIPSTMHQ 826
Query: 788 NFKFVKNGKLVTVHGEEAYLVS-------QLSSFSCIEA-GSAEGT-AFQGLTIEGMELK 838
+ + K+ V + + V+ ++ C+ G EG+ + T++ ++
Sbjct: 827 SLIQWQGDKIEIVPADRSVNVASADLALWEMDGIDCLSGKGVVEGSNVYTKDTVDDLD-D 885
Query: 839 KTGTAMASLKDAQK 852
K G S D ++
Sbjct: 886 KQGQGFMSADDLEE 899
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,123,667,915
Number of Sequences: 2790947
Number of extensions: 95734137
Number of successful extensions: 362123
Number of sequences better than 10.0: 20111
Number of HSP's better than 10.0 without gapping: 1962
Number of HSP's successfully gapped in prelim test: 18212
Number of HSP's that attempted gapping in prelim test: 351027
Number of HSP's gapped (non-prelim): 24302
length of query: 1235
length of database: 848,049,833
effective HSP length: 139
effective length of query: 1096
effective length of database: 460,108,200
effective search space: 504278587200
effective search space used: 504278587200
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 81 (35.8 bits)
Medicago: description of AC144656.13


