

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144618.8 + phase: 0 /pseudo
(558 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
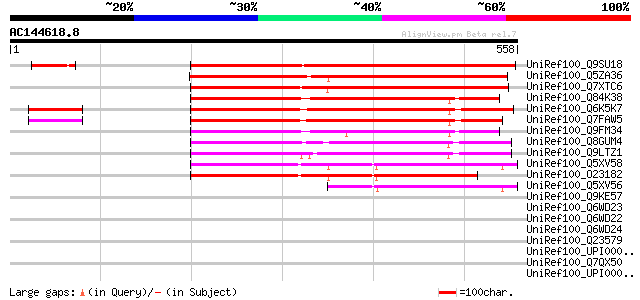
UniRef100_Q9SU18 Hypothetical protein T20K18.30 [Arabidopsis tha... 493 e-138
UniRef100_Q5ZA36 Hypothetical protein P0501G08.34 [Oryza sativa] 441 e-122
UniRef100_Q7XTC6 OSJNBa0064H22.21 protein [Oryza sativa] 283 9e-75
UniRef100_Q84K38 Hypothetical protein At5g40640 [Arabidopsis tha... 280 1e-73
UniRef100_Q6K5K7 Hypothetical protein OJ1212_D02.6 [Oryza sativa] 278 4e-73
UniRef100_Q7FAW5 OSJNBa0081L15.3 protein [Oryza sativa] 277 7e-73
UniRef100_Q9FM34 Dbj|BAA95714.1 [Arabidopsis thaliana] 273 1e-71
UniRef100_Q8GUM4 Hypothetical protein At3g27390 [Arabidopsis tha... 266 2e-69
UniRef100_Q9LTZ1 Emb|CAB40985.1 [Arabidopsis thaliana] 255 2e-66
UniRef100_Q5XV58 Hypothetical protein [Arabidopsis thaliana] 250 7e-65
UniRef100_O23182 Hypothetical protein C7A10.330 [Arabidopsis tha... 224 7e-57
UniRef100_Q5XV56 Hypothetical protein [Arabidopsis thaliana] 121 6e-26
UniRef100_Q9KE57 BH1001 protein [Bacillus halodurans] 46 0.003
UniRef100_Q6WD23 Chitin synthase variant 1 [Tribolium castaneum] 39 0.52
UniRef100_Q6WD22 Chitin synthase variant 2 [Tribolium castaneum] 39 0.52
UniRef100_Q6WD24 Chitin synthase CHS2 [Tribolium castaneum] 37 1.5
UniRef100_Q23579 Helix loop helix protein 10 [Caenorhabditis ele... 37 2.0
UniRef100_UPI00002A73EA UPI00002A73EA UniRef100 entry 36 3.4
UniRef100_Q7QX50 GLP_192_30681_21532 [Giardia lamblia ATCC 50803] 36 3.4
UniRef100_UPI0000432841 UPI0000432841 UniRef100 entry 35 4.4
>UniRef100_Q9SU18 Hypothetical protein T20K18.30 [Arabidopsis thaliana]
Length = 590
Score = 493 bits (1270), Expect = e-138
Identities = 241/359 (67%), Positives = 292/359 (81%), Gaps = 3/359 (0%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
DV+LIT++A++KSPYML +GWKRLLEDLVGR GPFLE+ CVPFAGLAI+LWPLAV GAV+
Sbjct: 211 DVLLITAVAVYKSPYMLLKGWKRLLEDLVGREGPFLESVCVPFAGLAILLWPLAVAGAVI 270
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
A+++ SFFL LY GV+VHQEDS +MG YI++ VSLFDEYVNDLLYLREG+ LPRP YR
Sbjct: 271 ASVLSSFFLGLYSGVIVHQEDSFRMGLNYIIAAVSLFDEYVNDLLYLREGTSLPRPCYRT 330
Query: 320 NVKHAVERKKLEGIDHN--LKNRRDSSQNSKHTLQQSRSMKWKIQQYKPVQVWDWFFKSC 377
+ V K++ G N LK++R SS SK +QSR++K I YKPVQVW+W FKSC
Sbjct: 331 KTE-TVHGKRILGESKNVDLKSKRSSSLGSKLVSEQSRTLKKAITLYKPVQVWEWLFKSC 389
Query: 378 EVNGRIVLRDGLISVKEIEECIFKGNCKKLSIKLPAWSLLQCLLTSAKSNSDGLVISDDI 437
EVNGRI+LRDGLI VK++EEC+ KGN KKL IKLPAW++LQCLL SAKSNS GLVI+D +
Sbjct: 390 EVNGRILLRDGLIDVKDVEECLVKGNSKKLYIKLPAWTVLQCLLASAKSNSSGLVITDGV 449
Query: 438 ELTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVMRSKNDIPEDWDSTGF 497
ELT +N P+D+VF W +GPLLIMKEQ+KNL+L E EE CL++LVM KN+ EDWD+TGF
Sbjct: 450 ELTELNSPRDKVFVWLVGPLLIMKEQIKNLKLTEDEEFCLRKLVMVCKNERTEDWDNTGF 509
Query: 498 PSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKILYIEALQASASAKEGNNI 556
PS D VR+AQLQAIIRRLQG+ +SMSR+PTFRR+F NLVK+LYIEAL+ AS I
Sbjct: 510 PSSDTVRKAQLQAIIRRLQGMVASMSRIPTFRRRFMNLVKVLYIEALEMGASGNRAGGI 568
Score = 65.5 bits (158), Expect = 4e-09
Identities = 29/48 (60%), Positives = 38/48 (78%), Gaps = 1/48 (2%)
Query: 25 LIGPIAFAIMVVGNSAVIIGLWTAHVFWTYYCVARLYFLSKFFVICFI 72
+IGPI+ AI++VGNS VIIGLW AH WTYYC+AR + KF ++CF+
Sbjct: 34 IIGPISSAIILVGNSCVIIGLWPAHFIWTYYCLARCIPI-KFEILCFL 80
>UniRef100_Q5ZA36 Hypothetical protein P0501G08.34 [Oryza sativa]
Length = 477
Score = 441 bits (1133), Expect = e-122
Identities = 209/356 (58%), Positives = 273/356 (75%), Gaps = 10/356 (2%)
Query: 199 FDVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAV 258
FDV +I+ +A+WKSP ML +GW+RL EDLVGR GPFLET CVPFAGL+IILWPLAV+GAV
Sbjct: 83 FDVFMISGVALWKSPCMLLKGWQRLCEDLVGREGPFLETVCVPFAGLSIILWPLAVIGAV 142
Query: 259 LAAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYR 318
+A+ + SFF + G++ +QE SL+MG AY+VS V+LFDEY ND+LYLREGSC PRP YR
Sbjct: 143 VASFLSSFFFGMRAGLIAYQEASLQMGLAYMVSAVALFDEYTNDMLYLREGSCFPRPKYR 202
Query: 319 KNVKHAVERKKLEGIDHNLKNRRDSSQNSKH------TLQQSRSMKWKIQQYKPVQVWDW 372
K + E G ++ ++N KH LQ+S++ IQ+ +P+Q+WDW
Sbjct: 203 KTDRMNNET----GQNNEVRNATSPLGEKKHHHKTMKALQRSKTFMEAIQRLRPIQIWDW 258
Query: 373 FFKSCEVNGRIVLRDGLISVKEIEECIFKGNCKKLSIKLPAWSLLQCLLTSAKSNSDGLV 432
F+SCE+NGRI+L +GLIS +++EECI KG CKKLSIKLPAW +LQCL+ SAK +S GL+
Sbjct: 259 LFRSCELNGRILLGEGLISAEDMEECIIKGKCKKLSIKLPAWCILQCLIRSAKHDSHGLL 318
Query: 433 ISDDIELTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVMRSKNDIPEDW 492
ISDD+E+T N PKD+VF+W +GPLL++KEQ+K LEL E EE CL+ L+M + ND P DW
Sbjct: 319 ISDDVEVTNFNWPKDKVFDWMLGPLLVIKEQMKQLELTEDEELCLRRLIMTNNNDKPSDW 378
Query: 493 DSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKILYIEALQASA 548
D GFPS DN+RRA+LQAIIRRLQGI ++S +P+FRR+F +LVK LY+EA++A A
Sbjct: 379 DDCGFPSSDNIRRARLQAIIRRLQGIVVNLSWVPSFRRRFIDLVKALYLEAVEAGA 434
>UniRef100_Q7XTC6 OSJNBa0064H22.21 protein [Oryza sativa]
Length = 583
Score = 283 bits (724), Expect = 9e-75
Identities = 152/358 (42%), Positives = 232/358 (64%), Gaps = 9/358 (2%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
D+ L T IA+ KSPYMLF+GW+RLL DL+ R GPFLET CVP AGLAI+ WPL V+G+VL
Sbjct: 207 DIPLYTVIALIKSPYMLFKGWQRLLHDLISREGPFLETVCVPIAGLAILFWPLVVVGSVL 266
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
AI+ S F+ LYG VVV+QE S + G +Y+V++V+ FDEY ND LYLREG+ LP+P YRK
Sbjct: 267 LAIVSSIFVGLYGAVVVYQEKSFQRGVSYVVAMVAEFDEYTNDWLYLREGTVLPKPSYRK 326
Query: 320 NVKHAVERKKLEGIDHNLKNRRDSSQNSK-----HTLQQSRSMKWKIQQYKPVQVWDWFF 374
K + + + ++K S N TL +RS++ IQ+ K VQ+W+
Sbjct: 327 R-KSSSSTEFSVRTNASVKGGDQPSSNEAPAMLVPTLAPARSVREAIQEVKMVQIWENMM 385
Query: 375 KSCEVNGRIVLRDGLISVKEIEECI--FKGNCKKLSIKLPAWSLLQCLLTSAKSNSDGLV 432
K+CE+ GR +L +I+ ++ E + + + + + +P++SLL +L S K+ S GL+
Sbjct: 386 KNCELRGRDLLNLNVITTVDLTEWLRTKESGHEAIGLGVPSYSLLCMILHSIKAGSGGLL 445
Query: 433 ISDDIELTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVMRSKN-DIPED 491
I + IE+ + N P+DR+ +WF+ P+L++K+Q++ L++ E E L++L + N +
Sbjct: 446 IGNGIEINQYNRPQDRLIDWFLHPVLVLKDQIQALKMTEEEVRFLEKLTLFIGNSERANG 505
Query: 492 WDSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKILYIEALQASAS 549
WD+ +D VR Q+QAI RRL GI SMS+ PT+RR+ R+++K+L +++ S
Sbjct: 506 WDNGAEIPQDPVRAGQIQAISRRLVGIVRSMSKFPTYRRRHRHVMKLLVTYSVEKEGS 563
Score = 44.3 bits (103), Expect = 0.009
Identities = 18/39 (46%), Positives = 29/39 (74%)
Query: 21 LAGALIGPIAFAIMVVGNSAVIIGLWTAHVFWTYYCVAR 59
+ A++GP+A A+MV+GN VI+ L+ AHV+WT Y + +
Sbjct: 41 IKAAVVGPVAAALMVLGNVGVILLLFPAHVWWTIYSLIK 79
>UniRef100_Q84K38 Hypothetical protein At5g40640 [Arabidopsis thaliana]
Length = 586
Score = 280 bits (715), Expect = 1e-73
Identities = 157/347 (45%), Positives = 210/347 (60%), Gaps = 20/347 (5%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
D +I+ +A+ KSPYMLF+GW RL DL+GR GPFLET CVP AGL I+LWPLAV+GAVL
Sbjct: 195 DFPVISLLALCKSPYMLFKGWHRLFHDLIGREGPFLETMCVPIAGLVILLWPLAVVGAVL 254
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
+++ S FL YGGVV +QE S G Y+V+ VS++DEY ND+L + EGSC PRPIYR+
Sbjct: 255 GSVVSSVFLGAYGGVVSYQESSFFFGLCYVVASVSIYDEYSNDVLDMPEGSCFPRPIYRR 314
Query: 320 NVKHAVERKKLEGIDHNLKNRRDSSQNSKHTLQQSRSMKWKIQQYKPVQVWDWFFKSCEV 379
N EG + K T + S K + KP+ + + F C
Sbjct: 315 NE---------EGASTAFSGGLSRPNSFKTTPSRGGSNKGPMIDLKPLDLLEALFVECRR 365
Query: 380 NGRIVLRDGLISVKEIEECIFKGNCKKLSIKLPAWSLLQCLLTSAKSNSDGLVISDDI-E 438
+G I++ G+I+ K+IEE + +S LPA+SLL LL S KSNS GL++ D + E
Sbjct: 366 HGEIMVTKGIINSKDIEEAKSSKGSQVISFGLPAYSLLHELLRSIKSNSTGLLLGDGVTE 425
Query: 439 LTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVM------RSKNDIPEDW 492
+T N PKD F+WF+ P LI+K+Q++ L E EE L +LV+ R K+ I E
Sbjct: 426 ITTRNRPKDAFFDWFLNPFLILKDQIEAANLSEEEEEYLGKLVLLFGDSERLKSSIVE-- 483
Query: 493 DSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKIL 539
+ P +R+A+L A RRLQG+ S+SR PTFRR F LVK L
Sbjct: 484 --SESPPLTELRKAELDAFARRLQGLTKSVSRYPTFRRHFVELVKKL 528
Score = 38.9 bits (89), Expect = 0.40
Identities = 18/53 (33%), Positives = 28/53 (51%), Gaps = 1/53 (1%)
Query: 21 LAGALIGPIAFAIMVVGNSAVIIGLWTAHVFWTYYCVARLYFLSKFFVICFIC 73
+ G ++ P+ + +GNSA+I+GL H WT Y +A L I F+C
Sbjct: 30 IKGIVLCPLICLTVAIGNSAIILGLLPVHAIWTLYSIASAKQLGPILKI-FLC 81
>UniRef100_Q6K5K7 Hypothetical protein OJ1212_D02.6 [Oryza sativa]
Length = 585
Score = 278 bits (710), Expect = 4e-73
Identities = 157/363 (43%), Positives = 220/363 (60%), Gaps = 20/363 (5%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
D ++ T IA +K P MLF+GWKRL+EDLVGR GPFLET CVPFAGLAI+LWP AV GA L
Sbjct: 196 DGIMFTLIAFYKFPVMLFKGWKRLIEDLVGREGPFLETACVPFAGLAILLWPFAVFGAFL 255
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
A+II S L + VVV+QE SL MG Y++S V++FDEY ND+L + GSC PR YRK
Sbjct: 256 ASIISSIPLGAFAAVVVYQESSLIMGLNYVISSVAIFDEYTNDVLDMAPGSCFPRFKYRK 315
Query: 320 NVKHAVERKKLEGIDHNLKNRRDSSQNSKHTLQQSRSMKWKIQQYKPVQVWDWFFKSCEV 379
N EG + Q+ K + S K ++ P ++ D F+ C+
Sbjct: 316 N------EASTEGGSLSRPASFKDKQDGKKAPSRVTSFKGSFDEFNPFKLLDHLFEECKH 369
Query: 380 NGRIVLRDGLISVKEIEEC-IFKGNCKKLSIKLPAWSLLQCLLTSAKSNSDGLVISDDIE 438
G +++ +G+I+ K+IEE K L++ LPA+ +L L+ SAK+NSDGL++SD E
Sbjct: 370 RGEVLVAEGVITPKDIEETKSGKIGIGVLNVGLPAYVILHALIRSAKANSDGLILSDGSE 429
Query: 439 LTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVM------RSKNDIPEDW 492
+T N PK+ +F+WF PL+++KEQ+K E EE LK+ V+ R K +P
Sbjct: 430 ITSDNRPKNTIFDWFFDPLMVIKEQIKAQNFTEEEEAYLKKRVLLTSDPKRLKEVVPH-- 487
Query: 493 DSTGFPSKDNVRR-AQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKILYIEALQASASAK 551
PS N R+ A++ A+ RRLQGI S+SR PT +R+F +LV+ L E + ++
Sbjct: 488 ----LPSSLNERKQAEIDALSRRLQGITRSISRYPTAKRRFDDLVRSLSEELERTMGGSQ 543
Query: 552 EGN 554
G+
Sbjct: 544 SGS 546
Score = 56.2 bits (134), Expect = 2e-06
Identities = 23/60 (38%), Positives = 38/60 (63%)
Query: 21 LAGALIGPIAFAIMVVGNSAVIIGLWTAHVFWTYYCVARLYFLSKFFVICFICSNSISLI 80
+ GAL+ P A+ IM++G SA+++GLW HV WTYYC+ R + + + + S+ L+
Sbjct: 30 IKGALLFPWAWLIMMIGISALVLGLWPMHVIWTYYCIIRSKLVGPVVKLLLLVAASVILV 89
>UniRef100_Q7FAW5 OSJNBa0081L15.3 protein [Oryza sativa]
Length = 588
Score = 277 bits (708), Expect = 7e-73
Identities = 157/350 (44%), Positives = 214/350 (60%), Gaps = 18/350 (5%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
D ++ T IAI+K P MLF+GWKRL++D++GR GPFLET CVPFAGLAI+LWP AV+GAVL
Sbjct: 196 DGIMFTLIAIYKCPVMLFKGWKRLIQDMIGREGPFLETACVPFAGLAILLWPFAVVGAVL 255
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
A+I+ S L +G VV +QE SLKMG +Y+VS VS+FDEY ND+L + GSC PR YRK
Sbjct: 256 ASILSSIPLGAFGAVVAYQESSLKMGLSYVVSSVSIFDEYTNDVLDMAPGSCFPRLKYRK 315
Query: 320 NVKHAVERKKLEGIDHNLKNRRDSSQNSKHTLQQSRSMKWKIQQYKPVQVWDWFFKSCEV 379
E G + Q K + S K I+ + P ++ + F C+
Sbjct: 316 R-----EDSSHGGSLSRPTSFNKEKQEGKKPPARVTSFKNSIEDFNPFKLLEHLFVECKH 370
Query: 380 NGRIVLRDGLISVKEIEEC-IFKGNCKKLSIKLPAWSLLQCLLTSAKSNSDGLVISDDIE 438
G ++ +G+I++K+IEE K L++ LPA+ +L LL SAK+NS GL++SD E
Sbjct: 371 QGETLVNEGVITMKDIEETKSGKVGTGVLNVGLPAYVILNALLRSAKANSVGLLLSDGSE 430
Query: 439 LTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVM------RSKNDIPEDW 492
+T N PK ++EWF PLL++KEQ+K E EE LK V+ R K +P+
Sbjct: 431 ITSDNRPKHTLYEWFFDPLLVIKEQIKAENFTEEEEKYLKMRVLLIGGPDRVKGSLPD-- 488
Query: 493 DSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKILYIE 542
PS D ++A++ A RRLQGI S+SR PT +R+F LVK L E
Sbjct: 489 ----VPSLDERKKAEIDAFARRLQGITKSISRYPTAKRRFDILVKQLLSE 534
Score = 53.1 bits (126), Expect = 2e-05
Identities = 24/60 (40%), Positives = 35/60 (58%)
Query: 21 LAGALIGPIAFAIMVVGNSAVIIGLWTAHVFWTYYCVARLYFLSKFFVICFICSNSISLI 80
+ G LI P A IM +G SA+I+GLW HV WTYYC+ R + + + + + +LI
Sbjct: 30 IKGILICPWACLIMAIGLSALILGLWPMHVIWTYYCIIRTKLVGPVVKLLLLIAATATLI 89
>UniRef100_Q9FM34 Dbj|BAA95714.1 [Arabidopsis thaliana]
Length = 595
Score = 273 bits (698), Expect = 1e-71
Identities = 158/356 (44%), Positives = 210/356 (58%), Gaps = 29/356 (8%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
D +I+ +A+ KSPYMLF+GW RL DL+GR GPFLET CVP AGL I+LWPLAV+GAVL
Sbjct: 195 DFPVISLLALCKSPYMLFKGWHRLFHDLIGREGPFLETMCVPIAGLVILLWPLAVVGAVL 254
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
+++ S FL YGGVV +QE S G Y+V+ VS++DEY ND+L + EGSC PRPIYR+
Sbjct: 255 GSVVSSVFLGAYGGVVSYQESSFFFGLCYVVASVSIYDEYSNDVLDMPEGSCFPRPIYRR 314
Query: 320 NVKHAVERKKLEGIDHNLKNRRDSSQNSKHTLQQSRSMKWKIQQYKPVQV---------W 370
N EG + K T + S K + KP+ V
Sbjct: 315 NE---------EGASTAFSGGLSRPNSFKTTPSRGGSNKGPMIDLKPLDVLTSHTLLLLL 365
Query: 371 DWFFKSCEVNGRIVLRDGLISVKEIEECIFKGNCKKLSIKLPAWSLLQCLLTSAKSNSDG 430
+ F C +G I++ G+I+ K+IEE + +S LPA+SLL LL S KSNS G
Sbjct: 366 EALFVECRRHGEIMVTKGIINSKDIEEAKSSKGSQVISFGLPAYSLLHELLRSIKSNSTG 425
Query: 431 LVISDDI-ELTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVM------R 483
L++ D + E+T N PKD F+WF+ P LI+K+Q++ L E EE L +LV+ R
Sbjct: 426 LLLGDGVTEITTRNRPKDAFFDWFLNPFLILKDQIEAANLSEEEEEYLGKLVLLFGDSER 485
Query: 484 SKNDIPEDWDSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKIL 539
K+ I E + P +R+A+L A RRLQG+ S+SR PTFRR F LVK L
Sbjct: 486 LKSSIVE----SESPPLTELRKAELDAFARRLQGLTKSVSRYPTFRRHFVELVKKL 537
Score = 38.9 bits (89), Expect = 0.40
Identities = 18/53 (33%), Positives = 28/53 (51%), Gaps = 1/53 (1%)
Query: 21 LAGALIGPIAFAIMVVGNSAVIIGLWTAHVFWTYYCVARLYFLSKFFVICFIC 73
+ G ++ P+ + +GNSA+I+GL H WT Y +A L I F+C
Sbjct: 30 IKGIVLCPLICLTVAIGNSAIILGLLPVHAIWTLYSIASAKQLGPILKI-FLC 81
>UniRef100_Q8GUM4 Hypothetical protein At3g27390 [Arabidopsis thaliana]
Length = 588
Score = 266 bits (679), Expect = 2e-69
Identities = 158/363 (43%), Positives = 216/363 (58%), Gaps = 25/363 (6%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
D +I+ +AI KSPYMLF+GW RL DL+GR GPFLET CVP AGLAI+LWPLAV GAV+
Sbjct: 195 DPPVISLVAICKSPYMLFKGWHRLFHDLIGREGPFLETMCVPIAGLAILLWPLAVTGAVI 254
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
++I S FL Y GVV +QE S G YIV+ VS++DEY D+L L EGSC PRP YR+
Sbjct: 255 GSVISSIFLGAYAGVVSYQESSFYYGLCYIVASVSIYDEYSTDILDLPEGSCFPRPKYRR 314
Query: 320 NVKHAVERKKLEGIDHNLKNRRDSSQNSKHTLQQSRSMKWKIQQYKPVQVWDWFFKSCEV 379
+ E G L + +++S + S++ + KP+ + + F C
Sbjct: 315 KDE---EPTPFSGPVPRLGSVKNASS------MRGGSVRVPMIDIKPLDLLNELFVECRR 365
Query: 380 NGRIVLRDGLISVKEIEECIFKGNCKKLSIKLPAWSLLQCLLTSAKSNSDGLVISDDI-E 438
G ++ GLI+ K+IEE + +S+ LPA+ LL +L S K+NS GL++SD + E
Sbjct: 366 YGEVLATKGLINSKDIEEARSSKGSQVISVGLPAYGLLYEILRSVKANSSGLLLSDGVTE 425
Query: 439 LTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELV--------MRSKNDIPE 490
+T MN PKD F+WF+ P LI+KEQ+K L E EE L LV ++S N I
Sbjct: 426 ITTMNRPKDVFFDWFLNPFLILKEQMKATNLSEEEEKYLGRLVLLFGDPERLKSSNAI-- 483
Query: 491 DWDSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKILYIEA-LQASAS 549
+ P +RA+L A RR+QG+ ++SR PTFRR F LVK L + L+ + S
Sbjct: 484 ----SASPPLTERKRAELDAFARRMQGLTKTVSRYPTFRRHFVALVKKLSEDLDLKDNNS 539
Query: 550 AKE 552
AK+
Sbjct: 540 AKD 542
Score = 42.4 bits (98), Expect = 0.036
Identities = 19/65 (29%), Positives = 34/65 (52%), Gaps = 1/65 (1%)
Query: 21 LAGALIGPIAFAIMVVGNSAVIIGLWTAHVFWTYYCVARLYFLSKFFVICFICSNSISLI 80
+ G ++ P+ ++ +GNSAVI+ L H+ WT+Y + + I F+C + I
Sbjct: 30 IKGIVLCPLVCLVVTIGNSAVILSLLPVHIVWTFYSIVSAKQVGPILKI-FLCLCLPAAI 88
Query: 81 NCWPL 85
WP+
Sbjct: 89 ILWPI 93
>UniRef100_Q9LTZ1 Emb|CAB40985.1 [Arabidopsis thaliana]
Length = 545
Score = 255 bits (652), Expect = 2e-66
Identities = 160/384 (41%), Positives = 220/384 (56%), Gaps = 40/384 (10%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
D +I+ +AI KSPYMLF+GW RL DL+GR GPFLET CVP AGLAI+LWPLAV GAV+
Sbjct: 125 DPPVISLVAICKSPYMLFKGWHRLFHDLIGREGPFLETMCVPIAGLAILLWPLAVTGAVI 184
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
++I S FL Y GVV +QE S G YIV+ VS++DEY D+L L EGSC PRP YR+
Sbjct: 185 GSVISSIFLGAYAGVVSYQESSFYYGLCYIVASVSIYDEYSTDILDLPEGSCFPRPKYRR 244
Query: 320 ----------------NVKHAVERK----KLEGIDHNLKNRRDSSQNSKHTLQQSRSMKW 359
+VK+A + ++ ID D S + T ++ +
Sbjct: 245 KDEEPTPFSGPVPRLGSVKNASSMRGGSVRVPMID---IKPLDVSPHLIFTKPNVKNQSF 301
Query: 360 KIQQYKP-VQVWDWFFKSCEVNGRIVLRDGLISVKEIEECIFKGNCKKLSIKLPAWSLLQ 418
+ + +Q+ + F C G ++ GLI+ K+IEE + +S+ LPA+ LL
Sbjct: 302 VVLDFDVCMQLLNELFVECRRYGEVLATKGLINSKDIEEARSSKGSQVISVGLPAYGLLY 361
Query: 419 CLLTSAKSNSDGLVISDDI-ELTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCL 477
+L S K+NS GL++SD + E+T MN PKD F+WF+ P LI+KEQ+K L E EE L
Sbjct: 362 EILRSVKANSSGLLLSDGVTEITTMNRPKDVFFDWFLNPFLILKEQMKATNLSEEEEEYL 421
Query: 478 KELV--------MRSKNDIPEDWDSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFR 529
LV ++S N I + P +RA+L A RR+QG+ ++SR PTFR
Sbjct: 422 GRLVLLFGDPERLKSSNAI------SASPPLTERKRAELDAFARRMQGLTKTVSRYPTFR 475
Query: 530 RKFRNLVKILYIEA-LQASASAKE 552
R F LVK L + L+ + SAK+
Sbjct: 476 RHFVALVKKLSEDLDLKDNNSAKD 499
>UniRef100_Q5XV58 Hypothetical protein [Arabidopsis thaliana]
Length = 569
Score = 250 bits (639), Expect = 7e-65
Identities = 138/376 (36%), Positives = 224/376 (58%), Gaps = 20/376 (5%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
D+ L T+IA+ KSPY+L +GW RL +D + R GPFLE C+P AGL ++LWP+ V+G +L
Sbjct: 196 DIPLFTAIAVIKSPYLLLKGWYRLAQDAINREGPFLEIACIPVAGLTVLLWPIVVIGFIL 255
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
I S F+ LYG VVV QE S + G +Y+++VV FDEY ND LYLREG+ P+P YR
Sbjct: 256 VTIFSSIFVGLYGAVVVFQERSFRRGVSYVIAVVGEFDEYTNDWLYLREGTIFPKPRYR- 314
Query: 320 NVKHAVERKKLEGIDHNLKNRRDSSQNSKH-------TLQQSRSMKWKIQQYKPVQVWDW 372
+ ++ I H R +S S +L S S++ IQ+ + VQ+W+
Sbjct: 315 -MGRGSFSSEVSVIVHPSDVTRVNSSGSVDAPAMLVPSLVHSVSVREAIQEVRMVQIWEH 373
Query: 373 FFKSCEVNGRIVLRDGLISVKEIEECIFKG----NCKKLSIKLPAWSLLQCLLTSAKSNS 428
E+ G+ +L +++ ++ E + KG +++ LP+++LL LL+S K+
Sbjct: 374 MMGWFEMQGKELLDAEVLTPTDLYESL-KGRHGNESSIINVGLPSYALLHTLLSSIKAGV 432
Query: 429 DGLVISDDIELTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVMRSKND- 487
G+++ D E+T +N P+D+ +W P++ +K+Q++ L+L ESE L+++V ++
Sbjct: 433 HGVLLLDGSEVTHLNRPQDKFLDWVFNPIMALKDQIRALKLGESEVKYLEKVVFFGNHEQ 492
Query: 488 IPEDWDSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKILYI-----E 542
E WD+ P ++N+R AQ+Q I RR+ G+ S+S++PT+RR+FR +VK L +
Sbjct: 493 RMEAWDNHSNPPQENLRTAQIQGISRRMMGMVRSVSKLPTYRRRFRQVVKALITYYSEKQ 552
Query: 543 ALQASASAKEGNNIDE 558
L + S G+ I+E
Sbjct: 553 GLNRTGSMNSGDFIEE 568
Score = 44.3 bits (103), Expect = 0.009
Identities = 19/39 (48%), Positives = 27/39 (68%)
Query: 21 LAGALIGPIAFAIMVVGNSAVIIGLWTAHVFWTYYCVAR 59
+ G ++GPIA ++VGN VI+ L+ AHV WT Y VA+
Sbjct: 30 IKGLIVGPIAGLTLIVGNVGVILCLFPAHVTWTIYAVAK 68
>UniRef100_O23182 Hypothetical protein C7A10.330 [Arabidopsis thaliana]
Length = 519
Score = 224 bits (570), Expect = 7e-57
Identities = 121/327 (37%), Positives = 197/327 (60%), Gaps = 15/327 (4%)
Query: 200 DVVLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVL 259
D+ L T+IA+ KSPY+L +GW RL +D + R GPFLE C+P AGL ++LWP+ V+G +L
Sbjct: 196 DIPLFTAIAVIKSPYLLLKGWYRLAQDAINREGPFLEIACIPVAGLTVLLWPIVVIGFIL 255
Query: 260 AAIIVSFFLSLYGGVVVHQEDSLKMGFAYIVSVVSLFDEYVNDLLYLREGSCLPRPIYRK 319
I S F+ LYG VVV QE S + G +Y+++VV FDEY ND LYLREG+ P+P YR
Sbjct: 256 VTIFSSIFVGLYGAVVVFQERSFRRGVSYVIAVVGEFDEYTNDWLYLREGTIFPKPRYR- 314
Query: 320 NVKHAVERKKLEGIDHNLKNRRDSSQNSKH-------TLQQSRSMKWKIQQYKPVQVWDW 372
+ ++ I H R +S S +L S S++ IQ+ + VQ+W+
Sbjct: 315 -MGRGSFSSEVSVIVHPSDVTRVNSSGSVDAPAMLVPSLVHSVSVREAIQEVRMVQIWEH 373
Query: 373 FFKSCEVNGRIVLRDGLISVKEIEECIFKG----NCKKLSIKLPAWSLLQCLLTSAKSNS 428
E+ G+ +L +++ ++ E + KG +++ LP+++LL LL+S K+
Sbjct: 374 MMGWFEMQGKELLDAEVLTPTDLYESL-KGRHGNESSIINVGLPSYALLHTLLSSIKAGV 432
Query: 429 DGLVISDDIELTRMNGPKDRVFEWFIGPLLIMKEQLKNLELEESEETCLKELVMRSKND- 487
G+++ D E+T +N P+D+ +W P++++K+Q++ L+L ESE L+++V+ ++
Sbjct: 433 HGVLLLDGSEVTHLNRPQDKFLDWVFNPIMVLKDQIRALKLGESEVKYLEKVVLFGNHEQ 492
Query: 488 IPEDWDSTGFPSKDNVRRAQLQAIIRR 514
E WD+ P ++N+R AQ+Q I RR
Sbjct: 493 RMEAWDNHSNPPQENLRTAQIQGISRR 519
Score = 44.3 bits (103), Expect = 0.009
Identities = 19/39 (48%), Positives = 27/39 (68%)
Query: 21 LAGALIGPIAFAIMVVGNSAVIIGLWTAHVFWTYYCVAR 59
+ G ++GPIA ++VGN VI+ L+ AHV WT Y VA+
Sbjct: 30 IKGLIVGPIAGLTLIVGNVGVILCLFPAHVTWTIYAVAK 68
>UniRef100_Q5XV56 Hypothetical protein [Arabidopsis thaliana]
Length = 255
Score = 121 bits (303), Expect = 6e-26
Identities = 69/219 (31%), Positives = 130/219 (58%), Gaps = 11/219 (5%)
Query: 350 TLQQSRSMKWKIQQYKPVQVWDWFFKSCEVNGRIVLRDGLISVKEIEECIFKGN----CK 405
+L S S++ IQ+ + VQ+W+ E+ G+ +L +++ ++ E + KG
Sbjct: 37 SLVHSVSVREAIQEVRMVQIWEHMMGWFEMQGKELLDAEVLTPTDLYESL-KGRHGNESS 95
Query: 406 KLSIKLPAWSLLQCLLTSAKSNSDGLVISDDIELTRMNGPKDRVFEWFIGPLLIMKEQLK 465
+++ LP+++LL LL+S K+ G+++ D E+T +N P+D+ +W P++ +K+Q++
Sbjct: 96 IINVGLPSYALLHTLLSSIKAGVHGVLLLDGSEVTHLNRPQDKFLDWVFNPIMALKDQIR 155
Query: 466 NLELEESEETCLKELVMRSKND-IPEDWDSTGFPSKDNVRRAQLQAIIRRLQGIGSSMSR 524
L+L ESE L+++V ++ E WD+ P ++N+R AQ+Q I RR+ G+ S+S+
Sbjct: 156 ALKLGESEVKYLEKVVFFGNHEQRMEAWDNHSNPPQENLRTAQIQGISRRMMGMVRSVSK 215
Query: 525 MPTFRRKFRNLVKILYI-----EALQASASAKEGNNIDE 558
+PT+RR+FR +VK L + L + S G+ I+E
Sbjct: 216 LPTYRRRFRQVVKALITYYSEKQGLNRTGSMNSGDFIEE 254
>UniRef100_Q9KE57 BH1001 protein [Bacillus halodurans]
Length = 448
Score = 45.8 bits (107), Expect = 0.003
Identities = 42/134 (31%), Positives = 63/134 (46%), Gaps = 10/134 (7%)
Query: 369 VWDWFFKSCEVNG--RIVLRDGLISVKEIEECIFKGNCKKLSIKLPAWS---LLQCLLTS 423
V +WF S EVNG R +L + L + K +EE GN + K W +L+ L
Sbjct: 171 VGEWF--STEVNGVIRPILEENLQNAKLVEEIEIDGNLFETREKNEEWVDKFILEKALLY 228
Query: 424 AKSNSDGLVISDDIELTRMNGPKDRVFEWFIGPL--LIMKEQLKNLELEESEETCLKELV 481
K N+DG IS++ + KD F+ FI L + E++K EE ET ++E +
Sbjct: 229 MKDNTDGEYISEEDSWIYLQLDKDLFFQSFIHALHEVEANERMKE-AFEEDGETTVEEAI 287
Query: 482 MRSKNDIPEDWDST 495
+ + E D T
Sbjct: 288 KELEQSLEEIEDYT 301
>UniRef100_Q6WD23 Chitin synthase variant 1 [Tribolium castaneum]
Length = 1558
Score = 38.5 bits (88), Expect = 0.52
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 9/76 (11%)
Query: 14 IHYKVIHLAGALIGPIAFAIMVVGN--SAVIIGLWTAHVFWTYYCVARLYFLSKFFVICF 71
I Y+++ + G ++GP +M+VG +A I WT+ YY + + F F ++CF
Sbjct: 910 IGYQILLMGGTILGPGTIFLMLVGAFVAAFQIDNWTSF----YYNIIPILF---FMLVCF 962
Query: 72 ICSNSISLINCWPLKT 87
C ++I LI L T
Sbjct: 963 TCKSNIQLIVAQILST 978
>UniRef100_Q6WD22 Chitin synthase variant 2 [Tribolium castaneum]
Length = 1558
Score = 38.5 bits (88), Expect = 0.52
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 9/76 (11%)
Query: 14 IHYKVIHLAGALIGPIAFAIMVVGN--SAVIIGLWTAHVFWTYYCVARLYFLSKFFVICF 71
I Y+++ + G ++GP +M+VG +A I WT+ YY + + F F ++CF
Sbjct: 910 IGYQILLMGGTILGPGTIFLMLVGAFVAAFQIDNWTSF----YYNIIPILF---FMLVCF 962
Query: 72 ICSNSISLINCWPLKT 87
C ++I LI L T
Sbjct: 963 TCKSNIQLIVAQILST 978
>UniRef100_Q6WD24 Chitin synthase CHS2 [Tribolium castaneum]
Length = 1464
Score = 37.0 bits (84), Expect = 1.5
Identities = 25/87 (28%), Positives = 44/87 (49%), Gaps = 16/87 (18%)
Query: 2 IIINVECIVKVN-------IHYKVIHLAGALIGPIAFAIMVVGNSAVIIGL--WTAHVFW 52
++++ E VK+N I Y++I + G +IGP +M+VG GL W++ +W
Sbjct: 864 LLMDYEHTVKINENISMLYIGYQIILMIGTVIGPGTIFLMLVGAFVAAFGLDQWSS-FYW 922
Query: 53 TYYCVARLYFLSKFFVICFICSNSISL 79
L ++ F ++C CS+ I L
Sbjct: 923 ------NLLPIAVFILVCATCSSDIQL 943
>UniRef100_Q23579 Helix loop helix protein 10 [Caenorhabditis elegans]
Length = 170
Score = 36.6 bits (83), Expect = 2.0
Identities = 26/96 (27%), Positives = 48/96 (49%), Gaps = 4/96 (4%)
Query: 446 KDRVFEWFIGPLLIMKEQLKNLELEESEETCLKE--LVMRSKNDIPEDWDSTGFPSKDNV 503
+ +F WF+ +L L +E E KE LV ++K+++ ++ +ST P++++
Sbjct: 12 QQNLFAWFLQSMLEASASQPQLTQDEPPENDTKENDLVKQNKSEVNDENESTPSPTQNSR 71
Query: 504 RRAQLQAIIRRLQGIGSSMSRMPTFRRKFRNLVKIL 539
RR I RR+ +G +R + RN V+ L
Sbjct: 72 RRTSTGKIDRRM--VGKMCTRRYEANARERNRVQQL 105
>UniRef100_UPI00002A73EA UPI00002A73EA UniRef100 entry
Length = 640
Score = 35.8 bits (81), Expect = 3.4
Identities = 25/126 (19%), Positives = 59/126 (45%), Gaps = 13/126 (10%)
Query: 202 VLITSIAIWKSPYMLFRGWKRLLEDLVGRRGPFLETECVPFAGLAIILWPLAVLGAVLAA 261
V++TS W+ +++F G R+ E+ + + E+ V L I+ G + +
Sbjct: 432 VVMTSFYSWRLMFLVFNGKTRMAEEDLSK---VHESSSVMLIPLIILAIGALFFGFLFKS 488
Query: 262 IIVSFFLSLY--GGVVVHQED--SLKMGFAYIVSVVSLFDEYVNDLLYLREGSC------ 311
+ ++ ++ G ++VH ED S+ + Y+ +++ + + +Y+R+
Sbjct: 489 YFIGYYSDVFWNGSIIVHHEDHHSIPLLIYYLPAILGIVGFIIAWFMYIRKSDMAKNIAE 548
Query: 312 LPRPIY 317
L +P+Y
Sbjct: 549 LNKPLY 554
>UniRef100_Q7QX50 GLP_192_30681_21532 [Giardia lamblia ATCC 50803]
Length = 3049
Score = 35.8 bits (81), Expect = 3.4
Identities = 23/88 (26%), Positives = 48/88 (54%), Gaps = 5/88 (5%)
Query: 144 KTNFIIASLMVVGQQFKQAVQWFRMS--QTFVFTPIFHTWMN*EKI*TLKKSHLISTFDV 201
+++++I S + Q Q V + R++ + + +PIF W+ ++ ++K+ L+ T+D
Sbjct: 343 RSSYLIHSFVFDIQYRVQVVLYIRLTLLKYTLISPIFVAWLTTIRMLRIRKTFLLLTYDK 402
Query: 202 VLITSIAIWKSPYMLFRGWKRLLEDLVG 229
+LIT+ W L RG +L+ +G
Sbjct: 403 LLITAFYRWSD---LARGRHKLMTKYLG 427
>UniRef100_UPI0000432841 UPI0000432841 UniRef100 entry
Length = 850
Score = 35.4 bits (80), Expect = 4.4
Identities = 24/95 (25%), Positives = 47/95 (49%), Gaps = 16/95 (16%)
Query: 2 IIINVECIVKVN-------IHYKVIHLAGALIGPIAFAIMVVGN--SAVIIGLWTAHVFW 52
++++ + +K+N I Y+++ + G ++GP +M+VG +A I WT+
Sbjct: 678 LLMDAKRTIKINDNISLPYISYQILLMGGTILGPGTIFLMLVGAFVAAFKIDNWTSF--- 734
Query: 53 TYYCVARLYFLSKFFVICFICSNSISLINCWPLKT 87
YY + + F ++CF C +I L+ L T
Sbjct: 735 -YYNIIPILL---FMLVCFTCKANIQLLCAQILST 765
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.333 0.144 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 858,805,378
Number of Sequences: 2790947
Number of extensions: 34135750
Number of successful extensions: 103640
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 103577
Number of HSP's gapped (non-prelim): 40
length of query: 558
length of database: 848,049,833
effective HSP length: 133
effective length of query: 425
effective length of database: 476,853,882
effective search space: 202662899850
effective search space used: 202662899850
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 77 (34.3 bits)
Medicago: description of AC144618.8


