

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.4 - phase: 0
(412 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
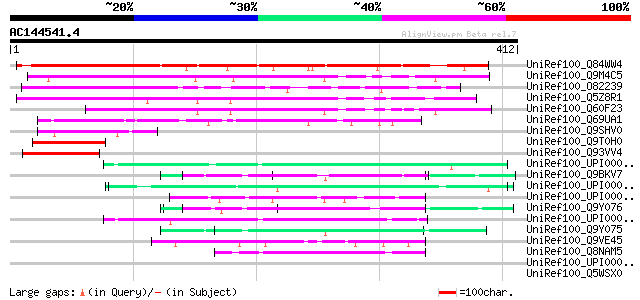
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84WW4 Hypothetical protein At3g05545 [Arabidopsis tha... 342 9e-93
UniRef100_Q9M4C5 VIP2 protein [Avena fatua] 250 4e-65
UniRef100_O82239 Hypothetical protein At2g47700 [Arabidopsis tha... 235 2e-60
UniRef100_Q5Z8R1 Putative VIP2 protein [Oryza sativa] 221 4e-56
UniRef100_Q60F23 Putative transcription factor VIP2 [Oryza sativa] 184 5e-45
UniRef100_Q69UA1 Putative VIP2 protein [Oryza sativa] 179 1e-43
UniRef100_Q9SHV0 Hypothetical protein At2g15260 [Arabidopsis tha... 89 2e-16
UniRef100_Q9T0H0 Hypothetical protein T6G15.40 [Arabidopsis thal... 82 2e-14
UniRef100_Q93VV4 ORF162 [Marchantia polymorpha] 65 3e-09
UniRef100_UPI0000363AC1 UPI0000363AC1 UniRef100 entry 60 9e-08
UniRef100_Q9BKV7 Ppg3 [Leishmania major] 56 2e-06
UniRef100_UPI00003641A6 UPI00003641A6 UniRef100 entry 51 7e-05
UniRef100_UPI000049A2DE UPI000049A2DE UniRef100 entry 50 1e-04
UniRef100_Q9Y076 Proteophosphoglycan precursor [Leishmania major] 50 1e-04
UniRef100_UPI000036004A UPI000036004A UniRef100 entry 49 2e-04
UniRef100_Q9Y075 Proteophosphoglycan [Leishmania major] 49 3e-04
UniRef100_Q9VE45 CG7709-PA [Drosophila melanogaster] 48 4e-04
UniRef100_Q8NAM5 Hypothetical protein FLJ35107 [Homo sapiens] 47 8e-04
UniRef100_UPI000043211F UPI000043211F UniRef100 entry 47 0.001
UniRef100_Q5WSX0 Hypothetical protein [Legionella pneumophila st... 47 0.001
>UniRef100_Q84WW4 Hypothetical protein At3g05545 [Arabidopsis thaliana]
Length = 425
Score = 342 bits (878), Expect = 9e-93
Identities = 199/412 (48%), Positives = 255/412 (61%), Gaps = 53/412 (12%)
Query: 6 DDGDGDTRHRKSSDSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCP 65
D+GDG SV CSICLE V NGDR++A LQC HQFHLDCIGSAFNAKG MQCP
Sbjct: 27 DEGDG-------FGSVACSICLETVVKNGDRAWANLQCDHQFHLDCIGSAFNAKGVMQCP 79
Query: 66 NCRKIEKGQWLYANGSRSYPEFNMEDWTRDDDVYDL-SYSEMSLGVHWCPFGNFAQLPSS 124
NCRK+EKGQWLYANG RSYPEFN+EDW ++D+YD+ +YSEMS GVHWCPFG+ A+LP S
Sbjct: 80 NCRKVEKGQWLYANGCRSYPEFNVEDWVHEEDIYDIGAYSEMSFGVHWCPFGSSARLP-S 138
Query: 125 FEEREYSPIPYHDAPGPM--FTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGT--VSEVS 180
FE+ E+SP YHD G +TE +A ++G HPCPY+ Y GPVH +S+SGG VS+ S
Sbjct: 139 FEDGEFSPSSYHDLLGQQGYYTEPAAPTAG-HPCPYVTYFGPVHSPSSSSGGAAGVSDSS 197
Query: 181 NF-NHWN-ARPIHGDMSTSYTVPAVVLHYHSWDHH------SSHFSSGSSHLGAADQPSV 232
+F +HWN + G++ T Y P V HYH WD+H HFS+ +H+G+ QP+
Sbjct: 198 SFSSHWNTGSSVSGEVPTPYGFP-VDPHYHGWDYHPPPPPPPQHFSASGAHVGSPTQPTP 256
Query: 233 SQSNQRPTR--GSE--ATRSGSYMHPYPVGHSSVARAGNSAAS-SMIPPYPGSNARARDR 287
+ R +R GS+ R + P+ GHSS RAG+S AS PP+PGSNAR RDR
Sbjct: 257 PPAAARTSRANGSDMIRPRPPHFTRPFH-GHSSSGRAGSSVASVPRTPPFPGSNARTRDR 315
Query: 288 VEALQAYYHQ-----QQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSYIYL 342
++ALQAYY Q QP + R PV S RR + ++ + S SS DQ+G ++
Sbjct: 316 MQALQAYYQQSSAQSHQPDSPIVSRGPVFPSGRRPA-RGIASGMGSTSSSSDQAGGSGFI 374
Query: 343 PGRNFQEETRLPSHFHAWERDHLPSS----SLNQVGRESS-WRAYHQTASGT 389
F+ WERD S S+NQ+ RE + W + SG+
Sbjct: 375 -------------RFNIWERDPYMQSQQAYSVNQMDREPNIWTSSFNEGSGS 413
>UniRef100_Q9M4C5 VIP2 protein [Avena fatua]
Length = 442
Score = 250 bits (639), Expect = 4e-65
Identities = 151/394 (38%), Positives = 208/394 (52%), Gaps = 38/394 (9%)
Query: 15 RKSSDSVPCSICLEIV--SDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRKIEK 72
++ +V CSICL+ V + GDRS A+LQCGH+FHLDCIGSAFNAKG MQCPNCRKIEK
Sbjct: 25 KEEKTAVSCSICLDAVVAAAGGDRSTARLQCGHEFHLDCIGSAFNAKGVMQCPNCRKIEK 84
Query: 73 GQWLYANGSRSYPEFNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQLPSSFEEREYS- 131
G WLYANGSR + NM++W D+D+YD+SYSEM HWCPFG AQLPS FEE E S
Sbjct: 85 GNWLYANGSRPSQDVNMDEWAHDEDLYDVSYSEMPFRFHWCPFGRLAQLPSFFEEGESSP 144
Query: 132 PIPYHDAPGP-MFTEHSAV--SSGSHPCPYIAYVGPVHPSTSNSGGTVSEVS-----NFN 183
P+ +HD G +F EH +V + G+HPCPY+AY+ P+ S+S V E + +
Sbjct: 145 PVTFHDFMGQHVFPEHLSVPAAPGAHPCPYVAYLQPLPSLASSSSSHVPERTMDSSVYHD 204
Query: 184 HWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTR-G 242
HWN D +V H++ W H ++ +S+ G +QP V R R
Sbjct: 205 HWNHLAGPSDGRPLQSVQPTDFHHNHWGHLPHSYAQSNSNNGVTEQPGVPFGTMRAARVD 264
Query: 243 SEATRSGSYMHP--YPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQP 300
++ R GS P + G S +RA N +PP RA + + Q P
Sbjct: 265 GDSQRRGSVFSPSYFSNGSGSRSRAPN------VPPLVPQFMRAHSSLNE----QYLQNP 314
Query: 301 PNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSYIYLPG----RNFQEETRLPSH 356
+S + + A RS G + P P+ ++ PG + + + +
Sbjct: 315 SSS------LFAGAHRSGGMRPAPP----PPQPENPTFCLFPPGSSGHSSMETDDAGGNR 364
Query: 357 FHAWERDHLPSSSLNQVGRESSWRAYHQTASGTD 390
F+AWERD L V E+SW + Q+ ++
Sbjct: 365 FYAWERDRFAPYPLMPVDCETSWWSSQQSHGASE 398
>UniRef100_O82239 Hypothetical protein At2g47700 [Arabidopsis thaliana]
Length = 358
Score = 235 bits (599), Expect = 2e-60
Identities = 150/362 (41%), Positives = 185/362 (50%), Gaps = 52/362 (14%)
Query: 10 GDTRHRKSSDSVPCSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRK 69
G+ S V CSICLE V D+G RS AKLQCGHQFHLDCIGSAFN KGAMQCPNCR
Sbjct: 25 GNPEDSSSPVEVSCSICLESVLDDGTRSKAKLQCGHQFHLDCIGSAFNMKGAMQCPNCRN 84
Query: 70 IEKGQWLYANGS-RSYPEFNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQLPSSFEER 128
+EKGQWLYANGS R +PEF+MEDW ++D+Y LSY EM VHWCPFG +Q +SFEE
Sbjct: 85 VEKGQWLYANGSTRPFPEFSMEDWIPEEDLYGLSYPEMQYRVHWCPFGELSQAAASFEEL 144
Query: 129 EYSPIPYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNAR 188
E + YH H+A + S Y+AYVGP +T + S+ N +
Sbjct: 145 EPATTTYHT---EFHGHHAAAVNHS----YLAYVGPGPAATPRT-------SDNNSTDDH 190
Query: 189 PIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHL--GAADQPSVSQSNQRPTRGSEAT 246
P + + + V YH HHS FS ++H+ G D S A
Sbjct: 191 PWNSHSNDHFHQLPVAPQYH---HHSPSFSLPAAHVVDGEVD--------------SSAA 233
Query: 247 RSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQP--PNST 304
R Y HP+ H S N +S I Y GS+ + R E AY HQ+Q N
Sbjct: 234 RGLPYAHPFLFSHRS-----NQRSSPAINSYQGSSTQMR---EQHHAYNHQRQQHHANGP 285
Query: 305 TVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWERDH 364
T+ P+ S RR P PDQ+ + PG + + ET HAWERD
Sbjct: 286 TLASPLISMTRRGLPPPPPPP-----PMPDQNVGFFIYPGGHHEPET---DQIHAWERDW 337
Query: 365 LP 366
P
Sbjct: 338 FP 339
>UniRef100_Q5Z8R1 Putative VIP2 protein [Oryza sativa]
Length = 455
Score = 221 bits (562), Expect = 4e-56
Identities = 141/391 (36%), Positives = 200/391 (51%), Gaps = 39/391 (9%)
Query: 6 DDGDGDTRHRKSSDSVPCSICLE-IVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQC 64
D D D ++ VPCSICL+ +V+ GDRS A+LQCGH+FHLDCIGSAFNAKG MQC
Sbjct: 34 DTADDDDSGESAAAVVPCSICLDAVVAGGGDRSTARLQCGHEFHLDCIGSAFNAKGVMQC 93
Query: 65 PNCRKIEKGQWLYANGSRSYPEFNMEDWTRDDDVYDLSYSEMSLGV------HWCPFGNF 118
PNCR+IE+G WLYANGSR + + +DW D+D YD + E S V WCP G
Sbjct: 94 PNCRQIERGNWLYANGSRPSQDVSNDDWGHDEDFYDANQPETSRSVFLPFRFQWCPIGRL 153
Query: 119 AQLPSSFEEREYS-PIPYHDAPGPMFT-EHSAVSS--GSHPCPYIAYVGPVHPSTSNSGG 174
AQLPS F+E E + P+ +HD G FT EH VS+ + P PYIAY P+ S S+S
Sbjct: 154 AQLPSVFDEGESAPPVTFHDFMGQNFTSEHLPVSAPGATPPGPYIAYFQPLQSSASSSSS 213
Query: 175 TVSE-----VSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQ 229
V+E + +HWN P D TV + H++ W H + +S +S+ G A+Q
Sbjct: 214 HVTERTMDGTTYHDHWNPLPGPSDGRPLATVHPIDFHHNHWTHLPNSYSQPNSNNGVAEQ 273
Query: 230 PSVSQSNQR-PTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRV 288
++ R S++ + GS Y G S +R + +PP R +
Sbjct: 274 MAIPVVPMRVGGLDSDSQQRGSLPSVYGNGSGSRSRIPS------VPPMAPQFMRPHGNI 327
Query: 289 EALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSYIYLPGRNFQ 348
++Q NS+++ A+ RR++ + + P +G + +
Sbjct: 328 -------NEQYQQNSSSL---YAAPQRRTAVQAVQDSMNFTLFPQAPTGP------NSME 371
Query: 349 EETRLPSHFHAWERDHLPSSSLNQVGRESSW 379
E + F+AWERD L V E++W
Sbjct: 372 TEDAGGNQFYAWERDRFAPYPLMPVDSEANW 402
>UniRef100_Q60F23 Putative transcription factor VIP2 [Oryza sativa]
Length = 365
Score = 184 bits (466), Expect = 5e-45
Identities = 124/348 (35%), Positives = 176/348 (49%), Gaps = 38/348 (10%)
Query: 62 MQCPNCRKIEKGQWLYANGSRSYPEFNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQL 121
MQCPNCRKIEKG WLYANGSR + NM++W D+D+YD+SYSEM HWCPFG AQL
Sbjct: 1 MQCPNCRKIEKGNWLYANGSRPTQDVNMDEWAHDEDLYDVSYSEMPFRFHWCPFGRLAQL 60
Query: 122 PSSFEEREYS-PIPYHDAPGP-MFTEH-SAVSS---GSHPCPYIAYVGPVHPSTSNSGGT 175
PS FEE E S P+ +HD G MFTEH +AVSS +HPCPY+AY+ P+ S+S
Sbjct: 61 PSFFEEGESSPPVTFHDFMGQHMFTEHVAAVSSAPGAAHPCPYVAYLHPLPSLASSSSSH 120
Query: 176 VSE-----VSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQP 230
V E + + W+ D +V H++ W H + + +++ G A+Q
Sbjct: 121 VPERTMDGPAYHDPWHPLAGPSDGRPLQSVQPADFHHNHWAHVPNSYPQPNNNNGVAEQQ 180
Query: 231 SVSQSNQRPTR-GSEATRSGSYMHP--YPVGHSSVARAGNSAASSMIPPYPGSNARARDR 287
V R R + R GS + P + G S +RA N +PP RA
Sbjct: 181 GVPFGTTRAARVDGDTQRRGSSISPSYFSNGSGSRSRAPN------VPPMVPQFMRAHGS 234
Query: 288 VEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSYIYLPG--- 344
+ Q Q +S++ + + A RS G R+AP L P+ ++ PG
Sbjct: 235 IS------EQYQQSSSSS----LFAGAHRSGG-MRTAPPPPL---PENPAFCLFPPGSSG 280
Query: 345 -RNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTASGTDP 391
+ + + + F+AWERD L V E++W + Q+ ++P
Sbjct: 281 HNSMETDDAGGNRFYAWERDRFAPYPLMPVDCETNWWSSQQSHGTSEP 328
>UniRef100_Q69UA1 Putative VIP2 protein [Oryza sativa]
Length = 424
Score = 179 bits (454), Expect = 1e-43
Identities = 126/338 (37%), Positives = 167/338 (49%), Gaps = 42/338 (12%)
Query: 23 CSICLEIVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRKIEKGQWLYANGSR 82
CSICL+ V G RS A+LQCGH+FHLDCIGSAFNAKGAMQCPNCRKIEKG+WLYA+G
Sbjct: 25 CSICLDPVVAGG-RSVARLQCGHEFHLDCIGSAFNAKGAMQCPNCRKIEKGRWLYASGHH 83
Query: 83 SYPEFNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQLPSSFEEREYSPIPYHDAPGPM 142
P+ ++ W + YD++ S++ G WCPF F QL S FEE E YH
Sbjct: 84 PSPDIDIGGWV-TGETYDIT-SDIPFGFQWCPFSGFTQLASVFEEGEAEQTSYHTV---- 137
Query: 143 FTEHSAVSSGSHPCPYIA---YVGPVH-PSTSNSGGTVSEVSNFNHWNARPIHGDMSTSY 198
+HS +S S CPY+A ++ PVH PS+S+SG +E S+F H + + G +
Sbjct: 138 -GDHSNAASSSLVCPYLALRGFLHPVHVPSSSSSG---AENSSF-HRHPTSLEGHAAHDL 192
Query: 199 TVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTR--GSEATRSGSYMHPYP 256
+ V S +H + H + + SV+ R S R+ Y H P
Sbjct: 193 SNTQVFHATESRNHDNDHRYMSNLPVSGIPDHSVAPFGIGLPRYDSSSQQRTRPYAHHRP 252
Query: 257 VGHSSVARAGNSAASSMIPP---YPGSNARARDRVEALQAYYHQQQ---------PPNST 304
+ H R G S+M+ P P A R + + +QQ PP S
Sbjct: 253 LVHRPTPRNG----SNMVTPLGSVPAVMAETRGHGHGARGHMYQQSMHSLQSSPFPPTSR 308
Query: 305 TVRPP--------VASSARRSSGHSRSAPLASLSSPPD 334
VRP ASS+ GH AP + S+ D
Sbjct: 309 RVRPRALTITSFIAASSSAEIGGHHGFAPPVNRSNSSD 346
>UniRef100_Q9SHV0 Hypothetical protein At2g15260 [Arabidopsis thaliana]
Length = 362
Score = 89.0 bits (219), Expect = 2e-16
Identities = 46/103 (44%), Positives = 62/103 (59%), Gaps = 7/103 (6%)
Query: 23 CSICLEIVSDNGD--RSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRKIEKGQWLYANG 80
CSIC + + D R+ L+C H+FHLDCIGSA+NAKG M+CPNCR IE G W +++G
Sbjct: 123 CSICRGALVNENDVQRTLVTLKCVHKFHLDCIGSAYNAKGFMECPNCRNIEPGHWQFSDG 182
Query: 81 SRSYPE---FNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQ 120
+ P N E+ D+D S+S+M + CPFG Q
Sbjct: 183 THFNPNGMIANNEEEEEDND--PGSFSQMIVKSEVCPFGCLGQ 223
>UniRef100_Q9T0H0 Hypothetical protein T6G15.40 [Arabidopsis thaliana]
Length = 226
Score = 82.4 bits (202), Expect = 2e-14
Identities = 33/61 (54%), Positives = 47/61 (76%), Gaps = 1/61 (1%)
Query: 19 DSVPCSICLE-IVSDNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRKIEKGQWLY 77
D C++CLE + +D +R+ KL+C H+FHLDC+GS+FN K M+CP CR+IEKG+WL+
Sbjct: 18 DDDDCAVCLEPLANDADERTVVKLRCSHKFHLDCVGSSFNIKNKMECPCCRQIEKGKWLF 77
Query: 78 A 78
A
Sbjct: 78 A 78
>UniRef100_Q93VV4 ORF162 [Marchantia polymorpha]
Length = 162
Score = 65.1 bits (157), Expect = 3e-09
Identities = 31/64 (48%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Query: 11 DTRHRKSSDSVPCSICLEIVS-DNGDRSFAKLQCGHQFHLDCIGSAFNAKGAMQCPNCRK 69
D + ++++ CSICL++V GDRS KL C H FH CI SAF AKG QCPNC
Sbjct: 79 DAGTSQEAETLTCSICLDVVLVQGGDRSITKLVCEHWFHFYCIVSAFIAKGTKQCPNCLA 138
Query: 70 IEKG 73
+G
Sbjct: 139 CREG 142
>UniRef100_UPI0000363AC1 UPI0000363AC1 UniRef100 entry
Length = 588
Score = 60.5 bits (145), Expect = 9e-08
Identities = 73/336 (21%), Positives = 134/336 (39%), Gaps = 17/336 (5%)
Query: 77 YANGSRSYPEFNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQLPSSFEEREYSPIPYH 136
Y++ S SY + ++ Y S S S +++ SS+ YS Y
Sbjct: 25 YSSSSSSYS--SSSSYSSSSSSYSSSSSSSSYSSSSYSSSSYSSSSSSYSSSSYSSSSYS 82
Query: 137 DAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMST 196
+ + S+ SS S +Y+ S+S S + S S+ + +++ + S+
Sbjct: 83 SSSYSSSSYSSSSSSSSSSSSSSSYI-----SSSYSSSSYSSSSSSSSYSSSSSYSSSSS 137
Query: 197 SYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSY-MHPY 255
SY+ + S + SS +SS SS ++ S S S+ + S + S SY Y
Sbjct: 138 SYSSSS--YSSSSSSYSSSSYSSSSSSYSSSSSYSSSSSSYSSSSSSSSYSSSSYSSSSY 195
Query: 256 PVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASS-A 314
SS + + S++S Y S++ + + + Y +S+ +SS +
Sbjct: 196 SSSSSSYSSSSYSSSSYSSSSYSSSSSSSSSSYSSSSSSYISSSYSSSSYSSSSSSSSYS 255
Query: 315 RRSSGHSRSAPLASLSSPPDQSGSYIYLPGRNFQEETRLPSHF------HAWERDHLPSS 368
SS +S S+ +S SS S SY ++ + S + ++ SS
Sbjct: 256 SSSSSYSSSSSYSSSSSSSCSSSSYSSSSSSSYSSSSSSSSSYSSSYSSSSYSSSSYSSS 315
Query: 369 SLNQVGRESSWRAYHQTASGTDPGFRSSSFRLRSDS 404
S + S +Y ++S + + SSS+ S S
Sbjct: 316 SYSSSSSSYSSSSYSSSSSSSSSSYSSSSYSSSSYS 351
Score = 47.0 bits (110), Expect = 0.001
Identities = 57/250 (22%), Positives = 104/250 (40%), Gaps = 12/250 (4%)
Query: 160 AYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSS 219
+Y + S+S+S + S S+ + +++ + S+SY+ + Y S + SS +SS
Sbjct: 7 SYSSSSYSSSSSSYSSSSYSSSSSSYSSSSSYSSSSSSYSSSSSSSSYSSSSYSSSSYSS 66
Query: 220 GSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPG 279
SS ++ S S S+ + S ++ S S SS + + +S SS
Sbjct: 67 SSSSYSSSSYSSSSYSSSSYSSSSYSSSSSS---------SSSSSSSSSYISSSYSSSSY 117
Query: 280 SNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSY 339
S++ + + +Y +S++ +SS SS S S+ +S SS S SY
Sbjct: 118 SSSSSSSSYSSSSSYSSSSSSYSSSSYSSS-SSSYSSSSYSSSSSSYSSSSSYSSSSSSY 176
Query: 340 IYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTASGTDPGFRSSSFR 399
+ + S ++ SSS + SS +Y ++S + + SSS
Sbjct: 177 SSSSSSSSYSSSSYSSSSYSSSSSSYSSSSYSSSSYSSS--SYSSSSSSSSSSYSSSSSS 234
Query: 400 LRSDSERTPS 409
S S + S
Sbjct: 235 YISSSYSSSS 244
Score = 46.6 bits (109), Expect = 0.001
Identities = 70/342 (20%), Positives = 128/342 (36%), Gaps = 30/342 (8%)
Query: 77 YANGSRSYPEFNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQLPSSFEEREYSPIPYH 136
Y++ S SY + + Y S S S +++ SS+ YS Y
Sbjct: 195 YSSSSSSYSSSSYSSSSYSSSSYSSSSSSSS--------SSYSSSSSSYISSSYSSSSYS 246
Query: 137 DAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMST 196
S+ SS S+ +Y S+S+S S + + ++ S+
Sbjct: 247 ----------SSSSSSSYSSSSSSYSSSSSYSSSSSSSCSSSSYSSSSSSSYSSSSSSSS 296
Query: 197 SYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPY- 255
SY+ Y S + SS +SS SS ++ S S S+ S + S + Y
Sbjct: 297 SYSSSYSSSSYSSSSYSSSSYSSSSSSYSSSSYSSSSSSSSSSYSSSSYSSSSYSISSYS 356
Query: 256 ------PVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPP 309
S ++ + +S++SS Y S+ + + + + +S++
Sbjct: 357 SSSYSSSSSSSYISSSYSSSSSSSSSSYSSSSYSSSSYISSSSSSSSSYISSSSSSSSSY 416
Query: 310 VASSARRSSGHSRSAPLASLSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSS 369
++SS SS S+ +S SS S SY ++ S + + + SSS
Sbjct: 417 ISSSYSSSSSSYSSSSYSSSSSYISSSSSYSSSSSSSYSS-----SSYSSSSSSYSSSSS 471
Query: 370 LNQVGRESSWRAYHQTASGTDPGFRSSSFRLRSDSERTPSQN 411
+ SS + + ++S + + SSS S S + S +
Sbjct: 472 SSSYISSSSSSSSYSSSSYSSSSYSSSSSSYSSSSYSSSSSS 513
Score = 36.2 bits (82), Expect = 1.7
Identities = 44/200 (22%), Positives = 79/200 (39%), Gaps = 4/200 (2%)
Query: 212 HHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAAS 271
H +S +SS S ++ S S S+ + S ++ S S Y SS + + +S +S
Sbjct: 3 HQNSSYSSSSYSSSSSSYSSSSYSSSSSSYSSSSSYSSS-SSSYSSSSSSSSYSSSSYSS 61
Query: 272 SMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSS 331
S Y S++ + +Y +S + +SS+ SS + S+ +S S
Sbjct: 62 SS---YSSSSSSYSSSSYSSSSYSSSSYSSSSYSSSSSSSSSSSSSSSYISSSYSSSSYS 118
Query: 332 PPDQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTASGTDP 391
S SY + + S + + + SS + SS +Y ++S
Sbjct: 119 SSSSSSSYSSSSSYSSSSSSYSSSSYSSSSSSYSSSSYSSSSSSYSSSSSYSSSSSSYSS 178
Query: 392 GFRSSSFRLRSDSERTPSQN 411
SSS+ S S + S +
Sbjct: 179 SSSSSSYSSSSYSSSSYSSS 198
>UniRef100_Q9BKV7 Ppg3 [Leishmania major]
Length = 1325
Score = 56.2 bits (134), Expect = 2e-06
Identities = 76/289 (26%), Positives = 111/289 (38%), Gaps = 34/289 (11%)
Query: 123 SSFEEREYSPIPYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNF 182
SS S P + P + SA SS S P + PS+S+S + S S
Sbjct: 774 SSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSA--PSSSSSAPSASSSSA- 830
Query: 183 NHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRG 242
S+S + P+ SS SS SS A+ + S S+ P+
Sbjct: 831 -----------PSSSSSAPSA--------SSSSAPSSSSSAPSASSSSAPSSSSSAPSAS 871
Query: 243 SEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPN 302
S + S S P S+ + + +SA S+ P S++ + + A P+
Sbjct: 872 SSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSA-------PS 924
Query: 303 STTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWER 362
S++ P ASS+ S S SAP AS SS P S S P + S +
Sbjct: 925 SSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSS---APSASSSSAPSSSSSAPSASS 981
Query: 363 DHLPSSSLNQVGRESSWRAYHQTASGTDPGFRSSSFRLRSDSERTPSQN 411
PSSS + SS A ++S + P SSS S S + S +
Sbjct: 982 SSAPSSSSSSAPSASSSSA--PSSSSSAPSASSSSAPSSSSSAPSASSS 1028
Score = 52.4 bits (124), Expect = 2e-05
Identities = 57/222 (25%), Positives = 86/222 (38%), Gaps = 12/222 (5%)
Query: 123 SSFEEREYSPIPYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNF 182
SS S P + P + SA SS S + P S++ S + S S+
Sbjct: 713 SSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSS 772
Query: 183 NHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRG 242
+ ++S + P+ S S+ SS SS A+ + S S+ P+
Sbjct: 773 SS------SAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSAS 826
Query: 243 SEATRSGSYMHPY------PVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYH 296
S + S S P P SS A +S+A S P +++ + +
Sbjct: 827 SSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSAS 886
Query: 297 QQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGS 338
P+S++ P ASS+ S S SAP AS SS P S S
Sbjct: 887 SSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSS 928
Score = 47.4 bits (111), Expect = 8e-04
Identities = 54/200 (27%), Positives = 84/200 (42%), Gaps = 15/200 (7%)
Query: 141 PMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTV 200
P + SA SS S P A S+S+S + S S + ++ P ++S +
Sbjct: 915 PSASSSSAPSSSSSSAPS-ASSSSAPSSSSSSAPSASSSSAPSSSSSAP----SASSSSA 969
Query: 201 PAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHS 260
P+ S S+ SS SS A+ + S S+ P+ S + S S S
Sbjct: 970 PSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSS---------S 1020
Query: 261 SVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGH 320
S A +S+A S P +++ + + P+S++ P ASS+ S
Sbjct: 1021 SAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSS- 1079
Query: 321 SRSAPLASLSSPPDQSGSYI 340
S SAP AS SS P S S++
Sbjct: 1080 SSSAPSASSSSAPSSSSSFL 1099
Score = 47.4 bits (111), Expect = 8e-04
Identities = 56/204 (27%), Positives = 79/204 (38%), Gaps = 12/204 (5%)
Query: 214 SSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPY------PVGHSSVARAGN 267
SS SS SS A+ + S S+ P+ S + S S P P SS A +
Sbjct: 691 SSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASS 750
Query: 268 SAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLA 327
S+A S P +++ + + A S++ P ASS+ S S SAP A
Sbjct: 751 SSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSA 810
Query: 328 SLSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTAS 387
S SS P S S P + S + PSSS + SS ++S
Sbjct: 811 SSSSAPSSSSS---APSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSS---SAPSSS 864
Query: 388 GTDPGFRSSSFRLRSDSERTPSQN 411
+ P SSS S S + S +
Sbjct: 865 SSAPSASSSSAPSSSSSAPSASSS 888
Score = 44.3 bits (103), Expect = 0.006
Identities = 48/198 (24%), Positives = 82/198 (41%), Gaps = 6/198 (3%)
Query: 141 PMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTV 200
P + SA SS S P + S+S++ S + + ++ P ++S +
Sbjct: 899 PSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAP----SASSSSA 954
Query: 201 PAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHS 260
P+ S S+ SS S+ ++ S S+ P+ S + S S P S
Sbjct: 955 PSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAP-SASSS 1013
Query: 261 SVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGH 320
S + +SA S+ P S++ A + A P++++ P +SS+ S
Sbjct: 1014 SAPSSSSSAPSASSSSAPSSSSSAPS-ASSSSAPSSSSSAPSASSSSAPSSSSSSAPSAS 1072
Query: 321 SRSAPLASLSSPPDQSGS 338
S SAP +S S+P S S
Sbjct: 1073 SSSAPSSSSSAPSASSSS 1090
Score = 43.9 bits (102), Expect = 0.008
Identities = 51/198 (25%), Positives = 79/198 (39%), Gaps = 7/198 (3%)
Query: 214 SSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSM 273
SS SS SS A+ + S S+ P+ S + S S P SS + +SA S+
Sbjct: 676 SSASSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAP-SASSSSAPSSSSSAPSAS 734
Query: 274 IPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPP 333
P S++ A + A P++++ P +SS+ S S SAP +S S+P
Sbjct: 735 SSSAPSSSSSAPS-ASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPS 793
Query: 334 DQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTASGTDPGF 393
S S P + S PS+S + SS + +S + P
Sbjct: 794 ASSSS---APSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSS--SAPSASSSSAPSS 848
Query: 394 RSSSFRLRSDSERTPSQN 411
SS+ S S + S +
Sbjct: 849 SSSAPSASSSSAPSSSSS 866
Score = 42.0 bits (97), Expect = 0.032
Identities = 51/190 (26%), Positives = 73/190 (37%), Gaps = 10/190 (5%)
Query: 227 ADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARD 286
A +P S+ P+ S + S S P SS + +SA S+ P S++ A
Sbjct: 659 ACKPETESSSSAPSASSSSASSSSSSAP-SASSSSAPSSSSSAPSASSSSAPSSSSSAPS 717
Query: 287 RVEALQAYYHQQQPPNSTTVRPPVASSARRSS-----GHSRSAPLASLSSPPDQSGSYIY 341
+ P S++ P +SSA +S S SAP AS SS P S S
Sbjct: 718 ASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSS--S 775
Query: 342 LPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTASGTDPGFRSSSFRLR 401
P + S + PSSS + SS A ++S + P SSS
Sbjct: 776 APSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSA--PSSSSSAPSASSSSAPSS 833
Query: 402 SDSERTPSQN 411
S S + S +
Sbjct: 834 SSSAPSASSS 843
Score = 40.8 bits (94), Expect = 0.071
Identities = 50/193 (25%), Positives = 76/193 (38%), Gaps = 7/193 (3%)
Query: 219 SGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYP 278
S SS A+ + S S+ P+ S + S S P SS + +SA S+ P
Sbjct: 666 SSSSAPSASSSSASSSSSSAPSASSSSAPSSSSSAP-SASSSSAPSSSSSAPSASSSSAP 724
Query: 279 GSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGS 338
S++ A + P S++ P +SSA S S SAP +S SS P S S
Sbjct: 725 SSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSA--PSASSSSAPSSSSSSAPSASSS 782
Query: 339 YIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTASGTDPGFRSSSF 398
P + + S + PS+S + SS + +S + P SS+
Sbjct: 783 --SAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSS--SAPSASSSSAPSSSSSAP 838
Query: 399 RLRSDSERTPSQN 411
S S + S +
Sbjct: 839 SASSSSAPSSSSS 851
>UniRef100_UPI00003641A6 UPI00003641A6 UniRef100 entry
Length = 476
Score = 50.8 bits (120), Expect = 7e-05
Identities = 68/334 (20%), Positives = 128/334 (37%), Gaps = 26/334 (7%)
Query: 81 SRSYPEFNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQLPSSFEEREYSPIPYHDAPG 140
S SY + ++ Y S S + + + SS+ YS Y +
Sbjct: 123 SSSYSSSSSSSYSSSSSSYSSSSS----------YSSSSSYSSSYSSSSYSSSSYSSSSS 172
Query: 141 PMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTV 200
++ S+ SS S Y + S+ +S + S S+++ ++ + S+SY+
Sbjct: 173 SSYSSSSSYSSSSSSSSYSSSSSSYSSSSYSSSSSYSSSSSYSS-SSSSSYSSSSSSYSS 231
Query: 201 PAVVLHYHSWDHHSSH--------FSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYM 252
+ + S + SS+ +SS SS+ + S S+ + S + S SY
Sbjct: 232 SSSSSYSSSSSYSSSYSSSSSYSXYSSSSSYSSSYSSSSSCSSSSYSSSSSSSYSSSSYS 291
Query: 253 HPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVAS 312
S + + +S +SS S++ + + +Y +S + +S
Sbjct: 292 SSSYSSSSYSSSSSSSYSSSSYSSSYSSSSYSSSSYSSSSSYSSSSYSSSSYSSSSYSSS 351
Query: 313 SARRSSGHSRSAPLASLSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQ 372
S+ SS +S S+ +S SS S S Y ++ S + + + SSS +
Sbjct: 352 SSYSSSSYSSSSYSSSYSSSSYSSSSSSYSSSSSYSS-----SSYSSSSSSYSSSSSYSS 406
Query: 373 VGRESSWRAYHQTAS--GTDPGFRSSSFRLRSDS 404
SS +Y ++S + + SSS S S
Sbjct: 407 SSYSSSSSSYSSSSSSYSSSSSYSSSSSYSSSSS 440
Score = 49.3 bits (116), Expect = 2e-04
Identities = 71/331 (21%), Positives = 121/331 (36%), Gaps = 29/331 (8%)
Query: 79 NGSRSYPEFNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQLPSSFEEREYSPIPYHDA 138
+ S SY + ++ + Y S S S + + SS+ YS Y +
Sbjct: 10 SSSSSYSSSSSSSYSSSNSSYSSSSSSSSSSYSSSNSSSSSSYSSSYSS-SYSSSSYSSS 68
Query: 139 PGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSY 198
+ S+ SS S Y S+S S + S SN + S+SY
Sbjct: 69 SSYSSSYSSSSSSSS-------YSSSSSSSSSYSSSSYSSSSNSS-----------SSSY 110
Query: 199 TVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVG 258
S +SS S SS ++ S S S+ + S + S SY Y
Sbjct: 111 ----------SSSSYSSSSSYSSSSYSSSSSSSYSSSSSSYSSSSSYSSSSSYSSSYSSS 160
Query: 259 HSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSS 318
S + +S++SS S++ + + + Y +S++ + S+ SS
Sbjct: 161 SYSSSSYSSSSSSSYSSSSSYSSSSSSSSYSSSSSSYSSSSYSSSSSYSSSSSYSSSSSS 220
Query: 319 GHSRSAPLASLSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESS 378
+S S+ S SS S S Y + S ++ + SSS + SS
Sbjct: 221 SYSSSSSSYSSSSSSSYSSSSSYSSSYSSSSSYSXYSSSSSYSSSYSSSSSCSSSSYSSS 280
Query: 379 WRAYHQTASGTDPGFRSSSFRLRSDSERTPS 409
+ + ++S + + SSS+ S S + S
Sbjct: 281 SSSSYSSSSYSSSSYSSSSYSSSSSSSYSSS 311
Score = 39.7 bits (91), Expect = 0.16
Identities = 55/246 (22%), Positives = 93/246 (37%), Gaps = 21/246 (8%)
Query: 166 HPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLG 225
HPS+S+S + S S+ + S+SY+ S+ SS SS S
Sbjct: 2 HPSSSSSYSSSSSYSSSS-----------SSSYSSSN-----SSYSSSSSSSSSSYSSSN 45
Query: 226 AADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARAR 285
++ S S S S + S SY Y SS + + +S++SS S++
Sbjct: 46 SSSSSSYSSSYSSSYSSSSYSSSSSYSSSYSSSSSSSSYSSSSSSSSSY-----SSSSYS 100
Query: 286 DRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSYIYLPGR 345
+ + Y +S++ SS+ SS S S+ +S SS S
Sbjct: 101 SSSNSSSSSYSSSSYSSSSSYSSSSYSSSSSSSYSSSSSSYSSSSSYSSSSSYSSSYSSS 160
Query: 346 NFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTASGTDPGFRSSSFRLRSDSE 405
++ + S ++ SSS + SS +Y ++ + + SSS S S
Sbjct: 161 SYSSSSYSSSSSSSYSSSSSYSSSSSSSSYSSSSSSYSSSSYSSSSSYSSSSSYSSSSSS 220
Query: 406 RTPSQN 411
S +
Sbjct: 221 SYSSSS 226
>UniRef100_UPI000049A2DE UPI000049A2DE UniRef100 entry
Length = 492
Score = 50.1 bits (118), Expect = 1e-04
Identities = 63/221 (28%), Positives = 93/221 (41%), Gaps = 28/221 (12%)
Query: 131 SPIPYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPST---SNSGGTVSEVSNFNHWNA 187
S +P H + P+ + + V S SHP ++ PVH S+ +S V S+ H ++
Sbjct: 168 SSVPIHSSSHPVHSSSTPVQSSSHPVHSSSH--PVHSSSHPVHSSSVPVHSSSHPVHSSS 225
Query: 188 RPIHGDMSTSYTVPAVVLHYHSWDH--HSSHFS-SGSSHLGAADQPSVSQSNQRPTRGSE 244
P+H S+S+ V + + HS H HSS SSH + V S+ S
Sbjct: 226 VPVH---SSSHPVHSSSVPVHSSSHPVHSSSTPVQSSSHPVHSSSVPVHSSSTPVQSSSH 282
Query: 245 ATRSGSYMHP-----YPVGHSSVARAGNSAA--SSMIPPYPGSNARARDRVEALQAYYHQ 297
S S HP +PV SSV +S SS +P + S+ V + H
Sbjct: 283 PVHSSS--HPVQSSSHPVHSSSVPVHSSSVPVHSSSVPVHSSSHP-----VHSSSVPVHS 335
Query: 298 QQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGS 338
P ++ P +SS HS S P+ S S+P S S
Sbjct: 336 SSHPVHSSSTPVQSSS---HPVHSSSVPVQSSSTPVHSSSS 373
Score = 35.0 bits (79), Expect = 3.9
Identities = 39/139 (28%), Positives = 56/139 (40%), Gaps = 14/139 (10%)
Query: 123 SSFEEREYSPIPYHDAPGPMFTEHSAVSSGSHPC-----PYIAYVGPVHPSTSNSGGTVS 177
SS + S P H + P+ + + V S SHP P + PVH +S V
Sbjct: 251 SSSTPVQSSSHPVHSSSVPVHSSSTPVQSSSHPVHSSSHPVQSSSHPVH----SSSVPVH 306
Query: 178 EVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDH--HSSHFSSGSSHLGAADQPSVSQS 235
S H ++ P+H S+S+ V + + HS H HSS SS QS
Sbjct: 307 SSSVPVHSSSVPVH---SSSHPVHSSSVPVHSSSHPVHSSSTPVQSSSHPVHSSSVPVQS 363
Query: 236 NQRPTRGSEATRSGSYMHP 254
+ P S + + GS P
Sbjct: 364 SSTPVHSSSSPKHGSSSVP 382
>UniRef100_Q9Y076 Proteophosphoglycan precursor [Leishmania major]
Length = 873
Score = 50.1 bits (118), Expect = 1e-04
Identities = 57/216 (26%), Positives = 86/216 (39%), Gaps = 12/216 (5%)
Query: 123 SSFEEREYSPIPYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNF 182
SS S P + P + SA SS S P A S+S+S + S S
Sbjct: 636 SSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPS-ASSSSAPSSSSSSAPSASSSSAP 694
Query: 183 NHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRG 242
+ ++ P ++S + P+ S S+ SS SS A+ + S S+
Sbjct: 695 SSSSSAP----SASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSA 750
Query: 243 SEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPN 302
S ++ S SS + +SA S+ P S++ + + A P+
Sbjct: 751 SSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSA-------PS 803
Query: 303 STTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGS 338
S++ P ASS+ S S SAP AS SS P S S
Sbjct: 804 SSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSS 839
Score = 49.7 bits (117), Expect = 2e-04
Identities = 53/192 (27%), Positives = 76/192 (38%), Gaps = 6/192 (3%)
Query: 218 SSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPY 277
SS S+ ++ PS S S+ + S A + S P SS A +S+A S
Sbjct: 642 SSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSS-SSSSAPSASSSSAPSSSSSA 700
Query: 278 PGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSG 337
P +++ + + P+S++ P ASS+ S S SAP AS SS P S
Sbjct: 701 PSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSS 760
Query: 338 SYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTASGTDPGFRSSS 397
S P + S + PSSS + SS A ++S + SSS
Sbjct: 761 S---APSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSA--PSSSSSSAPSASSS 815
Query: 398 FRLRSDSERTPS 409
S S PS
Sbjct: 816 SAPSSSSSSAPS 827
Score = 49.3 bits (116), Expect = 2e-04
Identities = 54/198 (27%), Positives = 83/198 (41%), Gaps = 12/198 (6%)
Query: 141 PMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTV 200
P + SA SS S P + PS+S+S + S S + ++ P S+S
Sbjct: 670 PSASSSSAPSSSSSSAPSASSSSA--PSSSSSAPSASSSSAPSSSSSAP---SASSSSAP 724
Query: 201 PAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHS 260
+ S S+ SS SS A+ + S S+ P+ S + S S P S
Sbjct: 725 SSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSS 784
Query: 261 SVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGH 320
+ + + +SA S+ P S++ + + A P+S++ P ASS+ S
Sbjct: 785 APSSSSSSAPSASSSSAPSSSSSSAPSASSSSA-------PSSSSSSAPSASSSSAPSSS 837
Query: 321 SRSAPLASLSSPPDQSGS 338
S SAP AS SS S S
Sbjct: 838 SSSAPSASSSSASSSSSS 855
Score = 48.1 bits (113), Expect = 4e-04
Identities = 52/216 (24%), Positives = 84/216 (38%), Gaps = 11/216 (5%)
Query: 126 EEREYSPIPYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTS---NSGGTVSEVSNF 182
E S P + + SA S+ S P + P S+S +S + S+
Sbjct: 616 ETESSSSAPSASSSSASSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSS 675
Query: 183 NHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRG 242
+ ++ ++S + P+ S S+ SS S+ ++ S S+ P+
Sbjct: 676 SAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSAS 735
Query: 243 SEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPN 302
S + S S SS + +SA S+ P S++ A + P+
Sbjct: 736 SSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSS--------SAPS 787
Query: 303 STTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGS 338
S++ P ASS+ S S SAP AS SS P S S
Sbjct: 788 SSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSS 823
Score = 43.1 bits (100), Expect = 0.014
Identities = 44/158 (27%), Positives = 63/158 (39%), Gaps = 4/158 (2%)
Query: 254 PYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASS 313
P SS A +S+ASS P +++ + + P+S++ P ASS
Sbjct: 615 PETESSSSAPSASSSSASSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASS 674
Query: 314 ARRSSGHSRSAPLASLSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQV 373
+ S S SAP AS SS P S S P + S + PSSS +
Sbjct: 675 SSAPSSSSSSAPSASSSSAPSSSSS---APSASSSSAPSSSSSAPSASSSSAPSSSSSSA 731
Query: 374 GRESSWRAYHQTASGTDPGFRSSSFRLRSDSERTPSQN 411
SS A ++S + P SSS S S + S +
Sbjct: 732 PSASSSSA-PSSSSSSAPSASSSSAPSSSSSAPSASSS 768
>UniRef100_UPI000036004A UPI000036004A UniRef100 entry
Length = 302
Score = 49.3 bits (116), Expect = 2e-04
Identities = 61/266 (22%), Positives = 111/266 (40%), Gaps = 11/266 (4%)
Query: 77 YANGSRSYPEFNMEDWTRDDDVYDLSYSEMSLGVHWCPFGNFAQLPSSFEERE--YSPIP 134
Y++ S SY + ++ S S S + +++ SS+ YS
Sbjct: 24 YSSSSSSYSS-SSSSYSSSSSYSSSSSSYSSSSSSYSSSSSYSSSSSSYSSSSSSYSSSS 82
Query: 135 YHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDM 194
+ + ++ S+ SS S + S S+S + S S+ + +++
Sbjct: 83 SYSSSSSSYSSSSSYSSSSSYSSSSSSYSSSSSSYSSSYSSSSSYSSSSSYSSSSYSSSY 142
Query: 195 STS-YTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMH 253
S+S Y+ + Y S + SS +SS SS+ + S S S+ + S ++ S SY
Sbjct: 143 SSSSYSSSS---SYSSSSYSSSSYSSSSSYSSSYSSSSSSNSSSSSSSSSNSSSSSSYSS 199
Query: 254 PYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASS 313
Y +SS + +S++SS S++ + A Q Q +S++ +SS
Sbjct: 200 SYSSSYSSSSSYSSSSSSSSYSSSYSSSSSSSSAAAAAATAEQQLQLSSSSSYS---SSS 256
Query: 314 ARRSSGHSRSAPLASLSSPPDQSGSY 339
+ SS +S S+ +S SS S SY
Sbjct: 257 SYSSSSYSSSSCSSSYSS-SSYSSSY 281
Score = 33.9 bits (76), Expect = 8.6
Identities = 42/201 (20%), Positives = 80/201 (38%), Gaps = 6/201 (2%)
Query: 214 SSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSM 273
SS +SS SS ++ S S+ + S ++ S SY SS + + +S++ S
Sbjct: 21 SSSYSSSSSSYSSSSSSYSSSSSYSSSSSSYSSSSSSYSSSSSYSSSSSSYSSSSSSYSS 80
Query: 274 IPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPP 333
Y S++ + + + ++ +SS SS +S S+ +S S
Sbjct: 81 SSSYSSSSSSYSSSSSYSSSSSYSSSSSSYSSSSSSYSSSYSSSSSYSSSSSYSSSSYSS 140
Query: 334 DQSGSYIYLPGRNFQEETRLPSHF---HAWERDHLPSSSLNQVGRESSWRAYHQTASGTD 390
S S Y ++ + S + ++ + SSS N SS + + ++S +
Sbjct: 141 SYSSS-SYSSSSSYSSSSYSSSSYSSSSSYSSSYSSSSSSNSSSSSSS--SSNSSSSSSY 197
Query: 391 PGFRSSSFRLRSDSERTPSQN 411
SSS+ S + S +
Sbjct: 198 SSSYSSSYSSSSSYSSSSSSS 218
>UniRef100_Q9Y075 Proteophosphoglycan [Leishmania major]
Length = 383
Score = 48.5 bits (114), Expect = 3e-04
Identities = 52/214 (24%), Positives = 86/214 (39%), Gaps = 5/214 (2%)
Query: 123 SSFEEREYSPIPYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNF 182
SS S P + P + SA SS S P A S+S+S + S S
Sbjct: 23 SSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPS-ASSSSAPSSSSSSAPSASSSSAP 81
Query: 183 NHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRG 242
+ ++ P ++S + P+ S S+ SS S+ ++ S S+
Sbjct: 82 SSSSSAP----SASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASS 137
Query: 243 SEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPN 302
S A S S P S+ + + +SA S+ P S++ + + A P+
Sbjct: 138 SSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPS 197
Query: 303 STTVRPPVASSARRSSGHSRSAPLASLSSPPDQS 336
+++ P +SS+ S S SAP +S S+P S
Sbjct: 198 ASSSSAPSSSSSSAPSASSSSAPSSSSSAPSSSS 231
Score = 47.4 bits (111), Expect = 8e-04
Identities = 59/228 (25%), Positives = 91/228 (39%), Gaps = 13/228 (5%)
Query: 167 PSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGA 226
PS+S+S + S S + ++ P ++S + P+ S S+ SS SS A
Sbjct: 4 PSSSSSAPSASSSSAPSSSSSAP----SASSSSAPSSSSSAPSASSSSAPSSSSSSAPSA 59
Query: 227 ADQPSVSQSNQR-PTRGSEATRSGSYMHPY------PVGHSSVARAGNSAASSMIPPYPG 279
+ + S S+ P+ S + S S P P SS A +S+A S P
Sbjct: 60 SSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPS 119
Query: 280 SNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSY 339
+++ + + P+S++ P ASS+ S S SAP AS SS P S S
Sbjct: 120 ASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSS- 178
Query: 340 IYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTAS 387
P + S + PSSS + SS A ++S
Sbjct: 179 -SAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSS 225
Score = 38.1 bits (87), Expect = 0.46
Identities = 53/200 (26%), Positives = 78/200 (38%), Gaps = 22/200 (11%)
Query: 214 SSHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSM 273
SS SS SS A+ + S S+ P+ S + S S P S+ + + +SA S+
Sbjct: 1 SSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSAS 60
Query: 274 IPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPP 333
P S++ + P S++ P +SSA +S S SAP +S S+P
Sbjct: 61 SSSAPSSSSSSA--------------PSASSSSAPSSSSSAPSAS--SSSAPSSSSSAPS 104
Query: 334 DQS----GSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESSWRAYHQTASGT 389
S S P + S + PSSS + SS A ++S +
Sbjct: 105 ASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSA--PSSSSS 162
Query: 390 DPGFRSSSFRLRSDSERTPS 409
SSS S S PS
Sbjct: 163 SAPSASSSSAPSSSSSSAPS 182
>UniRef100_Q9VE45 CG7709-PA [Drosophila melanogaster]
Length = 950
Score = 48.1 bits (113), Expect = 4e-04
Identities = 69/245 (28%), Positives = 99/245 (40%), Gaps = 29/245 (11%)
Query: 116 GNFAQLPSSFEEREYSPI----PYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTS- 170
G++ PSS S PY AP ++ S+ S+ P P PS S
Sbjct: 614 GSYPSAPSSSYSAPSSSSSSGGPYASAPSSSYSAPSSGSNSGGPYPAAPSSSYSAPSASA 673
Query: 171 NSGGTVSEVSNFNHW-------NARPIHGDMSTSYTVPAVVLH----YHSWDHHSSHFSS 219
NSGG+ + ++ + P S+SY+ P+ + Y S S S
Sbjct: 674 NSGGSYPSAPSSSYSAPSPGSNSGGPYPAAPSSSYSAPSPSANSGGPYASAPSSSYSAPS 733
Query: 220 GSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPG 279
SS+ G + S S P S ++ SG PYP SS A +S+ SS PYP
Sbjct: 734 SSSNSGGPYAAAPSSSYSAP---SSSSSSGG---PYPSAPSSSYSAPSSSLSSG-GPYPS 786
Query: 280 --SNARARDRVEALQAYYHQQQPPN--STTVRPPVASSARRSSGHSR--SAPLASLSSPP 333
S++ A + + P N S + PP +S +SG S SAP +S +P
Sbjct: 787 APSSSYAAPSPSSNSGGPYPAAPSNSYSAPIAPPSSSYGAPASGPSPSFSAPSSSYGAPS 846
Query: 334 DQSGS 338
SGS
Sbjct: 847 TGSGS 851
Score = 44.7 bits (104), Expect = 0.005
Identities = 48/176 (27%), Positives = 65/176 (36%), Gaps = 24/176 (13%)
Query: 116 GNFAQLPSSFEEREYSPI----PYHDAPGPMFTEHSAVSSGSHPCPYI---AYVGPVHPS 168
G + PSS S + PY AP + S S+ P P +Y P+ P
Sbjct: 761 GPYPSAPSSSYSAPSSSLSSGGPYPSAPSSSYAAPSPSSNSGGPYPAAPSNSYSAPIAPP 820
Query: 169 TSNSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAAD 228
+S+ G S S P S+SY P S SS FSS SS A
Sbjct: 821 SSSYGAPASGPS--------PSFSAPSSSYGAP-------STGSGSSSFSSSSSSFSGAS 865
Query: 229 QPSVSQSNQRPTR--GSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNA 282
S + P+ G+ +T SG P S AG S++S P P ++
Sbjct: 866 SSSSAGYPSAPSSSYGAPSTGSGHSFSSAPSSSYSAPPAGGSSSSGPYPSAPSQDS 921
Score = 40.8 bits (94), Expect = 0.071
Identities = 78/288 (27%), Positives = 112/288 (38%), Gaps = 48/288 (16%)
Query: 122 PSSFEEREYSPIPYHDAPGPMFTEHSAV-----SSGSHPCPYIAYVGPVHPSTS------ 170
PSS P P + AP P + A S G+ P +Y P PS+S
Sbjct: 379 PSSSYGAPAPPSPSYGAPAPPSKSYGAPAPPSSSYGAPAAPSKSYGAPAPPSSSYGAPAP 438
Query: 171 --NSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAAD 228
+S G S S+ ++ +P S+SY P S+ +S S SS GA
Sbjct: 439 PSSSYGAPSAPSS-SYGPPKPAPAPPSSSYGAPPQA-PVSSYLPPASRPSKPSSSYGA-- 494
Query: 229 QPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRV 288
PSVS P+ S T G+ +G S + +S+ + + P S
Sbjct: 495 -PSVSSFVPLPSAPS--TNYGAPSKTQSLGSSGYSSGPSSSYEAPVAPPSSSYG------ 545
Query: 289 EALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSYIYLPGRNFQ 348
A + + PP+S+ P SS SS S SA +SL S P + S G +FQ
Sbjct: 546 -APSSSFQPISPPSSSYGAP---SSGSGSSSGSFSAAPSSLYSAPSKGSS-----GGSFQ 596
Query: 349 EETRLPSHFHAWERDHLPSSSLNQVGR-----ESSWRAYHQTASGTDP 391
PS ++ PS+S N G SS+ A ++S P
Sbjct: 597 S---APSSSYS-----APSASANSGGSYPSAPSSSYSAPSSSSSSGGP 636
Score = 39.7 bits (91), Expect = 0.16
Identities = 66/251 (26%), Positives = 94/251 (37%), Gaps = 51/251 (20%)
Query: 116 GNFAQLPSSFEEREYSPI----PYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTS- 170
G +A PSS S PY AP ++ S+ SS P P PS+S
Sbjct: 719 GPYASAPSSSYSAPSSSSNSGGPYAAAPSSSYSAPSSSSSSGGPYPSAPSSSYSAPSSSL 778
Query: 171 NSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGA-ADQ 229
+SGG + ++ P P+ + +S+ + SS GA A
Sbjct: 779 SSGGPYPSAPSSSYAAPSPSSNSGGPYPAAPS--------NSYSAPIAPPSSSYGAPASG 830
Query: 230 PSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVE 289
PS S S + G+ +T SGS SS + + + A+SS YP +
Sbjct: 831 PSPSFSAPSSSYGAPSTGSGS------SSFSSSSSSFSGASSSSSAGYPSA--------- 875
Query: 290 ALQAYYHQQQPPNSTTVRPPVASSARRSSGHS-RSAPLASLSSPP----DQSGSYIYLPG 344
P+S+ P SGHS SAP +S S+PP SG Y P
Sbjct: 876 -----------PSSSYGAPST------GSGHSFSSAPSSSYSAPPAGGSSSSGPYPSAPS 918
Query: 345 RNFQEETRLPS 355
++ E PS
Sbjct: 919 QDSGYEYNGPS 929
Score = 37.0 bits (84), Expect = 1.0
Identities = 54/226 (23%), Positives = 79/226 (34%), Gaps = 19/226 (8%)
Query: 122 PSSFEEREYSPIPYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPV-HPSTSNSGGTVSEVS 180
PSS +P + P P S+ P +Y+ P PS +S VS
Sbjct: 439 PSSSYGAPSAPSSSYGPPKPAPAPPSSSYGAPPQAPVSSYLPPASRPSKPSSSYGAPSVS 498
Query: 181 NFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPT 240
+F P+ ST+Y P+ + SS +SSG S A S S P+
Sbjct: 499 SFV-----PLPSAPSTNYGAPS-----KTQSLGSSGYSSGPSSSYEAPVAPPSSSYGAPS 548
Query: 241 RGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQP 300
+ S + P S + SAA S + P + A + Y
Sbjct: 549 SSFQPISPPSSSYGAPSSGSGSSSGSFSAAPSSLYSAPSKGSSGGSFQSAPSSSYSAPSA 608
Query: 301 --------PNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGS 338
P++ + SS+ S G SAP +S S+P S S
Sbjct: 609 SANSGGSYPSAPSSSYSAPSSSSSSGGPYASAPSSSYSAPSSGSNS 654
Score = 36.6 bits (83), Expect = 1.3
Identities = 60/224 (26%), Positives = 86/224 (37%), Gaps = 36/224 (16%)
Query: 128 REYSPIPYHDAPGPMFTEHSAVSS-----GSHPCPYIAYVGPVHPSTSNSGGTVSEVSNF 182
R +P + AP P + + A ++ G+ P +Y P PS+S G + S
Sbjct: 285 RPAAPSSSYGAPAPPSSSYGAPAAPSSSYGAPAAPSSSYGAPAAPSSSY--GAPAPPSKS 342
Query: 183 NHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQ---SNQRP 239
A P S+SY PA + + SS GA PS S + P
Sbjct: 343 YGAPAPP-----SSSYGAPAAPSKSYGAP------APPSSSYGAPAPPSSSYGAPAPPSP 391
Query: 240 TRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQ 299
+ G+ A S SY P P S A A S S P P S++ Y
Sbjct: 392 SYGAPAPPSKSYGAPAPPSSSYGAPAAPS--KSYGAPAPPSSS------------YGAPA 437
Query: 300 PPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGSYIYLP 343
PP+S+ P SS+ + + P +S +PP Q+ YLP
Sbjct: 438 PPSSSYGAPSAPSSSYGPPKPAPAPPSSSYGAPP-QAPVSSYLP 480
>UniRef100_Q8NAM5 Hypothetical protein FLJ35107 [Homo sapiens]
Length = 258
Score = 47.4 bits (111), Expect = 8e-04
Identities = 46/172 (26%), Positives = 75/172 (42%), Gaps = 12/172 (6%)
Query: 167 PSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGA 226
P+ SGG+ S S+ +R I S ++P+ S SSH SS S +
Sbjct: 27 PNALGSGGSRSSSSS-----SRSILSSSILSSSIPSS--SSSSSSPSSSHSSSSPSSSHS 79
Query: 227 ADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARD 286
+ PS S S P+ S ++ S S SS + +S++SS P S++ +
Sbjct: 80 SSSPSSSSSTSSPSSSSSSSSSSSPSSSNSSSSSSSSSPSSSSSSSSSSPSSSSSSPSSS 139
Query: 287 RVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGS 338
+ + P+S++ +SS+ SS S S+P +S SSP + S
Sbjct: 140 SSSSSSSPSSSSSSPSSSS-----SSSSSSSSSPSSSSPSSSGSSPSSSNSS 186
Score = 42.7 bits (99), Expect = 0.019
Identities = 53/220 (24%), Positives = 84/220 (38%), Gaps = 21/220 (9%)
Query: 119 AQLPSSFEEREYSPIPYHDAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSE 178
+ +PSS SP H + P + S+ S S PS+S+S + S
Sbjct: 53 SSIPSSSSSSS-SPSSSHSSSSPSSSHSSSSPSSSSSTS--------SPSSSSSSSSSSS 103
Query: 179 VSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQR 238
S+ N S+S + P+ S SS S SS ++ PS S S+
Sbjct: 104 PSSSN--------SSSSSSSSSPSSSSSSSSSSPSSSSSSPSSSSSSSSSSPSSSSSSPS 155
Query: 239 PTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQ 298
+ S ++ S S P S + NS+ SS S++ R + +
Sbjct: 156 SSSSSSSSSSSSPSSSSPSSSGSSPSSSNSSPSSSSSSPSSSSSSPSPRSSSPSSSSSST 215
Query: 299 QPPNSTTVRPPVASSARRSSGHSRSAPLASLSSPPDQSGS 338
P++++ +SS+ SS S S P A+L P S
Sbjct: 216 SSPSTSS----PSSSSPSSSSPSSSCPSAALGRRPQSPQS 251
>UniRef100_UPI000043211F UPI000043211F UniRef100 entry
Length = 937
Score = 46.6 bits (109), Expect = 0.001
Identities = 61/219 (27%), Positives = 84/219 (37%), Gaps = 25/219 (11%)
Query: 215 SHFSSGSSHLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSA----- 269
S S+G + +QPS S + +RP S + SGS G S + G S+
Sbjct: 503 SSSSTGGWFTSSTEQPS-SGNTERPPGSSTPSTSGSTGGSSSTGLPSSSSTGGSSGTNTQ 561
Query: 270 -ASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVAS--SARRSSGHSRSAPL 326
+SS PGS++ A+ QQP + TTVRP +S S S+G S S+
Sbjct: 562 QSSSSSTEQPGSSSTAQSSSTGGSFSSSTQQPSSGTTVRPSGSSTPSTSGSTGGSSSSNT 621
Query: 327 ASLSS-----PPDQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRESS--- 378
SS P S + PG + E S PS+S + G SS
Sbjct: 622 EQPSSRSTETPGSSSTAQSNSPGGSSSTEQSSSSSTERPSGSSTPSTSGSTGGSSSSNTE 681
Query: 379 --------WRAYHQTASGTDPGFRSSSFRLRSDSERTPS 409
TA + PG SS+ + S S PS
Sbjct: 682 QPSSRSTERPGSSSTAQSSSPGGSSSTEQSSSSSTERPS 720
Score = 45.4 bits (106), Expect = 0.003
Identities = 76/294 (25%), Positives = 116/294 (38%), Gaps = 54/294 (18%)
Query: 137 DAPGPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMST 196
+ PG T S+ + GS PS+S++GG+ + + ++ G ST
Sbjct: 272 EQPGSSSTAQSSSTGGSSSTGL--------PSSSSTGGSSGTNTQQSSSSSTEQPGSSST 323
Query: 197 SYTVPAVVLHYHSWDHHSSHFSSGSSHLGAA--DQPSVSQSNQRPTRGSEATRSGSYMHP 254
+ SSG++ +A +QPS S + +RP+ S + S S
Sbjct: 324 AQ-------------------SSGTAGWSSASTEQPS-SGNTERPSGSSTPSTSDSTGGS 363
Query: 255 YPVGHSSVARAGNSA------ASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRP 308
G S + G S+ +SS PGS++ A+ + +QP + +T RP
Sbjct: 364 SSTGLPSSSSTGGSSGTNTQQSSSSSTEQPGSSSTAQSSSTGGSSSASTEQPSSGSTERP 423
Query: 309 PVAS--SARRSSGHSRSAPLASLSS-------PPDQSGSYIYLPGRNFQEETRLPSHFHA 359
P +S S S+G S S L S SS S PG T S
Sbjct: 424 PGSSTPSTSGSTGGSSSTGLPSSSSTGGSGTNTQQSSSGSTEQPG---SSSTAQSSSTGG 480
Query: 360 WERDHLPSS-SLNQVGRESSWRAYHQTASGTDPGFRSSSFRLRS-DSERTPSQN 411
W PSS S Q G S+ Q++S T F SS+ + S ++ER P +
Sbjct: 481 WSSTEQPSSGSTEQPGSSST----AQSSSSTGGWFTSSTEQPSSGNTERPPGSS 530
Score = 44.7 bits (104), Expect = 0.005
Identities = 75/289 (25%), Positives = 111/289 (37%), Gaps = 33/289 (11%)
Query: 140 GPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNARPIH-------- 191
G TE S+ SS P G PSTS S G S SN ++R
Sbjct: 645 GSSSTEQSSSSSTERPS------GSSTPSTSGSTGGSSS-SNTEQPSSRSTERPGSSSTA 697
Query: 192 ------GDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLGAADQPSVSQSNQRPTRGSEA 245
G ST + + S+ S+G S + +QPS S + +RP+ S
Sbjct: 698 QSSSPGGSSSTEQSSSSSTERPSGSSTPSTSGSTGESSSSSTEQPS-SGNTERPSGSSTP 756
Query: 246 TRSGSYMHPYPVGHSSVARAGNSAASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTT 305
+ SGS G SS++ +S++S+ PGS++ A+ + +QP + T
Sbjct: 757 STSGS-----TGGSSSISTQQSSSSST---EQPGSSSTAQSSSSGGWSSSSTEQPSSGNT 808
Query: 306 VRPPVAS--SARRSSGHSRSAPLASLSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWERD 363
RP +S S S+G S S L +S S S + E+ S +
Sbjct: 809 ERPSGSSTPSTSGSTGGSSSTGLPFSNSTGGSSSSSTQQSSSSSTEQPGSSSTAQSSSTG 868
Query: 364 HLPSSSLNQVGRESSWR-AYHQTASGTDPGFRSSSFRLRSDSERTPSQN 411
SSS Q S+ R TA + G SS+ + S S S +
Sbjct: 869 GWSSSSTEQPSSGSTERPGGSSTAQSSSTGGSSSTEQPNSGSTEPGSSS 917
Score = 41.2 bits (95), Expect = 0.054
Identities = 51/195 (26%), Positives = 75/195 (38%), Gaps = 24/195 (12%)
Query: 163 GPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSS 222
G PSTS S T S+ W++ S + P+ S +S + GSS
Sbjct: 191 GSSTPSTSGSSSTAQSSSSTGGWSSISTEQSSSGNTERPS-----GSSTPSTSSSTEGSS 245
Query: 223 HLGAADQPSVSQSNQRPTRGSEATRS---------------GSYMHPYPVGHSSVARAGN 267
G S S+ T+ S +T + GS P S+ +G
Sbjct: 246 STGLPSSSSTGGSSGTNTQQSSSTNTEQPGSSSTAQSSSTGGSSSTGLPSSSSTGGSSGT 305
Query: 268 SA--ASSMIPPYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVAS--SARRSSGHSRS 323
+ +SS PGS++ A+ A + +QP + T RP +S S S+G S S
Sbjct: 306 NTQQSSSSSTEQPGSSSTAQSSGTAGWSSASTEQPSSGNTERPSGSSTPSTSDSTGGSSS 365
Query: 324 APLASLSSPPDQSGS 338
L S SS SG+
Sbjct: 366 TGLPSSSSTGGSSGT 380
Score = 37.0 bits (84), Expect = 1.0
Identities = 59/222 (26%), Positives = 87/222 (38%), Gaps = 30/222 (13%)
Query: 140 GPMFTEHSAVSSGSHPCPYIAYVGPVHPSTSNSGGTVSEVSNFNHWNA---RP------- 189
G TE S+ SS P G PSTS S G S S + RP
Sbjct: 704 GSSSTEQSSSSSTERPS------GSSTPSTSGSTGESSSSSTEQPSSGNTERPSGSSTPS 757
Query: 190 IHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSSHLG-----AADQPSVSQSNQRPTRGSE 244
G S ++ S + S ++ SS G + +QPS S + +RP+ S
Sbjct: 758 TSGSTGGSSSISTQQSSSSSTEQPGSSSTAQSSSSGGWSSSSTEQPS-SGNTERPSGSST 816
Query: 245 ATRSGSYMHPYPVGHSSVARAGNSAASSM------IPPYPGSNARARDRVEALQAYYHQQ 298
+ SGS G G S++SS PGS++ A+ + +
Sbjct: 817 PSTSGSTGGSSSTGLPFSNSTGGSSSSSTQQSSSSSTEQPGSSSTAQSSSTGGWSSSSTE 876
Query: 299 QPPNSTTVRPPVASSARRSS--GHSRSAPLASLSSPPDQSGS 338
QP + +T RP +S+A+ SS G S + S S+ P S +
Sbjct: 877 QPSSGSTERPGGSSTAQSSSTGGSSSTEQPNSGSTEPGSSST 918
>UniRef100_Q5WSX0 Hypothetical protein [Legionella pneumophila str. Lens]
Length = 395
Score = 46.6 bits (109), Expect = 0.001
Identities = 65/263 (24%), Positives = 95/263 (35%), Gaps = 26/263 (9%)
Query: 163 GPVHPSTSNSGGTVSEVSNFNHWNARPIHGDMSTSYTVPAVVLHYHSWDHHSSHFSSGSS 222
GP P S + T S+ N ++ S++ T P D SS + SS
Sbjct: 140 GPEEPKPSPNSSTQQSSSSSNSSSSSSSSPSSSSTTTTPTPTSSSSGSDSQSSSPPTDSS 199
Query: 223 HLGAADQPSVSQSNQRPTRGSEATRSGSYMHPYPVGHSSVARAGNSAASSMIP------- 275
G S SQS+ PT S ++ S S P SS +G+ + SS P
Sbjct: 200 SSG-----SDSQSSSPPTTSSSSSGSDSQSSSPPTDSSS---SGSDSQSSSPPTDSSSSG 251
Query: 276 -------PYPGSNARARDRVEALQAYYHQQQPPNSTTVRPPVASSARRSSGHSRSAPLAS 328
P S++ D + +S + PP SS+ S S S P S
Sbjct: 252 SDSQSSSPPTDSSSSGSDSQSSSPPTDSSSSGSDSQSSSPPTDSSSSGSDSQSSSPPTDS 311
Query: 329 LSSPPDQSGSYIYLPGRNFQEETRLPSHFHAWERDHLPSSSLNQVGRE-SSWRAYHQTAS 387
+S D S P + + S + + P S+ N + S Y +T+S
Sbjct: 312 SNSGSDSQSS---SPPTDSSNSSSSSSSDTSSQLSPPPDSNTNTSTPDLGSQTKYDETSS 368
Query: 388 GTDPGFRSSSFRLRSDSERTPSQ 410
++ G SS S TPSQ
Sbjct: 369 SSNSGSGQSSGSGSSSDVPTPSQ 391
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.128 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 756,957,080
Number of Sequences: 2790947
Number of extensions: 33176763
Number of successful extensions: 77439
Number of sequences better than 10.0: 1117
Number of HSP's better than 10.0 without gapping: 78
Number of HSP's successfully gapped in prelim test: 1053
Number of HSP's that attempted gapping in prelim test: 76116
Number of HSP's gapped (non-prelim): 1685
length of query: 412
length of database: 848,049,833
effective HSP length: 130
effective length of query: 282
effective length of database: 485,226,723
effective search space: 136833935886
effective search space used: 136833935886
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 76 (33.9 bits)
Medicago: description of AC144541.4


