

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144618.9 + phase: 0
(313 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
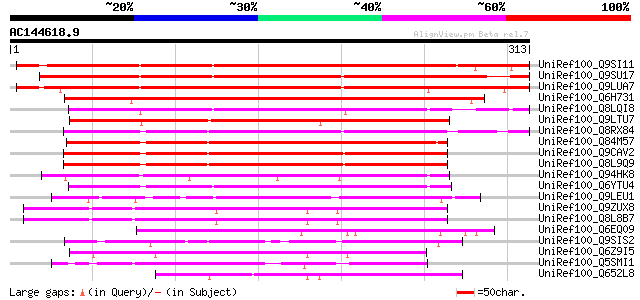
UniRef100_Q9SI11 Hypothetical protein At2g04220 [Arabidopsis tha... 388 e-106
UniRef100_Q9SU17 Hypothetical protein T20K18.40 [Arabidopsis tha... 353 3e-96
UniRef100_Q9LUA7 Arabidopsis thaliana genomic DNA, chromosome 5,... 308 2e-82
UniRef100_Q6H731 Hypothetical protein P0026H03.22 [Oryza sativa] 236 5e-61
UniRef100_Q8LQI8 P0456E05.14 protein [Oryza sativa] 228 2e-58
UniRef100_Q9LTU7 Emb|CAB40986.1 [Arabidopsis thaliana] 213 7e-54
UniRef100_Q8RX84 AT5g28150/T24G3_80 [Arabidopsis thaliana] 199 8e-50
UniRef100_Q84M57 Hypothetical protein OSJNBa0059E14.19 [Oryza sa... 193 4e-48
UniRef100_Q9CAV2 Hypothetical protein T9J14.19 [Arabidopsis thal... 189 1e-46
UniRef100_Q8L9Q9 Hypothetical protein [Arabidopsis thaliana] 187 2e-46
UniRef100_Q94HK8 Hypothetical protein [Oryza sativa] 181 2e-44
UniRef100_Q6YTU4 Hypothetical protein OJ1121_A05.24 [Oryza sativa] 177 2e-43
UniRef100_Q9LEU1 Hypothetical protein T30N20_270 [Arabidopsis th... 126 7e-28
UniRef100_Q9ZUX8 Hypothetical protein At2g27770 [Arabidopsis tha... 125 1e-27
UniRef100_Q8L8B7 Hypothetical protein At2g27770/F15K20.13 [Arabi... 117 5e-25
UniRef100_Q6EQ09 Hypothetical protein P0014G10.19-1 [Oryza sativa] 110 6e-23
UniRef100_Q9SIS2 Hypothetical protein At2g25200 [Arabidopsis tha... 107 4e-22
UniRef100_Q6Z9I5 Hypothetical protein P0524F03.23 [Oryza sativa] 99 2e-19
UniRef100_Q5SMI1 Hypothetical protein OSJNBb0019L07.28 [Oryza sa... 95 3e-18
UniRef100_Q652L8 Hypothetical protein OJ1155_H10.25 [Oryza sativa] 94 4e-18
>UniRef100_Q9SI11 Hypothetical protein At2g04220 [Arabidopsis thaliana]
Length = 307
Score = 388 bits (996), Expect = e-106
Identities = 195/314 (62%), Positives = 240/314 (76%), Gaps = 12/314 (3%)
Query: 5 LSFRSQSFSKQNLASLMSQRITEEPLPTKTAQSTVTCLYQTSIGGQWRNISVLWCKNLIN 64
+SFRS S A +++TE+P+ KTAQSTVTC+YQ I G WRN++VLW KNL+N
Sbjct: 1 MSFRSHSRGSSQFA----EKVTEDPVTYKTAQSTVTCIYQAHISGFWRNVTVLWSKNLMN 56
Query: 65 HTLNLKVDSARGEQFHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCP 124
H+L + V + G+ + CK+++KPW FWN+KGY+SFDV+GN ++VYWD RSAKF + P
Sbjct: 57 HSLMVMVTNVEGDM-NYCCKVDLKPWHFWNKKGYKSFDVEGNPVEVYWDFRSAKFTSS-P 114
Query: 125 EPCSDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKSFSTKAK 184
EP SD+YV LVS+EEVVLL+GDYKKKA+KR K R ALVEA L K+ENVF KK F+T+AK
Sbjct: 115 EPSSDFYVALVSEEEVVLLVGDYKKKAFKRTKSRPALVEAALFYKKENVFGKKCFTTRAK 174
Query: 185 FDEKRKESDIVVESSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFW 244
F +++KE +I+VESST+G K+PEMWISIDGIVLI +KNLQWKFRGNQTV+V+KQPV VFW
Sbjct: 175 FYDRKKEHEIIVESSTSGPKEPEMWISIDGIVLIQVKNLQWKFRGNQTVLVDKQPVQVFW 234
Query: 245 DVYDWLFSVSGSGPGLFIFKPGPTEVESDKEEKGL---EGESDDGSATSGYYSTKSYTPF 301
DVYDWLFS+ G+G GLFIFKPG TE +SD E G G D S S YYSTKS P+
Sbjct: 235 DVYDWLFSMPGTGHGLFIFKPGTTE-DSDMEGSGHGGGGGGESDTSTGSRYYSTKSSNPW 293
Query: 302 --ESCLVLCAYKLE 313
E CL L AYKLE
Sbjct: 294 PPEFCLFLHAYKLE 307
>UniRef100_Q9SU17 Hypothetical protein T20K18.40 [Arabidopsis thaliana]
Length = 285
Score = 353 bits (907), Expect = 3e-96
Identities = 173/295 (58%), Positives = 221/295 (74%), Gaps = 16/295 (5%)
Query: 19 SLMSQRITEEPLPTKTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQ 78
S +++ITE+P+ KTAQS+VTC+YQ + G WRN+ VLW KNL+NH+L + V S +G+
Sbjct: 7 SSTAEKITEDPVTYKTAQSSVTCIYQAHMVGFWRNVRVLWSKNLMNHSLTVMVTSVQGDM 66
Query: 79 FHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDE 138
+ CK+++KPW FW +KGY+SF+V+GNQ+DVYWD RSAKF G PEP SD+YV LVS+E
Sbjct: 67 -NYCCKVDLKPWHFWYKKGYKSFEVEGNQVDVYWDFRSAKFNGG-PEPSSDFYVALVSEE 124
Query: 139 EVVLLLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVES 198
EVVLLLGD+KKKA+KR K R +LV+A L K+ENVF KK FST+AKF ++++E +IVVES
Sbjct: 125 EVVLLLGDHKKKAFKRTKSRPSLVDAALFYKKENVFGKKIFSTRAKFHDRKREHEIVVES 184
Query: 199 STAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSGP 258
ST G K+PEMWIS+DGIVL+ ++NLQWKFRGNQTV+V+K+PV VFWDVYDWLFS G+G
Sbjct: 185 ST-GAKEPEMWISVDGIVLVQVRNLQWKFRGNQTVLVDKEPVQVFWDVYDWLFSTPGTGH 243
Query: 259 GLFIFKPGPTEVESDKEEKGLEGESDDGSATSGYYSTKSYTPFESCLVLCAYKLE 313
GLFIFKP E E+ E K S S+ E CL L A+KLE
Sbjct: 244 GLFIFKPESGESETSNETKNCSASSSSSSS-------------EFCLFLYAWKLE 285
>UniRef100_Q9LUA7 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MIF21
[Arabidopsis thaliana]
Length = 322
Score = 308 bits (788), Expect = 2e-82
Identities = 168/325 (51%), Positives = 225/325 (68%), Gaps = 20/325 (6%)
Query: 5 LSFRSQSFSKQNLASLMSQRITEEP--------LPTKTAQSTVTCLYQTSIGGQWRNISV 56
LSFR S Q A + +IT+ P + + STVTC YQ + G +RN++V
Sbjct: 2 LSFRMPSIGSQPTA--FAAKITDNPKTVAQLKSAASPSPHSTVTCGYQAHVAGFFRNVTV 59
Query: 57 LWCKNLINHTLNLKVDSARGEQFHQNCKIE-VKPWCFWNRKGYRSFDVDGNQIDVYWDLR 115
LW KNL+NH+L + V S + + CKI+ VKPW FW+++G +SFDV+GN ++V+WDLR
Sbjct: 60 LWSKNLMNHSLTVMVSSLDNDM-NYCCKIDLVKPWQFWSKRGSKSFDVEGNFVEVFWDLR 118
Query: 116 SAKFAGN-CPEPCSDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEATLLVKRENVF 174
SAK AGN PEP SDYYV +VSDEEVVLLLGD K+KAYKR K R ALVE + K+E++F
Sbjct: 119 SAKLAGNGSPEPVSDYYVAVVSDEEVVLLLGDLKQKAYKRTKSRPALVEGFIYFKKESIF 178
Query: 175 AKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVM 234
KK+FST+A+FDE+RKE ++VVESS G +PEMWIS+DGIV++++KNLQWKFRGNQ VM
Sbjct: 179 GKKTFSTRARFDEQRKEHEVVVESSN-GAAEPEMWISVDGIVVVNVKNLQWKFRGNQMVM 237
Query: 235 VNKQPVHVFWDVYDWLF--SVSGSGPGLFIFKPGPTEVESDKE-EKGLEGESDDGSATSG 291
V++ PV V++DV+DWLF S + + GLF+FKP P D+ EG+S GS+
Sbjct: 238 VDRTPVMVYYDVHDWLFASSETAASSGLFLFKPVPVGAMVDESFSDAEEGDSGGGSSPLS 297
Query: 292 YYSTKS--YTPF-ESCLVLCAYKLE 313
Y++ S + P + CL L A+KLE
Sbjct: 298 RYNSASSGFGPLHDFCLFLYAWKLE 322
>UniRef100_Q6H731 Hypothetical protein P0026H03.22 [Oryza sativa]
Length = 328
Score = 236 bits (603), Expect = 5e-61
Identities = 110/263 (41%), Positives = 177/263 (66%), Gaps = 10/263 (3%)
Query: 34 TAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVD------SARGEQFHQNCKIEV 87
T+ + +Y+ I G R+++ +W + LIN + + +D + G+ + K+E+
Sbjct: 54 TSHCSSVSVYRAKINGAPRHVTAVWHRTLINQSFTISIDGGGGGGAGAGDDGALSHKVEL 113
Query: 88 KPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDY 147
KPW FW+++G ++ DVDG+++D+ WDLRSAKF + PEP + YYV LVS +EVVLLLGD
Sbjct: 114 KPWPFWSKRGAKTLDVDGDRLDIVWDLRSAKFPASSPEPAAGYYVALVSRDEVVLLLGDG 173
Query: 148 KKKAYKRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAGNKDPE 207
KK A+KR + R +L +A L+ +RE+V +++F+ +A RK+ +IVV+S+ AG ++PE
Sbjct: 174 KKDAFKRTRSRPSLDDAVLVSRRESVSGRRTFAARAPLAAGRKDHEIVVDSAIAGPREPE 233
Query: 208 MWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSGPGLFIFKPGP 267
M I++DG+VL+H+++LQWKFRGN+TV+V++ PV V WDV+DW+F+ + +F+FKPG
Sbjct: 234 MRITVDGVVLVHVRSLQWKFRGNETVIVDQSPVQVLWDVHDWIFAGGPAAQAVFVFKPGA 293
Query: 268 TEVESDKEEK----GLEGESDDG 286
D+ + G G D+G
Sbjct: 294 PPPGGDRCGRRGGAGAGGIGDEG 316
>UniRef100_Q8LQI8 P0456E05.14 protein [Oryza sativa]
Length = 324
Score = 228 bits (581), Expect = 2e-58
Identities = 126/309 (40%), Positives = 177/309 (56%), Gaps = 49/309 (15%)
Query: 36 QSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGE------------------ 77
QS VT +Y+ I G R ++V WC+NL+ H L++ ++ + G
Sbjct: 34 QSAVTLVYRAEISGHRRLVTVTWCRNLLTHGLSVSIEGSAGNGKDKIGREYGEAAVAATA 93
Query: 78 ----------QFHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPC 127
+ CK+E++PW FW + G + F VDGN IDV WDLRSA+F+ PEP
Sbjct: 94 ADGGGGGGGGKSCSACKVEMQPWHFWRKYGAKQFQVDGNAIDVVWDLRSARFSDE-PEPL 152
Query: 128 SDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDE 187
SDYYV +V+ EEVVLLLG+ KK A++R R +L +A L+ K+E+VF+KK F TKA+F +
Sbjct: 153 SDYYVAVVAGEEVVLLLGNLKKDAFRRTGSRPSLQDAVLVCKKEHVFSKKRFVTKARFSD 212
Query: 188 KRKESDIVVESSTA---GNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFW 244
+ K DI +E S++ G D +M I IDG V + +++LQWKFRGN+ + +NK V V+W
Sbjct: 213 RGKLHDISIECSSSNLTGGTDVDMAIKIDGCVSVLVRHLQWKFRGNECISINKLKVQVYW 272
Query: 245 DVYDWLFSVSGSGPGLFIFKPGPTEVESDKEEKGLEGESDDGSATSGYYSTKSYTPFESC 304
D +DWLF +G LFIFKP P S S +ST Y+ F C
Sbjct: 273 DAHDWLFG-TGMRHALFIFKPEP--------------PSPSPPGASSEFSTDEYSDF--C 315
Query: 305 LVLCAYKLE 313
L L A+K+E
Sbjct: 316 LFLYAWKVE 324
>UniRef100_Q9LTU7 Emb|CAB40986.1 [Arabidopsis thaliana]
Length = 289
Score = 213 bits (541), Expect = 7e-54
Identities = 103/234 (44%), Positives = 151/234 (64%), Gaps = 6/234 (2%)
Query: 37 STVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQ--FHQNCKIEVKPWCFWN 94
S+V+ +Y I +N+ V W K +H+L +K+++ + EQ HQ KI++ FW
Sbjct: 6 SSVSLIYVVEIAKTPQNVDVTWSKTTSSHSLTIKIENVKDEQQNHHQPVKIDLSGSSFWA 65
Query: 95 RKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAYKR 154
+KG +S + +G ++DVYWD R AKF+ N PEP S +YV LVS VL +GD + +A KR
Sbjct: 66 KKGLKSLEANGTRVDVYWDFRQAKFS-NFPEPSSGFYVSLVSQNATVLTIGDLRNEALKR 124
Query: 155 MKMRQALVEATLLVKRENVFAKKSFSTKAKFD--EKRKESDIVVESSTAGNKDPEMWISI 212
K + EA L+ K+E+V K+ F T+ F E R+E+++V+E+S +G DPEMWI++
Sbjct: 125 TKKNPSATEAALVSKQEHVHGKRVFYTRTAFGGGESRRENEVVIETSLSGPSDPEMWITV 184
Query: 213 DGIVLIHIKNLQWKFRGNQTVMVNK-QPVHVFWDVYDWLFSVSGSGPGLFIFKP 265
DG+ I I NL W+FRGN+ V V+ + +FWDV+DWLF SGS GLF+FKP
Sbjct: 185 DGVPAIRIMNLNWRFRGNEVVTVSDGVSLEIFWDVHDWLFEPSGSSSGLFVFKP 238
>UniRef100_Q8RX84 AT5g28150/T24G3_80 [Arabidopsis thaliana]
Length = 289
Score = 199 bits (506), Expect = 8e-50
Identities = 111/281 (39%), Positives = 163/281 (57%), Gaps = 17/281 (6%)
Query: 33 KTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNCKIEVKPWCF 92
K AQ+ VTC+YQ I G+ I+V W KNL+ ++ + VD + + CK+E+KPW F
Sbjct: 26 KNAQNLVTCIYQCRIRGRNCLITVTWTKNLMGQSVTVGVDDSCNQSL---CKVEIKPWLF 82
Query: 93 WNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAY 152
RKG +S + IDV+WDL SAKF G+ PE +YV +V D+E+VLLLGD KK+A+
Sbjct: 83 TKRKGSKSLEAYSCNIDVFWDLSSAKF-GSGPEALGGFYVGVVVDKEMVLLLGDMKKEAF 141
Query: 153 KRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWISI 212
K+ + + A + K+E+VF K+ F+TKA+ K D+++E T DP + + +
Sbjct: 142 KKTNASPSSLGAVFIAKKEHVFGKRVFATKAQLFADGKFHDLLIECDT-NVTDPCLVVRV 200
Query: 213 DGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSGPGLFIFKPGPTEVES 272
DG L+ +K L+WKFRGN T++VNK V V WDV+ WLF + +G +F+F+
Sbjct: 201 DGKTLLQVKRLKWKFRGNDTIVVNKMTVEVLWDVHSWLFGLPTTGNAVFMFR------TC 254
Query: 273 DKEEKGLEGESDDGSATSGYYSTKSYTPFESCLVLCAYKLE 313
EK L D + S +S F L+L A+K E
Sbjct: 255 QSTEKSLSFSQDVTTTNSKSHS------FGFSLILYAWKSE 289
>UniRef100_Q84M57 Hypothetical protein OSJNBa0059E14.19 [Oryza sativa]
Length = 291
Score = 193 bits (491), Expect = 4e-48
Identities = 95/230 (41%), Positives = 145/230 (62%), Gaps = 5/230 (2%)
Query: 35 AQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNCKIEVKPWCFWN 94
AQ+ VTCLYQ G+ ISV W K+L+ L++ VD + CK ++KPW F
Sbjct: 30 AQNLVTCLYQAQFSGRPCVISVTWSKSLMGQGLSIGVDDLSNQCL---CKADIKPWLFSK 86
Query: 95 RKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAYKR 154
+KG + DV+ +I+++WDL AKF G PEP +YV +V D E++LLLGD KK AY++
Sbjct: 87 KKGSKRLDVEDGKIEIFWDLSGAKF-GAGPEPMEGFYVAVVFDLELILLLGDMKKDAYRK 145
Query: 155 MKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWISIDG 214
+ ++ A + +RE+++ KK ++ KA+F E + D+V+E T KDP + I +D
Sbjct: 146 TGANRPMLNAAFVARREHIYGKKIYTAKAQFCENGQFHDVVIECDTVSLKDPCLEIRVDK 205
Query: 215 IVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSGPGLFIFK 264
++ +K L WKFRGNQT++V+ PV VFWDV+ WLF ++ S +F+F+
Sbjct: 206 KPVMQVKRLAWKFRGNQTILVDGLPVEVFWDVHSWLFGLTTSN-AVFMFQ 254
>UniRef100_Q9CAV2 Hypothetical protein T9J14.19 [Arabidopsis thaliana]
Length = 289
Score = 189 bits (479), Expect = 1e-46
Identities = 96/234 (41%), Positives = 147/234 (62%), Gaps = 7/234 (2%)
Query: 33 KTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNCKIEVKPWCF 92
+ AQ+ V C+Y+ I G+ I+V W KNL+ + + VD + CK+E+KPW F
Sbjct: 26 RNAQNLVICIYRCRIRGRTCLITVTWTKNLMGQCVTVGVDDSCNRSL---CKVEIKPWLF 82
Query: 93 WNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAY 152
RKG ++ + IDV+WDL SAKF G+ PEP +YV +V D+E+VLLLGD KK+A+
Sbjct: 83 TKRKGSKTLEAYACNIDVFWDLSSAKF-GSSPEPLGGFYVGVVVDKEMVLLLGDMKKEAF 141
Query: 153 KRMKMR-QALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWIS 211
K+ + + A + K+E+VF K++F+TKA+F K D+V+E T+ DP + +
Sbjct: 142 KKTNAAPSSSLGAVFIAKKEHVFGKRTFATKAQFSGDGKTHDLVIECDTS-LSDPCLIVR 200
Query: 212 IDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSV-SGSGPGLFIFK 264
+DG +L+ ++ L WKFRGN T++VN+ V V WDV+ W F + S G +F+F+
Sbjct: 201 VDGKILMQVQRLHWKFRGNDTIIVNRISVEVLWDVHSWFFGLPSSPGNAVFMFR 254
>UniRef100_Q8L9Q9 Hypothetical protein [Arabidopsis thaliana]
Length = 289
Score = 187 bits (476), Expect = 2e-46
Identities = 96/234 (41%), Positives = 146/234 (62%), Gaps = 7/234 (2%)
Query: 33 KTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNCKIEVKPWCF 92
+ AQ+ V C+Y+ I G+ I+V W KNL+ + + VD + CK+E+KPW F
Sbjct: 26 RNAQNLVICIYRCRIRGRTCLITVTWTKNLMGQCVTVGVDDSCNRSL---CKVEIKPWLF 82
Query: 93 WNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAY 152
RKG ++ IDV+WDL SAKF G+ PEP +YV +V D+E+VLLLGD KK+A+
Sbjct: 83 TKRKGSKTLKAYACNIDVFWDLSSAKF-GSSPEPLGGFYVGVVVDKEMVLLLGDIKKEAF 141
Query: 153 KRMKMR-QALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWIS 211
K+ + + A + K+E+VF K++F+TKA+F K D+V+E T+ DP + +
Sbjct: 142 KKTNAAPSSSLGAVFIAKKEHVFGKRTFATKAQFSGDGKTHDLVIECDTS-LSDPCLIVR 200
Query: 212 IDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSV-SGSGPGLFIFK 264
+DG +L+ ++ L WKFRGN T++VN+ V V WDV+ W F + S G +F+F+
Sbjct: 201 VDGKILMQVQRLHWKFRGNDTIIVNRISVEVLWDVHSWFFGLPSSPGNAVFMFR 254
>UniRef100_Q94HK8 Hypothetical protein [Oryza sativa]
Length = 306
Score = 181 bits (460), Expect = 2e-44
Identities = 96/267 (35%), Positives = 152/267 (55%), Gaps = 24/267 (8%)
Query: 20 LMSQRITEEPLPT-------------KTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHT 66
++ QR ++P P+ K ++ +Y+ I G R ++V W +++++H+
Sbjct: 6 VVQQRSGDQPAPSCDIAADEIPVNGHKPGRAVTASVYRAKIAGHSRVLTVSWSRDMLSHS 65
Query: 67 LNLKVDSARGEQFHQNCKIEVKPWCFWNRKGYRSFDVDGNQ---IDVYWDLRSAKFAGNC 123
+ V G C+++++PW FW R G R ++ G + V WDLR A+F
Sbjct: 66 FAVSVTGVDGAS--AECRVDLRPWQFWRRAGSRRVELAGTAPATVRVMWDLRRARFGAGL 123
Query: 124 PEPCSDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQA--LVEATLLVKRENVFAKKSFST 181
PEP S YYV + + EVVL++GD +K A +R R A +A + +RE+VF K+ F+
Sbjct: 124 PEPRSGYYVAVEAAGEVVLVVGDMRKDALRRASPRAAPAACDAVPVARREHVFGKRRFAA 183
Query: 182 KAKFDEKRKESDIVVE---SSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQ 238
KA+F ++ DI +E G+ D EM I+IDG + +K+LQWKFRGNQ+V ++
Sbjct: 184 KARFHDQGTVHDIAIECGGGGEGGDADMEMTIAIDGEEAVQVKHLQWKFRGNQSVTFSRA 243
Query: 239 PVHVFWDVYDWLFSVSGSGPGLFIFKP 265
V ++WDV+DWLFS +G P LFIF+P
Sbjct: 244 KVEIYWDVHDWLFS-AGMRPALFIFRP 269
>UniRef100_Q6YTU4 Hypothetical protein OJ1121_A05.24 [Oryza sativa]
Length = 284
Score = 177 bits (450), Expect = 2e-43
Identities = 93/231 (40%), Positives = 135/231 (58%), Gaps = 5/231 (2%)
Query: 36 QSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNCKIEVKPWCFWNR 95
Q+ V C Y + G+ +++V W K + L++ VD + CK E+KPW F R
Sbjct: 24 QNMVQCTYLARLRGKSCSVTVTWSKMTMGQALSVAVDDSSNRCL---CKAEIKPWLFSKR 80
Query: 96 KGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAYKRM 155
KG ++ +VDG +D+ WDL SAKFA PEP +YV LV D E VL+LGD +K R+
Sbjct: 81 KGSKAMEVDGGALDIVWDLSSAKFAAG-PEPVEGFYVALVCDLEAVLVLGDMRKDGDHRV 139
Query: 156 KMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWISIDGI 215
A ++ ++E+V+ KK +S KA+F + + I +E T+G KDP + I I
Sbjct: 140 SSDVLASNAVMIARKEHVYGKKVYSAKARFLDIGQLHHITIECDTSGLKDPSLEIRIGKK 199
Query: 216 VLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSGPGLFIFKPG 266
++ +K L WKFRGNQTV V+ PV V WDV+DWLF S + +F+F+ G
Sbjct: 200 RVMQVKRLAWKFRGNQTVYVDGLPVEVLWDVHDWLFG-SSNKCAVFLFQSG 249
>UniRef100_Q9LEU1 Hypothetical protein T30N20_270 [Arabidopsis thaliana]
Length = 389
Score = 126 bits (317), Expect = 7e-28
Identities = 94/287 (32%), Positives = 139/287 (47%), Gaps = 42/287 (14%)
Query: 26 TEEPLPTKTAQSTVT-CLYQTS------------IGGQWRNISVLWCKNLINHTLNLKVD 72
++ P P + +T CLYQT +GG N+ L ++ NH+ L
Sbjct: 21 SQSPSPLSSGDPNLTTCLYQTDHGVFYLTWSRTFLGGHSVNL-FLHSQDYYNHSSPLSFS 79
Query: 73 SA-----RGEQFHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPC 127
SA FH N + FW ++G R +I V+WDL AKF EP
Sbjct: 80 SADLSLSSAVSFHLN----LNTLAFWKKRGSRFVSP---KIQVFWDLSKAKFDSGS-EPR 131
Query: 128 SDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEA-TLLVKRENVFAKKSFSTKAKFD 186
S +Y+ +V D E+ LL+GD K+AY R K + LL+++E+VF + F+TKA+F
Sbjct: 132 SGFYIAVVVDGEMGLLVGDSVKEAYARAKSAKPPTNPQALLLRKEHVFGARVFTTKARFG 191
Query: 187 EKRKESDIVVESSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDV 246
K +E I ++D ++ S+D ++ IK L+WKFRGN+ V ++ V + WDV
Sbjct: 192 GKNREISI----DCRVDEDAKLCFSVDSKQVLQIKRLRWKFRGNEKVEIDGVHVQISWDV 247
Query: 247 YDWLFSVSGSGPG---------LFIFKPGPTEVESDKEEKGLEGESD 284
Y+WLF SG G +F F+ P E E E K E E +
Sbjct: 248 YNWLFQSKSSGDGGGGGGHAVFMFRFESDP-EAEEVCETKRKEEEDE 293
>UniRef100_Q9ZUX8 Hypothetical protein At2g27770 [Arabidopsis thaliana]
Length = 320
Score = 125 bits (314), Expect = 1e-27
Identities = 82/267 (30%), Positives = 137/267 (50%), Gaps = 13/267 (4%)
Query: 9 SQSFSKQNLASLMSQRITEEPLPTKTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLN 68
S S S + + + + P + Q+++T +Y+ ++ + I V WC N+ L+
Sbjct: 20 SISSSSSSCSKYSTNNVCISPSLIPSIQTSITSIYRITLS-KHLIIKVTWCNPHNNNGLS 78
Query: 69 LKVDSARGEQFHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNC--PEP 126
+ V SA + K+ F +KG +S D D +I+V+WDL SAK+ N PEP
Sbjct: 79 ISVASA-DQNPSTTLKLNTSSRFFRKKKGNKSVDSDLGKIEVFWDLSSAKYDSNLCGPEP 137
Query: 127 CSDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKS--FSTKAK 184
+ +YV+++ D ++ LLLGD ++ ++ + LV R+ F + +STK +
Sbjct: 138 INGFYVIVLVDGQMGLLLGDSSEETLRKKGFSGDIGFDFSLVSRQEHFTGNNTFYSTKVR 197
Query: 185 FDEKRKESDIVV------ESSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQ 238
F E +IV+ E N P + + ID +I +K LQW FRGNQT+ ++
Sbjct: 198 FVETGDSHEIVIRCNKETEGLKQSNHYPVLSVCIDKKTVIKVKRLQWNFRGNQTIFLDGL 257
Query: 239 PVHVFWDVYDWLFSVSGS-GPGLFIFK 264
V + WDV+DW FS G+ G +F+F+
Sbjct: 258 LVDLMWDVHDWFFSNQGACGRAVFMFR 284
>UniRef100_Q8L8B7 Hypothetical protein At2g27770/F15K20.13 [Arabidopsis thaliana]
Length = 320
Score = 117 bits (292), Expect = 5e-25
Identities = 79/267 (29%), Positives = 133/267 (49%), Gaps = 13/267 (4%)
Query: 9 SQSFSKQNLASLMSQRITEEPLPTKTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLN 68
S S S + + + + P + Q+++T +Y+ ++ + I V WC N+ L+
Sbjct: 20 SISSSSSSCSKYSTNNVCISPSLIPSIQTSITSIYRITLS-KHLIIKVTWCNPHNNNGLS 78
Query: 69 LKVDSARGEQFHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNC--PEP 126
+ V SA + K+ F +KG +S D D +I+V+WDL SAK+ N P
Sbjct: 79 ISVASA-DQNPSTTLKLNTSSRFFRKKKGNKSVDSDLGKIEVFWDLSSAKYDSNLCGPGT 137
Query: 127 CSDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKS--FSTKAK 184
+ +YV+++ D + LLLGD ++ ++ + L R+ F + +STK +
Sbjct: 138 INGFYVIVLVDGSMGLLLGDSSEETLRKKGFSGDIGFDFSLXSRQEHFTGNNTFYSTKVR 197
Query: 185 FDEKRKESDIVV------ESSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQ 238
F E +IV+ E N P + + ID +I +K LQW FRGNQT+ ++
Sbjct: 198 FVETGDSHEIVIRCNKETEGLKQSNHYPVLSVCIDKKTVIKVKRLQWNFRGNQTIFLDGL 257
Query: 239 PVHVFWDVYDWLFSVSGS-GPGLFIFK 264
V + WDV+DW FS G+ G +F+F+
Sbjct: 258 LVDLMWDVHDWFFSNQGACGRAVFMFR 284
>UniRef100_Q6EQ09 Hypothetical protein P0014G10.19-1 [Oryza sativa]
Length = 444
Score = 110 bits (274), Expect = 6e-23
Identities = 80/259 (30%), Positives = 126/259 (47%), Gaps = 43/259 (16%)
Query: 77 EQFHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFA-GNCPEPCSDYYVMLV 135
E+ + V+PW W R+G + F V ++D+ WDL A+FA PEP S Y+V +V
Sbjct: 108 EEEEATVAVRVRPWLLWRRRGSKRFRVRDRRVDLAWDLTRARFACPGSPEPSSGYFVAVV 167
Query: 136 SDEEVVLLLGDYKKKAYKRMKMRQAL-VEATLLVKRENVF---AKKSFSTKAKFDEKRKE 191
D E+ L+ GD ++AY++ K R+ +A L+ +RE+V A K + + KE
Sbjct: 168 VDGEMALVAGDMAEEAYRKTKARRGPGPDAVLISRREHVSMRDAGHGRGHKTFVNVRGKE 227
Query: 192 SDIVVESSTAG-------NKDPE-------MWISIDGIVLIHIKNLQWKFRGNQTV-MVN 236
+I V+ + G +KD E M +++DG ++HI+ L+WKFRG + V +
Sbjct: 228 REISVDLVSRGHGKDRDKDKDKERDKADVGMSVTVDGERVLHIRRLRWKFRGTEKVDLGG 287
Query: 237 KQPVHVFWDVYDWLFSVSGSGP--------------GLFIFKPGPTEVESD-------KE 275
V V WD++ WLF + P +FIF+ ++ D K+
Sbjct: 288 GDGVQVSWDLHHWLFPNRDTAPADASAVTPPPQPAHAVFIFRFELADIAGDDRDSAEVKD 347
Query: 276 EKGLE--GESDDGSATSGY 292
E LE G G A +GY
Sbjct: 348 EHLLENAGSGGGGGAWAGY 366
>UniRef100_Q9SIS2 Hypothetical protein At2g25200 [Arabidopsis thaliana]
Length = 354
Score = 107 bits (267), Expect = 4e-22
Identities = 76/257 (29%), Positives = 122/257 (46%), Gaps = 34/257 (13%)
Query: 34 TAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNC---------- 83
+ +T TC Y T++G + + W ++ + +L+L S + +
Sbjct: 45 SGDATTTCQYHTNVGVFFLS----WSRSFLRRSLHLHFYSCNSTNCYLHSLDCYRHSIPF 100
Query: 84 --KIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVV 141
++E+KP FW + G + + I V WDL AKF G+ P+P S +YV + EV
Sbjct: 101 AFRLEIKPLTFWRKNGSKKLSRKPD-IRVVWDLTHAKF-GSGPDPESGFYVAVFVSGEVG 158
Query: 142 LLLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTA 201
LL+G K R RQ LV K+EN+F + +STK K +E I V+
Sbjct: 159 LLVGGGNLKQRPR---RQILVS-----KKENLFGNRVYSTKIMIQGKLREISIDVK---V 207
Query: 202 GNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSG---- 257
N D + S+D ++ I LQWKFRGN ++++ + + WDV++WLF
Sbjct: 208 VNDDASLRFSVDDKSVLKISQLQWKFRGNTKIVIDGVTIQISWDVFNWLFGGKDKVKPDK 267
Query: 258 -PGLFIFKPGPTEVESD 273
P +F+ + EVE +
Sbjct: 268 IPAVFLLRFENQEVEGN 284
>UniRef100_Q6Z9I5 Hypothetical protein P0524F03.23 [Oryza sativa]
Length = 406
Score = 98.6 bits (244), Expect = 2e-19
Identities = 72/247 (29%), Positives = 121/247 (48%), Gaps = 32/247 (12%)
Query: 37 STVTCLYQTSIGG---QWRNISVLWCKNLINHTLNLKVDSARGEQFHQNCKIE-----VK 88
S T +Y+T +G W S+ + + ++ G F ++ E V+
Sbjct: 29 SLATSVYETHLGLAALSWTRTSLGLSLRAVLRLSSPATAASVGTCFDEDADEETLAFRVR 88
Query: 89 PWCFWNRKGYRSFDVDGNQ-IDVYWDLRSAKFAGN-CPEPCSDYYVMLVSDEEVVLLLGD 146
PW W R+G R F G++ +D+ WDL A+F G+ PEP S ++V +V D E+VL GD
Sbjct: 89 PWLLWRRRGTRRFRAAGDRRVDLAWDLTRARFPGSGSPEPSSGFFVAVVVDGEMVLAAGD 148
Query: 147 YKKKAYKRMKMRQ-ALVEATLLVKRENVFAKKS----FSTKAKFDEKRKESDIVVESSTA 201
AY+R + R+ A LL +RE+V + + ++ + KE +I V+ +
Sbjct: 149 LSDAAYRRTRARRPAGPRPVLLSRREHVAMRDAGRGGRGHRSWVTVRGKEREISVDLVSR 208
Query: 202 G----------------NKDPEMWISIDGIVLIHIKNLQWKFRGNQTV-MVNKQPVHVFW 244
G D + +SIDG ++H++ L+WKFRG++ V + V + W
Sbjct: 209 GRGRDTGSSGSSSREKDRADVGLSVSIDGERVLHVRRLRWKFRGSERVDLGGGDRVQLSW 268
Query: 245 DVYDWLF 251
D+++WLF
Sbjct: 269 DLHNWLF 275
>UniRef100_Q5SMI1 Hypothetical protein OSJNBb0019L07.28 [Oryza sativa]
Length = 384
Score = 94.7 bits (234), Expect = 3e-18
Identities = 65/230 (28%), Positives = 108/230 (46%), Gaps = 28/230 (12%)
Query: 26 TEEPLPTKTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNCKI 85
+ P P +A T +Y T +G +++ W + + L ++ A GE +
Sbjct: 19 SSSPAPAASA----TSVYWTHLG----TVTLTWSRGQLGLVLAAELHLA-GEGAAPALRF 69
Query: 86 EVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLG 145
++P W R+G + F G+ + WD+ A+ AG PEP + Y + + D E+VL G
Sbjct: 70 LLRPLLPWRRRGCKRFAGGGHAVAFTWDMSRARLAGRRPEPVARYSLHVCVDGELVLAAG 129
Query: 146 DYKKKAYKRMKMRQALVEATLLVKRENVFAK---KSFSTKAKFDEKRKESDIVVESSTAG 202
D A LL +REN A ++++T R E I VE
Sbjct: 130 DLALLAPSA---------GFLLTRRENAVAAGGGEAYATTVAVAGGRHEVSIAVE----- 175
Query: 203 NKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFS 252
D MW++IDG + ++ L+WKFRG++ + + + V V WD++ WLF+
Sbjct: 176 --DAVMWVAIDGEKALQVRRLRWKFRGSERLDLPRGRVRVSWDLHGWLFA 223
>UniRef100_Q652L8 Hypothetical protein OJ1155_H10.25 [Oryza sativa]
Length = 493
Score = 94.4 bits (233), Expect = 4e-18
Identities = 63/203 (31%), Positives = 100/203 (49%), Gaps = 19/203 (9%)
Query: 89 PWCFWNRKGYRSFDVD-GNQIDVYWDLRSAKF---AGNCPEPCSDYYVMLVSDEEVVLLL 144
P ++G RSF + G + +YWD AK+ + PEP DYY+ +V+D E+ +LL
Sbjct: 105 PQFLHKKRGTRSFVTEAGTVVAIYWDTTDAKYPAAGSSSPEPTRDYYLAVVADGELAVLL 164
Query: 145 GDYKKKAYKRMKMRQALVEATLLVKRENVFAKKS-----------FSTKAKF--DEKRKE 191
G + A + + A LL +RE + A + ST+ +F D E
Sbjct: 165 GG-GEAARELARRFAAAPRRALLSRREQLRAAPASPAAMAAAAVAHSTRCRFRADGAEHE 223
Query: 192 SDIVVESSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQT-VMVNKQPVHVFWDVYDWL 250
+V G +D E+ +SIDG ++ + ++W FRGN+T V+ + V V WDV+DW
Sbjct: 224 VAVVCRGEEWGTRDGEVAVSIDGKKVVEARRVKWNFRGNRTAVLGDGAVVEVMWDVHDWW 283
Query: 251 FSVSGSGPGLFIFKPGPTEVESD 273
F+ G G F+ K + + D
Sbjct: 284 FAGGGGGGAQFMVKARDGDGDGD 306
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 532,129,761
Number of Sequences: 2790947
Number of extensions: 22487824
Number of successful extensions: 53473
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 53356
Number of HSP's gapped (non-prelim): 49
length of query: 313
length of database: 848,049,833
effective HSP length: 127
effective length of query: 186
effective length of database: 493,599,564
effective search space: 91809518904
effective search space used: 91809518904
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC144618.9


