

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.5 - phase: 0
(254 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
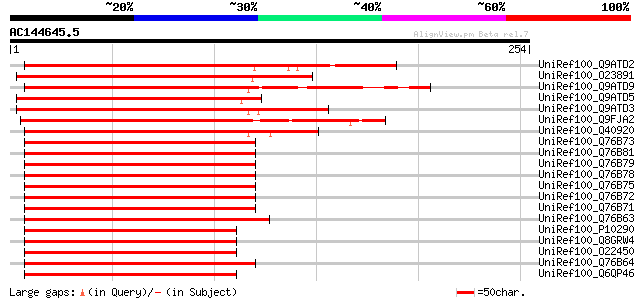
UniRef100_Q9ATD2 GHMYB38 [Gossypium hirsutum] 187 3e-46
UniRef100_O23891 OSMYB3 [Oryza sativa] 180 3e-44
UniRef100_Q9ATD9 BNLGHi233 [Gossypium hirsutum] 179 6e-44
UniRef100_Q9ATD5 GHMYB10 [Gossypium hirsutum] 178 1e-43
UniRef100_Q9ATD3 GHMYB36 [Gossypium hirsutum] 178 1e-43
UniRef100_Q9FJA2 TRANSPARENT TESTA 2 protein [Arabidopsis thaliana] 176 5e-43
UniRef100_Q40920 MYB-like transcriptional factor MBF1 [Picea mar... 175 1e-42
UniRef100_Q76B73 Myb protein [Oryza sativa] 172 7e-42
UniRef100_Q76B81 Myb protein [Oryza sativa] 172 7e-42
UniRef100_Q76B79 Myb protein [Oryza sativa] 172 7e-42
UniRef100_Q76B78 Myb protein [Oryza sativa] 172 7e-42
UniRef100_Q76B75 Myb protein [Oryza sativa] 172 7e-42
UniRef100_Q76B72 Myb protein [Oryza rufipogon] 172 7e-42
UniRef100_Q76B71 Myb protein [Oryza rufipogon] 172 7e-42
UniRef100_Q76B63 Myb protein [Oryza glumipatula] 172 1e-41
UniRef100_P10290 Anthocyanin regulatory C1 protein [Zea mays] 172 1e-41
UniRef100_Q8GRW4 PL transcription factor [Zea mays] 171 1e-41
UniRef100_O22450 PL transcription factor [Zea mays] 171 1e-41
UniRef100_Q76B64 Myb protein [Oryza glaberrima] 171 1e-41
UniRef100_Q6QP46 Anthocyanin biosynthesis regulatory protein Pl1... 171 1e-41
>UniRef100_Q9ATD2 GHMYB38 [Gossypium hirsutum]
Length = 254
Score = 187 bits (474), Expect = 3e-46
Identities = 98/185 (52%), Positives = 125/185 (66%), Gaps = 5/185 (2%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+NKGAW+ ED IL Y+ IHGEGKW+ + Q AGLKRCGKSCR RWLNYL+P IKRG+IS
Sbjct: 12 LNKGAWTANEDQILKNYITIHGEGKWRDLPQKAGLKRCGKSCRLRWLNYLRPDIKRGNIS 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTS-SSSLPSPS 126
+EE++IIRLH+LLGNRWSLIA RLPGRTDNEIKNYWNTNLSK+++ Q + S + P S
Sbjct: 72 IEEEELIIRLHKLLGNRWSLIAGRLPGRTDNEIKNYWNTNLSKRMEGQKRNISPNTPQVS 131
Query: 127 SVSLRHNHG-KCGH-VAPEAPKPRRLKAVHQYKILEKNSGSEYDQGSDETSIADFFIDFD 184
S ++ KC V + P+ V+ E S S ++ + + DF +DFD
Sbjct: 132 SHNVIWTKAVKCTKAVNVDTLDPKSETLVNSPS--ETPSSSAINEDNTNNNSMDFLVDFD 189
Query: 185 HQDQL 189
+ L
Sbjct: 190 IDELL 194
>UniRef100_O23891 OSMYB3 [Oryza sativa]
Length = 321
Score = 180 bits (457), Expect = 3e-44
Identities = 85/146 (58%), Positives = 110/146 (75%), Gaps = 1/146 (0%)
Query: 4 SPKEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKR 63
S + +N+GAW+ EDDIL Y+ HGEGKW + + AGLKRCGKSCR RWLNYL+PGIKR
Sbjct: 8 SKEGLNRGAWTAMEDDILVSYIAKHGEGKWGALPKRAGLKRCGKSCRLRWLNYLRPGIKR 67
Query: 64 GHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPT-SSSSL 122
G+IS DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LSK++ Q T +++S+
Sbjct: 68 GNISGDEEELILRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSKRVAMQRTAAATSM 127
Query: 123 PSPSSVSLRHNHGKCGHVAPEAPKPR 148
P+ ++ S + +P+PR
Sbjct: 128 PAAATTSSNADAAGAAARRRRSPEPR 153
>UniRef100_Q9ATD9 BNLGHi233 [Gossypium hirsutum]
Length = 247
Score = 179 bits (454), Expect = 6e-44
Identities = 96/207 (46%), Positives = 126/207 (60%), Gaps = 28/207 (13%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +G W+ EED++LS Y+ GEG+W+ + + AGL RCGKSCR RW+NYL+P +KRG I+
Sbjct: 23 LKRGPWTPEEDELLSNYINKEGEGRWRTLPKRAGLLRCGKSCRLRWMNYLRPSVKRGQIA 82
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ--------PTSS 119
DEED+I+RLHRLLGNRWSLIA R+PGRTDNEIKNYWNT+LSKKL Q P +
Sbjct: 83 PDEEDLILRLHRLLGNRWSLIAGRIPGRTDNEIKNYWNTHLSKKLISQGIDPRTHKPLNP 142
Query: 120 SSLPSPSSVSLRHNHGKCGHVAPEAPKPRRLKAVHQYKILEKNSGSEYDQGSDETSIADF 179
L SPS SL+ + +A KP + ++ N + YD GS+E
Sbjct: 143 QQL-SPSPPSLKPSPSSSSSMA----KPNNPPSPLPVHVVNANKQNYYDDGSNE------ 191
Query: 180 FIDFDHQDQLMVGDDESNSKIPQMEDH 206
DHQ +M +N Q +DH
Sbjct: 192 ----DHQGMIM-----NNDHYQQQQDH 209
>UniRef100_Q9ATD5 GHMYB10 [Gossypium hirsutum]
Length = 302
Score = 178 bits (451), Expect = 1e-43
Identities = 81/123 (65%), Positives = 100/123 (80%), Gaps = 3/123 (2%)
Query: 4 SPKEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKR 63
S + +N+GAW+ ED IL Y+ +HGEG+W+ + + AGLKRCGKSCR RWLNYL+P IKR
Sbjct: 8 SKEGLNRGAWTALEDKILKDYIKVHGEGRWRNLPKRAGLKRCGKSCRLRWLNYLRPDIKR 67
Query: 64 GHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL---QKQPTSSS 120
G+IS DEE++II+LH+LLGNRWSLIA RLPGRTDNEIKNYWNTNLSK++ QK P + S
Sbjct: 68 GNISPDEEELIIKLHKLLGNRWSLIAGRLPGRTDNEIKNYWNTNLSKRVSDRQKSPAAPS 127
Query: 121 SLP 123
P
Sbjct: 128 KKP 130
>UniRef100_Q9ATD3 GHMYB36 [Gossypium hirsutum]
Length = 271
Score = 178 bits (451), Expect = 1e-43
Identities = 90/162 (55%), Positives = 111/162 (67%), Gaps = 9/162 (5%)
Query: 4 SPKEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKR 63
S + +NKGAW+ ED IL+ Y+ +HGEGKW+ + + AGLKRCGKSCR RWLNYL+P IKR
Sbjct: 8 SKEGLNKGAWTALEDKILASYIHVHGEGKWRNLPKRAGLKRCGKSCRLRWLNYLRPDIKR 67
Query: 64 GHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ-------- 115
G+IS DEE++IIRLH LLGNRWSLIA RLPGRTDNEIKNYWNT L K+ + Q
Sbjct: 68 GNISHDEEELIIRLHNLLGNRWSLIAGRLPGRTDNEIKNYWNTTLGKRAKAQASIDAKTI 127
Query: 116 PTSSS-SLPSPSSVSLRHNHGKCGHVAPEAPKPRRLKAVHQY 156
PT S + PS SS + K + + P + A HQ+
Sbjct: 128 PTESRLNEPSKSSTKIEVIRTKAIRCSSKVMVPLQPPATHQH 169
>UniRef100_Q9FJA2 TRANSPARENT TESTA 2 protein [Arabidopsis thaliana]
Length = 258
Score = 176 bits (446), Expect = 5e-43
Identities = 93/186 (50%), Positives = 118/186 (63%), Gaps = 12/186 (6%)
Query: 6 KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGH 65
+E+N+GAW+ ED IL Y+ HGEGKW + AGLKRCGKSCR RW NYL+PGIKRG+
Sbjct: 12 EELNRGAWTDHEDKILRDYITTHGEGKWSTLPNQAGLKRCGKSCRLRWKNYLRPGIKRGN 71
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSP 125
IS+DEE++IIRLH LLGNRWSLIA RLPGRTDNEIKN+WN+NL K+L K T P
Sbjct: 72 ISSDEEELIIRLHNLLGNRWSLIAGRLPGRTDNEIKNHWNSNLRKRLPKTQTKQ---PKR 128
Query: 126 SSVSLRHNHGKCGHVAPEAPKPRRLKAVHQYKILEKNSGS-------EYDQGSDETSIAD 178
S + + C + +A + + + +K+S S E DQG + + D
Sbjct: 129 IKHSTNNENNVC-VIRTKAIRCSKTLLFSDLSLQKKSSTSPLPLKEQEMDQGG-SSLMGD 186
Query: 179 FFIDFD 184
DFD
Sbjct: 187 LEFDFD 192
>UniRef100_Q40920 MYB-like transcriptional factor MBF1 [Picea mariana]
Length = 388
Score = 175 bits (443), Expect = 1e-42
Identities = 85/149 (57%), Positives = 109/149 (73%), Gaps = 5/149 (3%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+NKGAWS EED +L KY+ HGEG W+ + + AGL+RCGKSCR RWLNYL+P IKRG+I+
Sbjct: 12 LNKGAWSAEEDSLLGKYIQTHGEGNWRSLPKKAGLRRCGKSCRLRWLNYLRPCIKRGNIT 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ---PTSSSSLPS 124
DEE++IIR+H LLGNRWS+IA R+PGRTDNEIKNYWNTNLSKKL + P + + +
Sbjct: 72 ADEEELIIRMHALLGNRWSIIAGRVPGRTDNEIKNYWNTNLSKKLAVRGIDPKTHKKVTT 131
Query: 125 PS--SVSLRHNHGKCGHVAPEAPKPRRLK 151
S S R N K G + + + +RL+
Sbjct: 132 DSINRASDRFNQRKGGKLYDCSQRSQRLE 160
>UniRef100_Q76B73 Myb protein [Oryza sativa]
Length = 272
Score = 172 bits (436), Expect = 7e-42
Identities = 74/113 (65%), Positives = 95/113 (83%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +GAW+ +EDD+L+ Y+ HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+I
Sbjct: 12 MKRGAWTSKEDDVLASYIKSHGEGKWREVRQRAGLRRCGKSCRLRWLNYLRPNIKRGNID 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSS 120
DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LS+K+ T+++
Sbjct: 72 DDEEELIVRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSRKIGTAATAAA 124
>UniRef100_Q76B81 Myb protein [Oryza sativa]
Length = 272
Score = 172 bits (436), Expect = 7e-42
Identities = 74/113 (65%), Positives = 95/113 (83%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +GAW+ +EDD+L+ Y+ HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+I
Sbjct: 12 MKRGAWTSKEDDVLASYIKSHGEGKWREVPQRAGLRRCGKSCRLRWLNYLRPNIKRGNID 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSS 120
DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LS+K+ T+++
Sbjct: 72 DDEEELIVRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSRKIGTAATAAA 124
>UniRef100_Q76B79 Myb protein [Oryza sativa]
Length = 272
Score = 172 bits (436), Expect = 7e-42
Identities = 74/113 (65%), Positives = 95/113 (83%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +GAW+ +EDD+L+ Y+ HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+I
Sbjct: 12 MKRGAWTSKEDDVLASYIKSHGEGKWREVPQRAGLRRCGKSCRLRWLNYLRPNIKRGNID 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSS 120
DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LS+K+ T+++
Sbjct: 72 DDEEELIVRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSRKIGTAATAAA 124
>UniRef100_Q76B78 Myb protein [Oryza sativa]
Length = 198
Score = 172 bits (436), Expect = 7e-42
Identities = 74/113 (65%), Positives = 95/113 (83%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +GAW+ +EDD+L+ Y+ HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+I
Sbjct: 12 MKRGAWTSKEDDVLASYIKSHGEGKWREVPQRAGLRRCGKSCRLRWLNYLRPNIKRGNID 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSS 120
DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LS+K+ T+++
Sbjct: 72 DDEEELIVRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSRKIGTAATAAA 124
>UniRef100_Q76B75 Myb protein [Oryza sativa]
Length = 272
Score = 172 bits (436), Expect = 7e-42
Identities = 74/113 (65%), Positives = 95/113 (83%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +GAW+ +EDD+L+ Y+ HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+I
Sbjct: 12 MKRGAWTSKEDDVLASYIKSHGEGKWREVPQRAGLRRCGKSCRLRWLNYLRPNIKRGNID 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSS 120
DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LS+K+ T+++
Sbjct: 72 DDEEELIVRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSRKIGTAATAAA 124
>UniRef100_Q76B72 Myb protein [Oryza rufipogon]
Length = 272
Score = 172 bits (436), Expect = 7e-42
Identities = 74/113 (65%), Positives = 95/113 (83%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +GAW+ +EDD+L+ Y+ HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+I
Sbjct: 12 MKRGAWTSKEDDVLASYIKSHGEGKWREVPQRAGLRRCGKSCRLRWLNYLRPNIKRGNID 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSS 120
DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LS+K+ T+++
Sbjct: 72 DDEEELIVRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSRKIGTAATAAA 124
>UniRef100_Q76B71 Myb protein [Oryza rufipogon]
Length = 272
Score = 172 bits (436), Expect = 7e-42
Identities = 74/113 (65%), Positives = 95/113 (83%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +GAW+ +EDD+L+ Y+ HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+I
Sbjct: 12 MKRGAWTSKEDDVLASYIKSHGEGKWREVPQRAGLRRCGKSCRLRWLNYLRPNIKRGNID 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSS 120
DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LS+K+ T+++
Sbjct: 72 DDEEELIVRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSRKIGTAATAAA 124
>UniRef100_Q76B63 Myb protein [Oryza glumipatula]
Length = 269
Score = 172 bits (435), Expect = 1e-41
Identities = 75/120 (62%), Positives = 95/120 (78%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +GAW+ +EDD+L+ Y+ HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+I
Sbjct: 12 MKRGAWTSKEDDVLASYIKSHGEGKWREVPQRAGLRRCGKSCRLRWLNYLRPNIKRGNID 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSPSS 127
DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LS+K+ S +P +
Sbjct: 72 DDEEELIVRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSRKIGTAAAGSRGGSTPDT 131
>UniRef100_P10290 Anthocyanin regulatory C1 protein [Zea mays]
Length = 273
Score = 172 bits (435), Expect = 1e-41
Identities = 76/104 (73%), Positives = 90/104 (86%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
V +GAW+ +EDD L+ YV HGEGKW++V Q AGL+RCGKSCR RWLNYL+P I+RG+IS
Sbjct: 12 VKRGAWTSKEDDALAAYVKAHGEGKWREVPQKAGLRRCGKSCRLRWLNYLRPNIRRGNIS 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKK 111
DEED+IIRLHRLLGNRWSLIA RLPGRTDNEIKNYWN+ L ++
Sbjct: 72 YDEEDLIIRLHRLLGNRWSLIAGRLPGRTDNEIKNYWNSTLGRR 115
>UniRef100_Q8GRW4 PL transcription factor [Zea mays]
Length = 271
Score = 171 bits (434), Expect = 1e-41
Identities = 75/104 (72%), Positives = 90/104 (86%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
V +GAW+ +EDD L+ YV HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+IS
Sbjct: 12 VKRGAWTAKEDDTLAAYVKAHGEGKWREVPQKAGLRRCGKSCRLRWLNYLRPNIKRGNIS 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKK 111
DEED+I+RLH+LLGNRWSLIA RLPGRTDNEIKNYWN+ L ++
Sbjct: 72 YDEEDLIVRLHKLLGNRWSLIAGRLPGRTDNEIKNYWNSTLGRR 115
>UniRef100_O22450 PL transcription factor [Zea mays]
Length = 267
Score = 171 bits (434), Expect = 1e-41
Identities = 75/104 (72%), Positives = 90/104 (86%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
V +GAW+ +EDD L+ YV HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+IS
Sbjct: 12 VKRGAWTAKEDDTLAAYVKAHGEGKWREVPQKAGLRRCGKSCRLRWLNYLRPNIKRGNIS 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKK 111
DEED+I+RLH+LLGNRWSLIA RLPGRTDNEIKNYWN+ L ++
Sbjct: 72 YDEEDLIVRLHKLLGNRWSLIAGRLPGRTDNEIKNYWNSTLGRR 115
>UniRef100_Q76B64 Myb protein [Oryza glaberrima]
Length = 270
Score = 171 bits (434), Expect = 1e-41
Identities = 74/113 (65%), Positives = 95/113 (83%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +GAW+ +EDD+L+ Y+ HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+I
Sbjct: 12 MKRGAWTSKEDDMLASYIKSHGEGKWREVPQRAGLRRCGKSCRLRWLNYLRPNIKRGNID 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSS 120
DEE++I+RLH LLGNRWSLIA RLPGRTDNEIKNYWN+ LS+K+ T+++
Sbjct: 72 DDEEELIVRLHTLLGNRWSLIAGRLPGRTDNEIKNYWNSTLSRKIGTAATAAA 124
>UniRef100_Q6QP46 Anthocyanin biosynthesis regulatory protein Pl1_B73 [Zea mays]
Length = 273
Score = 171 bits (434), Expect = 1e-41
Identities = 75/104 (72%), Positives = 90/104 (86%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
V +GAW+ +EDD L+ YV HGEGKW++V Q AGL+RCGKSCR RWLNYL+P IKRG+IS
Sbjct: 12 VKRGAWTAKEDDTLAAYVKAHGEGKWREVPQKAGLRRCGKSCRLRWLNYLRPNIKRGNIS 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKK 111
DEED+I+RLH+LLGNRWSLIA RLPGRTDNEIKNYWN+ L ++
Sbjct: 72 YDEEDLIVRLHKLLGNRWSLIAGRLPGRTDNEIKNYWNSTLGRR 115
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.131 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 469,522,223
Number of Sequences: 2790947
Number of extensions: 20313225
Number of successful extensions: 50842
Number of sequences better than 10.0: 1337
Number of HSP's better than 10.0 without gapping: 1210
Number of HSP's successfully gapped in prelim test: 128
Number of HSP's that attempted gapping in prelim test: 48350
Number of HSP's gapped (non-prelim): 1801
length of query: 254
length of database: 848,049,833
effective HSP length: 124
effective length of query: 130
effective length of database: 501,972,405
effective search space: 65256412650
effective search space used: 65256412650
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC144645.5


