

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144644.6 + phase: 0
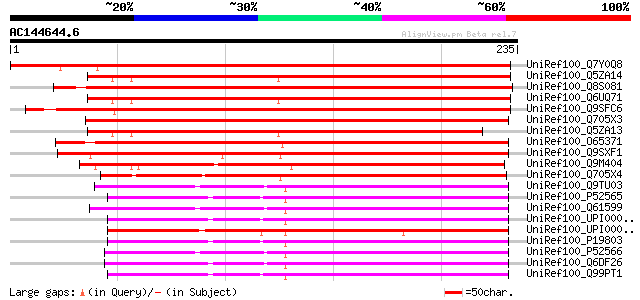
(235 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7Y0Q8 Rac GDP-dissociation inhibitor 1 [Oryza sativa] 275 7e-73
UniRef100_Q5ZA14 Putative Rho GDP dissociation inhibitor 2 [Oryz... 273 2e-72
UniRef100_Q8S081 Putative Rho GDP-dissociation inhibitor [Oryza ... 271 8e-72
UniRef100_Q6UQ71 Rho GDP dissociation inhibitor 2 [Oryza sativa] 270 2e-71
UniRef100_Q9SFC6 Rho GDP-dissociation inhibitor 1 [Arabidopsis t... 265 7e-70
UniRef100_Q705X3 Rho GDP dissociation inhibitor 2 [Medicago trun... 263 3e-69
UniRef100_Q5ZA13 Putative Rho GDP dissociation inhibitor 2 [Oryz... 238 1e-61
UniRef100_O65371 F12F1.5 protein [Arabidopsis thaliana] 237 2e-61
UniRef100_Q9SXF1 T3P18.2 [Arabidopsis thaliana] 231 2e-59
UniRef100_Q9M404 Putative Rho GDP dissociation inhibitor [Nicoti... 198 8e-50
UniRef100_Q705X4 Rho GDP dissociation inhibitor 1 [Medicago trun... 194 2e-48
UniRef100_Q9TU03 Rho GDP-dissociation inhibitor 2 [Bos taurus] 144 2e-33
UniRef100_P52565 Rho GDP-dissociation inhibitor 1 [Homo sapiens] 144 2e-33
UniRef100_Q61599 Rho GDP-dissociation inhibitor 2 [Mus musculus] 142 5e-33
UniRef100_UPI00003AF5BE UPI00003AF5BE UniRef100 entry 142 7e-33
UniRef100_UPI000025D5CD UPI000025D5CD UniRef100 entry 142 7e-33
UniRef100_P19803 Rho GDP-dissociation inhibitor 1 [Bos taurus] 142 7e-33
UniRef100_P52566 Rho GDP-dissociation inhibitor 2 [Homo sapiens] 142 9e-33
UniRef100_Q6DF26 Rho GDP dissociation inhibitor (GDI) beta [Xeno... 141 2e-32
UniRef100_Q99PT1 Rho GDP-dissociation inhibitor 1 [Mus musculus] 140 2e-32
>UniRef100_Q7Y0Q8 Rac GDP-dissociation inhibitor 1 [Oryza sativa]
Length = 254
Score = 275 bits (703), Expect = 7e-73
Identities = 138/251 (54%), Positives = 180/251 (70%), Gaps = 19/251 (7%)
Query: 1 MSAAVGAVSTTKDVSFNAHLEE--------------EHKNKGTNKVGVNYDEA-----EE 41
MS+AV A+S +K ++ E EH G EA E
Sbjct: 1 MSSAVDAISCSKGIAAPPTEEATKERVAVVGKNGGVEHGGDAATMNGKQCGEAPHCRKES 60
Query: 42 IVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEV 101
+ED+EE+ +DLGP+ S+K+QLEKDKDDESLR+WKEQLLG+VD+++VGE +EP+V
Sbjct: 61 NEEEEDDEEKAPKAIDLGPRVSIKDQLEKDKDDESLRRWKEQLLGSVDLNSVGETLEPDV 120
Query: 102 KIVSLTIICQGRPDLILPIPFTADSKKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVV 161
KI+SL I+ GRPD+ LP+P ++K FTLKEGS Y+LKF+F+VSNNIVSGLRYTN V
Sbjct: 121 KIMSLAILSPGRPDIFLPLPVEPNAKGVWFTLKEGSLYKLKFTFSVSNNIVSGLRYTNAV 180
Query: 162 WKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDA 221
WKTG++V++ K+MLGT+SPQ EPYTY EE TPSG+FARG+YSARTKF+DDDRKCYL+
Sbjct: 181 WKTGIKVDSHKEMLGTFSPQPEPYTYVTPEETTPSGMFARGSYSARTKFLDDDRKCYLEI 240
Query: 222 SYRFEIQKNWP 232
+Y F+I++ WP
Sbjct: 241 NYTFDIRREWP 251
>UniRef100_Q5ZA14 Putative Rho GDP dissociation inhibitor 2 [Oryza sativa]
Length = 270
Score = 273 bits (699), Expect = 2e-72
Identities = 131/202 (64%), Positives = 170/202 (83%), Gaps = 6/202 (2%)
Query: 37 DEAEEIVPDE---DEEEEPKLE--LDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVS 91
+E EE+V DE DEE+E K+ +DLGP+ S+KEQLE DK+DESLR+WKEQLLG+VD++
Sbjct: 66 EEEEEVVEDEEDDDEEDEGKVAEAIDLGPRVSIKEQLEMDKEDESLRRWKEQLLGSVDLN 125
Query: 92 AVGEKIEPEVKIVSLTIICQGRPDLILPIPFT-ADSKKSMFTLKEGSQYRLKFSFTVSNN 150
+VGE +EP+V+I SL I+ GRPD++LP+P ++SK+ FTLKEGS YRLKF+F+VS+N
Sbjct: 126 SVGESLEPDVRITSLCILSPGRPDVLLPLPVEPSNSKEPWFTLKEGSTYRLKFTFSVSSN 185
Query: 151 IVSGLRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKF 210
IVSGLRYTN VWK G+RV+ TK+MLGT+SPQ EPYTY EE TPSG+FARG+YSA+TKF
Sbjct: 186 IVSGLRYTNTVWKAGIRVDKTKEMLGTFSPQLEPYTYVTPEETTPSGVFARGSYSAKTKF 245
Query: 211 VDDDRKCYLDASYRFEIQKNWP 232
VDDDRKCYL+ +Y F+I+++WP
Sbjct: 246 VDDDRKCYLEINYTFDIRRDWP 267
>UniRef100_Q8S081 Putative Rho GDP-dissociation inhibitor [Oryza sativa]
Length = 217
Score = 271 bits (694), Expect = 8e-72
Identities = 132/213 (61%), Positives = 160/213 (74%), Gaps = 4/213 (1%)
Query: 21 EEEHKNKGTNKVGVNYDEAEEIVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKW 80
E+EH+ + + + ++ +E++EE K + LGPQ LKEQLE DKDDESLR+W
Sbjct: 7 EKEHEQTASGR----NPDVNDVEEEEEDEEGNKRAVVLGPQVPLKEQLELDKDDESLRRW 62
Query: 81 KEQLLGNVDVSAVGEKIEPEVKIVSLTIICQGRPDLILPIPFTADSKKSMFTLKEGSQYR 140
KEQLLG VD +GE EPEVK+++LTI+ RPDL+LPIPF D K F LK+GS Y
Sbjct: 63 KEQLLGQVDTEQLGETAEPEVKVLNLTILSPDRPDLVLPIPFVPDEKGYAFALKDGSTYS 122
Query: 141 LKFSFTVSNNIVSGLRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFA 200
+FSF VSNNIVSGL+YTN VWKTGVRVEN K MLGT+SPQ EPYTYE EEE TP+G+FA
Sbjct: 123 FRFSFIVSNNIVSGLKYTNTVWKTGVRVENQKVMLGTFSPQLEPYTYEGEEETTPAGMFA 182
Query: 201 RGTYSARTKFVDDDRKCYLDASYRFEIQKNWPT 233
RG+YSA+ KFVDDD KCYL+ SY FEI+K WPT
Sbjct: 183 RGSYSAKLKFVDDDGKCYLEMSYYFEIRKEWPT 215
>UniRef100_Q6UQ71 Rho GDP dissociation inhibitor 2 [Oryza sativa]
Length = 264
Score = 270 bits (691), Expect = 2e-71
Identities = 130/202 (64%), Positives = 169/202 (83%), Gaps = 6/202 (2%)
Query: 37 DEAEEIVPDE---DEEEEPKLE--LDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVS 91
+E EE+V DE DEE+E K+ +DLGP+ S+KEQLE DK+DESLR+WKEQLLG+VD++
Sbjct: 60 EEEEEVVEDEEDDDEEDEGKVAEAIDLGPRVSIKEQLEMDKEDESLRRWKEQLLGSVDLN 119
Query: 92 AVGEKIEPEVKIVSLTIICQGRPDLILPIPFT-ADSKKSMFTLKEGSQYRLKFSFTVSNN 150
+VGE +EP+V+I SL I+ GR D++LP+P ++SK+ FTLKEGS YRLKF+F+VS+N
Sbjct: 120 SVGESLEPDVRITSLCILSPGRTDVLLPLPVEPSNSKEPWFTLKEGSTYRLKFTFSVSSN 179
Query: 151 IVSGLRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKF 210
IVSGLRYTN VWK G+RV+ TK+MLGT+SPQ EPYTY EE TPSG+FARG+YSA+TKF
Sbjct: 180 IVSGLRYTNTVWKAGIRVDKTKEMLGTFSPQLEPYTYVTPEETTPSGVFARGSYSAKTKF 239
Query: 211 VDDDRKCYLDASYRFEIQKNWP 232
VDDDRKCYL+ +Y F+I+++WP
Sbjct: 240 VDDDRKCYLEINYTFDIRRDWP 261
>UniRef100_Q9SFC6 Rho GDP-dissociation inhibitor 1 [Arabidopsis thaliana]
Length = 240
Score = 265 bits (677), Expect = 7e-70
Identities = 131/240 (54%), Positives = 171/240 (70%), Gaps = 20/240 (8%)
Query: 8 VSTTKDVSFNAHLEEEHKNKGTNKVGVNYDEAEEIVPDED---------------EEEEP 52
VS +D+ F +++ N NK G + + + D+D EEE+
Sbjct: 4 VSGARDMGF-----DDNNNNKNNKDGDDENSSSRTRADDDALSRQMSESSLCATEEEEDD 58
Query: 53 KLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVSLTIICQG 112
+L LGPQ+++KE LEKDKDDESLRKWKEQLLG+VDV+ +GE ++PEV+I SL II G
Sbjct: 59 DSKLQLGPQYTIKEHLEKDKDDESLRKWKEQLLGSVDVTNIGETLDPEVRIDSLAIISPG 118
Query: 113 RPDLILPIPFTADSKKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVWKTGVRVENTK 172
RPD++L +P + K FTLKEGS+Y LKF+F V+NNIVSGLRYTN VWKTGV+V+ K
Sbjct: 119 RPDIVLLVPENGNPKGMWFTLKEGSKYNLKFTFHVNNNIVSGLRYTNTVWKTGVKVDRAK 178
Query: 173 KMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDASYRFEIQKNWP 232
+MLGT+SPQ EPY + + EE TPSG+FARG+YSARTKF+DDD KCYL+ +Y F+I+K WP
Sbjct: 179 EMLGTFSPQLEPYNHVMPEETTPSGMFARGSYSARTKFLDDDNKCYLEINYSFDIRKEWP 238
>UniRef100_Q705X3 Rho GDP dissociation inhibitor 2 [Medicago truncatula]
Length = 233
Score = 263 bits (672), Expect = 3e-69
Identities = 120/196 (61%), Positives = 160/196 (81%)
Query: 36 YDEAEEIVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGE 95
Y + E+E+++ +++LGPQ +LKEQLEKDKDDESLR+WKEQLLG+VD++AVGE
Sbjct: 37 YMSESSVAATEEEDDDEDRKIELGPQCTLKEQLEKDKDDESLRRWKEQLLGSVDINAVGE 96
Query: 96 KIEPEVKIVSLTIICQGRPDLILPIPFTADSKKSMFTLKEGSQYRLKFSFTVSNNIVSGL 155
+EPEVKI+SL I RPD++LPIP + K FTLKEGS+YRL F+F V++NIVSGL
Sbjct: 97 TLEPEVKILSLAIKAADRPDIVLPIPEDGNPKGLWFTLKEGSKYRLMFTFQVNHNIVSGL 156
Query: 156 RYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDR 215
+YTN VWKTG++V+++K+M+GT+SPQ E YT+E+ EE TPSG+FARG YSAR+KFVDDD
Sbjct: 157 KYTNTVWKTGIKVDSSKEMIGTFSPQTETYTHEMPEETTPSGMFARGAYSARSKFVDDDN 216
Query: 216 KCYLDASYRFEIQKNW 231
KCYL+ +Y F+I+K+W
Sbjct: 217 KCYLEINYTFDIRKDW 232
>UniRef100_Q5ZA13 Putative Rho GDP dissociation inhibitor 2 [Oryza sativa]
Length = 254
Score = 238 bits (606), Expect = 1e-61
Identities = 118/189 (62%), Positives = 153/189 (80%), Gaps = 6/189 (3%)
Query: 37 DEAEEIVPDE---DEEEEPKLE--LDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVS 91
+E EE+V DE DEE+E K+ +DLGP+ S+KEQLE DK+DESLR+WKEQLLG+VD++
Sbjct: 66 EEEEEVVEDEEDDDEEDEGKVAEAIDLGPRVSIKEQLEMDKEDESLRRWKEQLLGSVDLN 125
Query: 92 AVGEKIEPEVKIVSLTIICQGRPDLILPIPFT-ADSKKSMFTLKEGSQYRLKFSFTVSNN 150
+VGE +EP+V+I SL I+ GRPD++LP+P ++SK+ FTLKEGS YRLKF+F+VS+N
Sbjct: 126 SVGESLEPDVRITSLCILSPGRPDVLLPLPVEPSNSKEPWFTLKEGSTYRLKFTFSVSSN 185
Query: 151 IVSGLRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKF 210
IVSGLRYTN VWK G+RV+ TK+MLGT+SPQ EPYTY EE TPSG+FARG+YSA+TK
Sbjct: 186 IVSGLRYTNTVWKAGIRVDKTKEMLGTFSPQLEPYTYVTPEETTPSGVFARGSYSAKTKV 245
Query: 211 VDDDRKCYL 219
+ D +L
Sbjct: 246 MKSDTVIHL 254
>UniRef100_O65371 F12F1.5 protein [Arabidopsis thaliana]
Length = 223
Score = 237 bits (605), Expect = 2e-61
Identities = 118/212 (55%), Positives = 156/212 (72%), Gaps = 6/212 (2%)
Query: 22 EEHKNKGTNKVGVNYDEAEEIVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWK 81
EE K +G ++ + + P +D+EEE +L+LGP +LKEQLEKDKDDESLR+WK
Sbjct: 15 EEPKGQGLSRKNSH----SSMCPTDDDEEEEDKKLELGPMIALKEQLEKDKDDESLRRWK 70
Query: 82 EQLLGNVDVSAVGEKIEPEVKIVSLTIICQGRPDLILPIPFTAD--SKKSMFTLKEGSQY 139
EQLLG+VD+ VGE +P VKI++LTI R D++L IP SK FTLKEGS+Y
Sbjct: 71 EQLLGSVDLEEVGETPDPLVKILTLTIRSPDREDMVLTIPENGKPASKGPWFTLKEGSKY 130
Query: 140 RLKFSFTVSNNIVSGLRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLF 199
L F+F V+NNIVSGL+Y+N VWKTG++V + K+MLGT+SPQ EPYT+ + EE PSGL
Sbjct: 131 TLIFTFRVTNNIVSGLQYSNTVWKTGIKVYSRKEMLGTFSPQAEPYTHVMFEETAPSGLL 190
Query: 200 ARGTYSARTKFVDDDRKCYLDASYRFEIQKNW 231
RG+YS ++KFVDDD +CYL+ +Y F+I+KNW
Sbjct: 191 VRGSYSVKSKFVDDDNQCYLENNYTFDIRKNW 222
>UniRef100_Q9SXF1 T3P18.2 [Arabidopsis thaliana]
Length = 236
Score = 231 bits (588), Expect = 2e-59
Identities = 118/225 (52%), Positives = 158/225 (69%), Gaps = 16/225 (7%)
Query: 23 EHKNKGTNKVGVNY-DEAEEIVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWK 81
E + + K+G++ + + P ED+EE+ +L+LGP +LKEQLE+DKDDESLR+WK
Sbjct: 11 EASGETSEKMGLSRKNSGSSLSPTEDDEEDEDKKLELGPMIALKEQLERDKDDESLRRWK 70
Query: 82 EQLLGNVDVSAVGEKI-------------EPEVKIVSLTIICQGRPDLILPIPFTA--DS 126
EQLLG VD+ VG K +P VKI+ LTI R +++L IP +
Sbjct: 71 EQLLGVVDLEDVGGKPNSLLDLDSMVQTPDPVVKILDLTIRSPDREEMVLTIPEDGLPNP 130
Query: 127 KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVWKTGVRVENTKKMLGTYSPQQEPYT 186
K FT+KEGS+Y L F+F V+NNIVSGLRY N VWKTGV+V++TK MLGT+SPQ E Y
Sbjct: 131 KGPWFTIKEGSKYTLVFNFRVTNNIVSGLRYNNTVWKTGVKVDSTKAMLGTFSPQAESYQ 190
Query: 187 YELEEEITPSGLFARGTYSARTKFVDDDRKCYLDASYRFEIQKNW 231
+ + EE+TPSG+FARG+YSARTKF+DDD KCYL+ +Y F+I+K+W
Sbjct: 191 HVMPEEMTPSGMFARGSYSARTKFIDDDNKCYLEINYTFDIRKSW 235
>UniRef100_Q9M404 Putative Rho GDP dissociation inhibitor [Nicotiana tabacum]
Length = 226
Score = 198 bits (504), Expect = 8e-50
Identities = 105/210 (50%), Positives = 146/210 (69%), Gaps = 14/210 (6%)
Query: 33 GVNYDE---AEEIVPDEDEEEEPKLE--LDL-------GPQFSLKEQLEKDKDDESLRKW 80
G+N+ E EE+ + EE+E + D+ GP SLK+Q+EKDK+DESLR+W
Sbjct: 16 GLNFSEDKRKEELEEGDSEEDEHNQDSATDIITTSYVPGPLLSLKDQIEKDKEDESLRRW 75
Query: 81 KEQLLGNVDVSAVGEKIEPEVKIVSLTIICQGRPDLILPIPFTADSKKS-MFTLKEGSQY 139
KE+LLG ++ G+ +EPEVK S+ I+ ++ P+P KS +FTL+EGS+Y
Sbjct: 76 KEKLLGCLESDLNGQ-MEPEVKFHSVGILSSDFEEINTPLPVKEGQSKSVLFTLREGSEY 134
Query: 140 RLKFSFTVSNNIVSGLRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLF 199
RLK +F+V +NIVSGL YTN VWK G++V+ +K MLGT++PQ+EPY + LEEE TPSG
Sbjct: 135 RLKLTFSVLHNIVSGLAYTNTVWKAGLQVDQSKGMLGTFAPQREPYIHMLEEETTPSGAL 194
Query: 200 ARGTYSARTKFVDDDRKCYLDASYRFEIQK 229
ARGTY+A+ KFVDDD++C+L +Y FEI K
Sbjct: 195 ARGTYTAKLKFVDDDKRCHLVLNYSFEISK 224
>UniRef100_Q705X4 Rho GDP dissociation inhibitor 1 [Medicago truncatula]
Length = 219
Score = 194 bits (492), Expect = 2e-48
Identities = 92/189 (48%), Positives = 142/189 (74%), Gaps = 3/189 (1%)
Query: 43 VPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVK 102
V EDEE+E ++ + P SLKEQ+EKDK+DESLR+WKE+LLG+++ S + +++EPEVK
Sbjct: 32 VASEDEEDEGQVFVP-SPLLSLKEQIEKDKEDESLRRWKEKLLGSLE-SDLDDQLEPEVK 89
Query: 103 IVSLTIICQGRPDLILPIPFTA-DSKKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVV 161
S+ I+ + +++ P+P + + +FTL+EGS+Y+LK F+V +N+VSGL Y+N V
Sbjct: 90 FHSIGILSEDFGEIVTPLPVEEHQNSRMLFTLREGSRYQLKLQFSVMHNLVSGLTYSNTV 149
Query: 162 WKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDA 221
WK G++V+ +K MLGT++PQ+ PY Y L+E+ TP+G ARG YSA+ KF DDD++C+++
Sbjct: 150 WKGGLQVDQSKGMLGTFAPQKGPYVYALKEDTTPAGALARGVYSAKLKFEDDDKRCHMEL 209
Query: 222 SYRFEIQKN 230
Y FEI+K+
Sbjct: 210 KYLFEIKKS 218
>UniRef100_Q9TU03 Rho GDP-dissociation inhibitor 2 [Bos taurus]
Length = 200
Score = 144 bits (362), Expect = 2e-33
Identities = 80/195 (41%), Positives = 114/195 (58%), Gaps = 6/195 (3%)
Query: 40 EEIVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEP 99
E V ++D+E + KL PQ SLKE E DKDDESL K+K+ LLG D V + P
Sbjct: 7 EPHVEEDDDELDGKLNYKPPPQKSLKELQEMDKDDESLTKYKKTLLG--DGPVVADPTAP 64
Query: 100 EVKIVSLTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLR 156
V + LT++C+ P I + T D KK F LKEG +YR+K +F V+ +IVSGL+
Sbjct: 65 NVTVTRLTLVCESAPGPIT-MDLTGDLEALKKETFVLKEGVEYRVKINFKVNKDIVSGLK 123
Query: 157 YTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRK 216
Y ++TGV+V+ M+G+Y P+ E Y + E P G+ ARGTY ++ F DDD+
Sbjct: 124 YVQHTYRTGVKVDKATFMVGSYGPRPEEYEFLTPIEEAPKGMLARGTYHNKSFFTDDDKH 183
Query: 217 CYLDASYRFEIQKNW 231
+L + I+K+W
Sbjct: 184 DHLTWEWNLSIKKDW 198
>UniRef100_P52565 Rho GDP-dissociation inhibitor 1 [Homo sapiens]
Length = 204
Score = 144 bits (362), Expect = 2e-33
Identities = 78/189 (41%), Positives = 113/189 (59%), Gaps = 6/189 (3%)
Query: 46 EDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVS 105
E+EE+E + Q S++E E DKDDESLRK+KE LLG V VSA + P V +
Sbjct: 17 ENEEDEHSVNYKPPAQKSIQEIQELDKDDESLRKYKEALLGRVAVSA--DPNVPNVVVTG 74
Query: 106 LTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVW 162
LT++C P L + T D KK F LKEG +YR+K SF V+ IVSG++Y +
Sbjct: 75 LTLVCSSAPGP-LELDLTGDLESFKKQSFVLKEGVEYRIKISFRVNREIVSGMKYIQHTY 133
Query: 163 KTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDAS 222
+ GV+++ T M+G+Y P+ E Y + E P G+ ARG+YS +++F DDD+ +L
Sbjct: 134 RKGVKIDKTDYMVGSYGPRAEEYEFLTPVEEAPKGMLARGSYSIKSRFTDDDKTDHLSWE 193
Query: 223 YRFEIQKNW 231
+ I+K+W
Sbjct: 194 WNLTIKKDW 202
>UniRef100_Q61599 Rho GDP-dissociation inhibitor 2 [Mus musculus]
Length = 200
Score = 142 bits (359), Expect = 5e-33
Identities = 78/197 (39%), Positives = 116/197 (58%), Gaps = 6/197 (3%)
Query: 38 EAEEIVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKI 97
+A+ + + D++ + KL PQ SLKE E DKDDESL K+K+ LLG DV V +
Sbjct: 5 DAQPQLEEADDDLDSKLNYKPPPQKSLKELQEMDKDDESLTKYKKTLLG--DVPVVADPT 62
Query: 98 EPEVKIVSLTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSG 154
P V + L+++C P I + T D KK F LKEG +YR+K +F V+ +IVSG
Sbjct: 63 VPNVTVTRLSLVCDSAPGPIT-MDLTGDLEALKKDTFVLKEGIEYRVKINFKVNKDIVSG 121
Query: 155 LRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDD 214
L+Y ++TG+RV+ M+G+Y P+ E Y + E P G+ ARGTY ++ F DDD
Sbjct: 122 LKYVQHTYRTGMRVDKATFMVGSYGPRPEEYEFLTPVEEAPKGMLARGTYHNKSFFTDDD 181
Query: 215 RKCYLDASYRFEIQKNW 231
++ +L + I+K+W
Sbjct: 182 KQDHLTWEWNLAIKKDW 198
>UniRef100_UPI00003AF5BE UPI00003AF5BE UniRef100 entry
Length = 203
Score = 142 bits (358), Expect = 7e-33
Identities = 78/189 (41%), Positives = 113/189 (59%), Gaps = 6/189 (3%)
Query: 46 EDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVS 105
E+EE+E + Q S++E E DKDDESLRK+KE LLG V V+A + P V +
Sbjct: 18 ENEEDEHSVNYKPPAQKSIQEIQELDKDDESLRKYKEALLGAVTVTA--DPNAPNVVVTK 75
Query: 106 LTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVW 162
LT++C P L + T D KK F LKEG +YR+K SF V+ IVSGL+Y +
Sbjct: 76 LTLVCTTAPGP-LELDLTGDLESYKKQAFVLKEGVEYRIKISFRVNREIVSGLKYIQHTF 134
Query: 163 KTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDAS 222
+ GV+++ T+ M+G+Y P+ E Y + E P G+ ARG+Y+ ++KF DDD+ +L
Sbjct: 135 RKGVKIDKTEYMVGSYGPRAEEYEFLTPMEEAPKGMLARGSYNIKSKFTDDDKTDHLSWE 194
Query: 223 YRFEIQKNW 231
+ I+K W
Sbjct: 195 WNLTIKKEW 203
>UniRef100_UPI000025D5CD UPI000025D5CD UniRef100 entry
Length = 205
Score = 142 bits (358), Expect = 7e-33
Identities = 80/189 (42%), Positives = 117/189 (61%), Gaps = 5/189 (2%)
Query: 46 EDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVS 105
E +E+E ++ Q SL+E E DKDDESLRK+KE LLGNV S + + P V++
Sbjct: 17 EIKEDECQVNYKPPAQKSLQEIQELDKDDESLRKYKETLLGNV--STLTDPKLPNVQVTK 74
Query: 106 LTIICQGRPD-LILPIPFTADS-KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVWK 163
+T++C P L+L + DS KK+ F LKEG +Y++K SF V+ IVSGL+Y ++
Sbjct: 75 MTLVCDTAPGPLVLDLQGDLDSFKKNPFVLKEGVEYKIKISFKVNKEIVSGLKYVQQTFR 134
Query: 164 TGVRVENTKKMLGTYSPQ-QEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDAS 222
GV+V+ T M+G+Y P+ E Y Y E P G+ ARGTY+ ++KF DDD+ +L
Sbjct: 135 KGVKVDKTDYMVGSYGPRPAEEYDYLTTAEEAPKGMLARGTYNIKSKFTDDDKHDHLSWE 194
Query: 223 YRFEIQKNW 231
+ I+K+W
Sbjct: 195 WSLTIKKDW 203
>UniRef100_P19803 Rho GDP-dissociation inhibitor 1 [Bos taurus]
Length = 204
Score = 142 bits (358), Expect = 7e-33
Identities = 78/189 (41%), Positives = 112/189 (58%), Gaps = 6/189 (3%)
Query: 46 EDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVS 105
E+EE+E + Q S++E E DKDDESLRK+KE LLG V VSA + P V +
Sbjct: 17 ENEEDEHSVNYKPPAQKSIQEIQELDKDDESLRKYKEALLGRVAVSA--DPNVPNVVVTR 74
Query: 106 LTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVW 162
LT++C P L + T D KK F LKEG +YR+K SF V+ IVSG++Y +
Sbjct: 75 LTLVCSTAPGP-LELDLTGDLESFKKQSFVLKEGVEYRIKISFRVNREIVSGMKYIQHTY 133
Query: 163 KTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDAS 222
+ GV+++ T M+G+Y P+ E Y + E P G+ ARG+Y+ +++F DDDR +L
Sbjct: 134 RKGVKIDKTDYMVGSYGPRAEEYEFLTPMEEAPKGMLARGSYNIKSRFTDDDRTDHLSWE 193
Query: 223 YRFEIQKNW 231
+ I+K W
Sbjct: 194 WNLTIKKEW 202
>UniRef100_P52566 Rho GDP-dissociation inhibitor 2 [Homo sapiens]
Length = 201
Score = 142 bits (357), Expect = 9e-33
Identities = 79/190 (41%), Positives = 111/190 (57%), Gaps = 6/190 (3%)
Query: 45 DEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIV 104
D+D+E + KL PQ SLKE E DKDDESL K+K+ LLG D V + P V +
Sbjct: 13 DDDDELDSKLNYKPPPQKSLKELQEMDKDDESLIKYKKTLLG--DGPVVTDPKAPNVVVT 70
Query: 105 SLTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVV 161
LT++C+ P I + T D KK LKEGS+YR+K F V+ +IVSGL+Y
Sbjct: 71 RLTLVCESAPGPIT-MDLTGDLEALKKETIVLKEGSEYRVKIHFKVNRDIVSGLKYVQHT 129
Query: 162 WKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDA 221
++TGV+V+ M+G+Y P+ E Y + E P G+ ARGTY ++ F DDD++ +L
Sbjct: 130 YRTGVKVDKATFMVGSYGPRPEEYEFLTPVEEAPKGMLARGTYHNKSFFTDDDKQDHLSW 189
Query: 222 SYRFEIQKNW 231
+ I+K W
Sbjct: 190 EWNLSIKKEW 199
>UniRef100_Q6DF26 Rho GDP dissociation inhibitor (GDI) beta [Xenopus tropicalis]
Length = 200
Score = 141 bits (355), Expect = 2e-32
Identities = 79/190 (41%), Positives = 111/190 (57%), Gaps = 6/190 (3%)
Query: 45 DEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIV 104
++D+E + KL PQ SL+E E DKDDESL K+K+ LLG+ V+A + P V +
Sbjct: 12 EDDDELDGKLNYKPPPQKSLQEIQELDKDDESLAKYKKSLLGDGPVAA--DPSAPNVIVT 69
Query: 105 SLTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVV 161
LT++C P LI + T D KK F LKEG +YR+K F V+ IVSGL+Y +
Sbjct: 70 RLTLVCATAPKLIT-MDLTGDLTNLKKETFALKEGVEYRVKIHFKVTKEIVSGLKYDQYI 128
Query: 162 WKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDA 221
++ GVRV + M+G+Y P+QE Y + E P G+ ARGTY ++ F DDD +L
Sbjct: 129 YRAGVRVTKARFMVGSYGPRQEEYEFLTPLEEAPKGILARGTYLNKSHFTDDDNHNHLSW 188
Query: 222 SYRFEIQKNW 231
+ I+K W
Sbjct: 189 EWNLTIRKEW 198
>UniRef100_Q99PT1 Rho GDP-dissociation inhibitor 1 [Mus musculus]
Length = 204
Score = 140 bits (354), Expect = 2e-32
Identities = 77/189 (40%), Positives = 112/189 (58%), Gaps = 6/189 (3%)
Query: 46 EDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVS 105
E+EE+E + Q S++E E DKDDESLRK+KE LLG V VSA + P V +
Sbjct: 17 ENEEDEHSVNYKPPAQKSIQEIQELDKDDESLRKYKEALLGRVAVSA--DPNVPNVIVTR 74
Query: 106 LTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVW 162
LT++C P L + T D KK F LKEG +YR+K SF V+ IVSG++Y +
Sbjct: 75 LTLVCSTAPGP-LELDLTGDLESFKKQSFVLKEGVEYRIKISFRVNREIVSGMKYIQHTY 133
Query: 163 KTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDAS 222
+ GV+++ T M+G+Y P+ E Y + E P G+ ARG+Y+ +++F DDD+ +L
Sbjct: 134 RKGVKIDKTDYMVGSYGPRAEEYEFLTPMEEAPKGMLARGSYNIKSRFTDDDKTDHLSWE 193
Query: 223 YRFEIQKNW 231
+ I+K W
Sbjct: 194 WNLTIKKEW 202
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.132 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 415,002,428
Number of Sequences: 2790947
Number of extensions: 18953887
Number of successful extensions: 70106
Number of sequences better than 10.0: 303
Number of HSP's better than 10.0 without gapping: 122
Number of HSP's successfully gapped in prelim test: 191
Number of HSP's that attempted gapping in prelim test: 68134
Number of HSP's gapped (non-prelim): 1018
length of query: 235
length of database: 848,049,833
effective HSP length: 123
effective length of query: 112
effective length of database: 504,763,352
effective search space: 56533495424
effective search space used: 56533495424
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC144644.6


