

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.15 - phase: 0
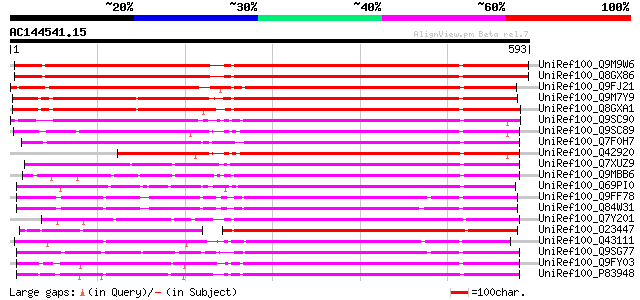
(593 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M9W6 Putative pectinesterase [Arabidopsis thaliana] 579 e-164
UniRef100_Q8GX86 Putative pectinesterase [Arabidopsis thaliana] 575 e-162
UniRef100_Q9FJ21 Pectin methylesterase [Arabidopsis thaliana] 522 e-146
UniRef100_Q9M7Y9 Pectin methylesterase, putative [Arabidopsis th... 478 e-133
UniRef100_Q8GXA1 Putative pectin methylesterase [Arabidopsis tha... 476 e-133
UniRef100_Q9SC90 Pectin methyl-esterase PER precursor [Medicago ... 460 e-128
UniRef100_Q9SC89 Pectin methyl-esterase PEF1 precursor [Medicago... 456 e-127
UniRef100_Q7F0H7 Putative pectin methylesterase [Oryza sativa] 455 e-126
UniRef100_Q42920 Pectinesterase precursor [Medicago sativa] 420 e-116
UniRef100_Q7XUZ9 OSJNBa0036B21.20 protein [Oryza sativa] 414 e-114
UniRef100_Q9MBB6 Pectin methylesterase [Salix gilgiana] 408 e-112
UniRef100_Q69PI0 Putative pectinesterase [Oryza sativa] 393 e-108
UniRef100_Q9FF78 Pectinesterase [Arabidopsis thaliana] 391 e-107
UniRef100_Q84W31 Hypothetical protein At5g04960 [Arabidopsis tha... 391 e-107
UniRef100_Q7Y201 Putative pectin methylesterase [Arabidopsis tha... 382 e-104
UniRef100_O23447 Pectinesterase [Arabidopsis thaliana] 382 e-104
UniRef100_Q43111 Pectinesterase 3 precursor [Phaseolus vulgaris] 381 e-104
UniRef100_Q9SG77 Putative pectinesterase [Arabidopsis thaliana] 377 e-103
UniRef100_Q9FY03 Putative pectin methylesterase precursor [Popul... 373 e-102
UniRef100_P83948 Pectinesterase 3 precursor [Citrus sinensis] 372 e-101
>UniRef100_Q9M9W6 Putative pectinesterase [Arabidopsis thaliana]
Length = 669
Score = 579 bits (1492), Expect = e-164
Identities = 288/590 (48%), Positives = 396/590 (66%), Gaps = 26/590 (4%)
Query: 6 DAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAP 65
++KR +R +I +S+ LL++MVVAVTV V N K G+ K + V+ SVKAVK +CAP
Sbjct: 8 ESKRKRRYIVITISSVLLISMVVAVTVGVSLN--KHDGDSKGK-AEVNASVKAVKDVCAP 64
Query: 66 TDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
TDY+K CED+LI + N T+P EL+K AFN+T+ +I++ KK+ + E +KD RT+ ALD
Sbjct: 65 TDYRKTCEDTLIKNGKNTTDPMELVKTAFNVTMKQITDAAKKSQTIMELQKDSRTRMALD 124
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDA 185
CK++M ++DE S E F+ + LD L +L++WLS AI+++ETCL+ F+ T +A
Sbjct: 125 QCKELMDYALDELSNSFEELGKFEFHLLDEALINLRIWLSAAISHEETCLEGFQGTQGNA 184
Query: 186 GKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPA--GRRLLKESVDGEEDVLGHGGDF 243
G+ MK+ L+T++ ++ NGL+II+++S +M+ P RRLL E
Sbjct: 185 GETMKKALKTAIELTHNGLAIISEMSNFVGQMQIPGLNSRRLLAEG-------------- 230
Query: 244 ELPEWVDDRAGVRKLLNKMTGRK-LQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIY 302
P WVD R RKLL ++ +VVA+DGSG + TI EAL+ VPKK FV++
Sbjct: 231 -FPSWVDQRG--RKLLQAAAAYSDVKPDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVH 287
Query: 303 IKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIG 362
IK G+YKEYV+V KTM+H+VFIGDG KT I+GNKN+ DG+ T++TA+VAI G++F+
Sbjct: 288 IKAGLYKEYVQVNKTMSHLVFIGDGPDKTIISGNKNYKDGITTYRTATVAIVGNYFIAKN 347
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
IGFEN+AG KHQAVA+RVQSD SIF+ CR DGYQDTLY H+ RQF+RDC ISGTIDF+F
Sbjct: 348 IGFENTAGAIKHQAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFLF 407
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
GD+ AV QNCT +VRKPL NQ C +TA GRK+ + TG + QG +I +P Y V+ +K
Sbjct: 408 GDAAAVFQNCTLLVRKPLPNQACPITAHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSK 467
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
AYL RPWK++SRTI ++T+I D + P+G+ PW G G T +Y E N GPGS +
Sbjct: 468 AYLGRPWKEYSRTIIMNTFIPDFVQPQGWQPW---LGDFGLKTLFYSEVQNTGPGSALAN 524
Query: 543 RVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPGATKTPPSRPT 592
RV W G+KT++ E F P ++ GDDWI VPY+ G P+ T
Sbjct: 525 RVTWAGIKTLSEEDILKFTPAQYIQGDDWIPGKGVPYTTGLLAGNPAAAT 574
>UniRef100_Q8GX86 Putative pectinesterase [Arabidopsis thaliana]
Length = 669
Score = 575 bits (1482), Expect = e-162
Identities = 286/590 (48%), Positives = 395/590 (66%), Gaps = 26/590 (4%)
Query: 6 DAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAP 65
++KR +R +I +S+ LL++MVVAVTV V N K G+ K + V+ SVKAVK +CAP
Sbjct: 8 ESKRKRRYIVITISSVLLISMVVAVTVGVSLN--KHDGDSKGK-AEVNASVKAVKDVCAP 64
Query: 66 TDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
TDY+K CED+LI + N T+P EL+K AFN+T+ +I++ KK+ + E +KD RT+ ALD
Sbjct: 65 TDYRKTCEDTLIKNGKNTTDPMELVKTAFNVTMKQITDAAKKSQTIMELQKDSRTRMALD 124
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDA 185
CK++M ++DE S E F+ + LD L +L++WLS AI+++ETCL+ F+ T +A
Sbjct: 125 QCKELMDYALDELSNSFEELGKFEFHLLDEALINLRIWLSAAISHEETCLEGFQGTQGNA 184
Query: 186 GKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPA--GRRLLKESVDGEEDVLGHGGDF 243
G+ MK+ L+T++ ++ NGL+II+++S +M+ P RRLL E
Sbjct: 185 GETMKKALKTAIELTHNGLAIISEMSNFVGQMQIPGLNSRRLLAEG-------------- 230
Query: 244 ELPEWVDDRAGVRKLLNKMTGRK-LQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIY 302
P WVD R RKLL ++ +VVA+DGSG + TI EAL+ VPKK FV++
Sbjct: 231 -FPSWVDQRG--RKLLQAAAAYSDVKPDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVH 287
Query: 303 IKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIG 362
IK G+YKEYV+V KTM+H+VFIGDG KT I+GNKN+ DG+ ++TA+VAI G++F+
Sbjct: 288 IKAGLYKEYVQVNKTMSHLVFIGDGPDKTIISGNKNYKDGITAYRTATVAIVGNYFIAKN 347
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
IGFEN+AG KHQAVA+RVQSD SIF+ CR DGYQ+TLY H+ RQF+RDC ISGTIDF+F
Sbjct: 348 IGFENTAGAIKHQAVAVRVQSDESIFFNCRFDGYQNTLYTHSHRQFFRDCTISGTIDFLF 407
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
GD+ AV QNCT +VRKPL NQ C +TA GRK+ + TG + QG +I +P Y V+ +K
Sbjct: 408 GDAAAVFQNCTLLVRKPLPNQACPITAHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSK 467
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
AYL RPWK++SRTI ++T+I D + P+G+ PW G G T +Y E N GPGS +
Sbjct: 468 AYLGRPWKEYSRTIIMNTFIPDFVQPQGWQPW---LGDFGLKTLFYSEVQNTGPGSALAN 524
Query: 543 RVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPGATKTPPSRPT 592
RV W G+KT++ E F P ++ GDDWI VPY+ G P+ T
Sbjct: 525 RVTWAGIKTLSEEDILKFTPAQYIQGDDWIPGKGVPYTTGLLAGNPAAAT 574
>UniRef100_Q9FJ21 Pectin methylesterase [Arabidopsis thaliana]
Length = 571
Score = 522 bits (1345), Expect = e-146
Identities = 275/588 (46%), Positives = 372/588 (62%), Gaps = 31/588 (5%)
Query: 1 MSGGGDAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVK 60
M G+ K+ K+ I GV T LLV MVVAV + N E + + AV+
Sbjct: 1 MGVDGELKK-KKCIIAGVITALLVLMVVAVGITTSRNTSHS---EKIVPVQIKTATTAVE 56
Query: 61 TLCAPTDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKT--HLLQEAEKDE 118
+CAPTDYK+ C +SL+ + + T+P +LIK+ FN+TI I + +KK L +A D+
Sbjct: 57 AVCAPTDYKETCVNSLMKASPDSTQPLDLIKLGFNVTIRSIEDSIKKASVELTAKAANDK 116
Query: 119 RTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAF 178
TK AL+ C+++M + D+ ++ L+ F F + ++ + L+VWLSG+I YQ+TC+D F
Sbjct: 117 DTKGALELCEKLMNDATDDLKKCLDNFDGFSIPQIEDFVEDLRVWLSGSIAYQQTCMDTF 176
Query: 179 ENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLG 238
E T + + M+++ +TS ++SNGL++I +S E +V G LG
Sbjct: 177 EETNSKLSQDMQKIFKTSRELTSNGLAMITNISNLLGEF-----------NVTGVTGDLG 225
Query: 239 H------GGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVP 292
+ +P WV R+L+ G K A+VVVA DGSG + TI EAL VP
Sbjct: 226 KYARKLLSAEDGIPSWVGPNT--RRLMATKGGVK--ANVVVAHDGSGQYKTINEALNAVP 281
Query: 293 KKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDG-VGTFKTASV 351
K N KPFVIYIK+GVY E V+VTK MTHV FIGDG KT+ITG+ N+ G V T+ TA+V
Sbjct: 282 KANQKPFVIYIKQGVYNEKVDVTKKMTHVTFIGDGPTKTKITGSLNYYIGKVKTYLTATV 341
Query: 352 AITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRD 411
AI GD F IGFEN+AGPE HQAVALRV +D ++FY C++DGYQDTLY H+ RQF+RD
Sbjct: 342 AINGDNFTAKNIGFENTAGPEGHQAVALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRD 401
Query: 412 CIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVAD 471
C +SGT+DF+FGD I VLQNC VVRKP+++Q C++TAQGR +K + TGL++Q I +
Sbjct: 402 CTVSGTVDFIFGDGIVVLQNCNIVVRKPMKSQSCMITAQGRSDKRESTGLVLQNCHITGE 461
Query: 472 PKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEY 531
P Y PV+ NKAYL RPWK+FSRTI + T I D+I P G++PW G +T YY EY
Sbjct: 462 PAYIPVKSINKAYLGRPWKEFSRTIIMGTTIDDVIDPAGWLPWN---GDFALNTLYYAEY 518
Query: 532 NNRGPGSDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPY 579
N GPGS+ QRVKW G+K ++ + A F P RF G+ WI RVPY
Sbjct: 519 ENNGPGSNQAQRVKWPGIKKLSPKQALRFTPARFLRGNLWIPPNRVPY 566
>UniRef100_Q9M7Y9 Pectin methylesterase, putative [Arabidopsis thaliana]
Length = 562
Score = 478 bits (1229), Expect = e-133
Identities = 254/584 (43%), Positives = 367/584 (62%), Gaps = 33/584 (5%)
Query: 4 GGDAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLC 63
G D + K+ + G + LV MVV+V V +K P ++ E+H+ ++ KAV+ +C
Sbjct: 2 GSDGDKKKKFIVAGSVSGFLVIMVVSVAV---VTSKHSPKDD---ENHIRKTTKAVQAVC 55
Query: 64 APTDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTH--LLQEAEKDERTK 121
APTD+K C +SL+ + + +P +LIK+ F +TI I+E L+K + +A+K+ K
Sbjct: 56 APTDFKDTCVNSLMGASPDSDDPVDLIKLGFKVTIKSINESLEKASGDIKAKADKNPEAK 115
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENT 181
A + C+++M +ID+ ++ ++ F ++ ++ + L+VWLSG+I +Q+TC+D+F
Sbjct: 116 GAFELCEKLMIDAIDDLKKCMDH--GFSVDQIEVFVEDLRVWLSGSIAFQQTCMDSFGEI 173
Query: 182 TTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPA--GRRLLKESVDGEEDVLGH 239
++ + M ++ +TS +SSN L+++ ++S A R+LL ED
Sbjct: 174 KSNLMQDMLKIFKTSRELSSNSLAMVTRISTLIPNSNLTAKYARKLLST-----ED---- 224
Query: 240 GGDFELPEWVDDRAGVRKLLNKMTGRK--LQAHVVVAKDGSGNFTTITEALKHVPKKNLK 297
+P WV A R+L+ G ++A+ VVA+DG+G F TIT+AL VPK N
Sbjct: 225 ----SIPTWVGPEA--RRLMAAQGGGPGPVKANAVVAQDGTGQFKTITDALNAVPKGNKV 278
Query: 298 PFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNF-IDGVGTFKTASVAITGD 356
PF+I+IKEG+YKE V VTK M HV FIGDG KT ITG+ NF I V TF TA++ I GD
Sbjct: 279 PFIIHIKEGIYKEKVTVTKKMPHVTFIGDGPNKTLITGSLNFGIGKVKTFLTATITIEGD 338
Query: 357 FFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISG 416
F IG EN+AGPE QAVALRV +D ++F+ C++DG+QDTLY H+ RQFYRDC +SG
Sbjct: 339 HFTAKNIGIENTAGPEGGQAVALRVSADYAVFHSCQIDGHQDTLYVHSHRQFYRDCTVSG 398
Query: 417 TIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYP 476
T+DF+FGD+ +LQNC VVRKP + Q C+VTAQGR + TGL++ G I DP Y P
Sbjct: 399 TVDFIFGDAKCILQNCKIVVRKPNKGQTCMVTAQGRSNVRESTGLVLHGCHITGDPAYIP 458
Query: 477 VRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGP 536
++ NKAYL RPWK+FSRTI + T I D+I P G++PW +G T YY E+ N GP
Sbjct: 459 MKSVNKAYLGRPWKEFSRTIIMKTTIDDVIDPAGWLPW---SGDFALKTLYYAEHMNTGP 515
Query: 537 GSDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYS 580
GS+ QRVKW G+K +T + A + RF GD WI T+VPY+
Sbjct: 516 GSNQAQRVKWPGIKKLTPQDALLYTGDRFLRGDTWIPQTQVPYT 559
>UniRef100_Q8GXA1 Putative pectin methylesterase [Arabidopsis thaliana]
Length = 568
Score = 476 bits (1225), Expect = e-133
Identities = 252/589 (42%), Positives = 368/589 (61%), Gaps = 31/589 (5%)
Query: 4 GGDAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLC 63
G D + K+ + G + LV MVV+V V +K P ++ E+H+ ++ KAV+ +C
Sbjct: 2 GSDGDKKKKFIVAGSVSGFLVIMVVSVAV---VTSKHSPKDD---ENHIRKTTKAVQAVC 55
Query: 64 APTDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTH--LLQEAEKDERTK 121
APTD+K C +SL+ + + +P +LIK+ F +TI I+E L+K + EA+K+ K
Sbjct: 56 APTDFKDTCVNSLMGASPDSDDPVDLIKLGFKVTIKSINESLEKASGDIKAEADKNPEAK 115
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENT 181
A + C+++M +ID+ ++ ++ F ++ ++ + L+VWLSG+I +Q+TC+D+F
Sbjct: 116 GAFELCEKLMIDAIDDLKKCMDH--GFSVDQIEVFVEDLRVWLSGSIAFQQTCMDSFGEI 173
Query: 182 TTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQP----AGRRLLKESVDGEEDVL 237
++ + M ++ +TS +SSN L+++ ++S A + ++ + E+ +
Sbjct: 174 KSNLMQDMLKIFKTSRELSSNSLAMVTRISTLIPNSNLTGLTGALAKYARKLLSTEDSI- 232
Query: 238 GHGGDFELPEWVDDRAGVRKLLNKMTGRK--LQAHVVVAKDGSGNFTTITEALKHVPKKN 295
P WV A R+L+ G ++A+ VVA+DG+G F TIT+AL VPK N
Sbjct: 233 --------PTWVGPEA--RRLMAAQGGGPGPVKANAVVAQDGTGQFKTITDALNAVPKGN 282
Query: 296 LKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNF-IDGVGTFKTASVAIT 354
PF+I+IKEG+YKE V VTK M HV FIGDG KT ITG+ NF I V TF TA++ I
Sbjct: 283 KVPFIIHIKEGIYKEKVTVTKKMPHVTFIGDGPNKTLITGSLNFGIGKVKTFLTATITIE 342
Query: 355 GDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCII 414
GD F IG EN+AGPE QAVALRV +D ++F+ C++DG+QDTLY H+ RQFYRDC +
Sbjct: 343 GDHFTAKNIGIENTAGPEGGQAVALRVSADYAVFHSCQIDGHQDTLYVHSHRQFYRDCTV 402
Query: 415 SGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKY 474
SGT+DF+FGD+ +LQNC VVRKP + Q C+VTAQGR + TGL++ G I DP Y
Sbjct: 403 SGTVDFIFGDAKCILQNCKIVVRKPNKGQTCMVTAQGRSNVRESTGLVLHGCHITGDPAY 462
Query: 475 YPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNR 534
P++ NKAYL RPWK+FSRTI + T I D+I P G++PW +G T YY E+ N
Sbjct: 463 IPMKSVNKAYLGRPWKEFSRTIIMKTTIDDVIDPAGWLPW---SGDFALKTLYYAEHMNT 519
Query: 535 GPGSDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPGA 583
GPGS+ QRVKW G+K +T + A + RF GD WI T+VPY+ A
Sbjct: 520 GPGSNQAQRVKWPGIKKLTPQDALLYTGDRFLRGDTWIPQTQVPYTAKA 568
>UniRef100_Q9SC90 Pectin methyl-esterase PER precursor [Medicago truncatula]
Length = 602
Score = 460 bits (1184), Expect = e-128
Identities = 251/588 (42%), Positives = 350/588 (58%), Gaps = 54/588 (9%)
Query: 2 SGGGDAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKT 61
+GG D ++KR A++GVS+ LLVAMV V + Q V+
Sbjct: 7 NGGHD--QSKRFALVGVSSILLVAMVATV-------------------ADAQQGQPNVQI 45
Query: 62 LCAPTDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTK 121
LC T Y++ C SL K+LIK AF+ T ++ + + + L+QE +D+ TK
Sbjct: 46 LCESTQYQQTCHQSLAKAPAETAGVKDLIKAAFSATSEELLKHINSSSLIQELGQDKMTK 105
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENT 181
QA++ C +V+ ++D +S+ FD+N + LKVWL+G +++Q+TCLD F NT
Sbjct: 106 QAMEVCNEVLDYAVDGIHKSVGAVDKFDINKIHEYSYDLKVWLTGTLSHQQTCLDGFANT 165
Query: 182 TTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGG 241
TT AG+ M L TS+ +SSN + +++ + A RRLL L +G
Sbjct: 166 TTKAGETMARALNTSIQLSSNAIDMVDAVYDLTN-----AKRRLLS---------LDNG- 210
Query: 242 DFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVI 301
P WV + G R+LL + T ++ +VVVA+DGSG F T+T+A+K VP N + FVI
Sbjct: 211 ---YPLWVSE--GQRRLLAEAT---VKPNVVVAQDGSGQFKTLTDAIKTVPANNAQNFVI 262
Query: 302 YIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGI 361
Y+KEGVY E V V K M V IGDG KT+ TG+ N+ DG+ + TA+ + G+ F+
Sbjct: 263 YVKEGVYNETVNVPKDMAFVTIIGDGPAKTKFTGSLNYADGLLPYNTATFGVNGENFMAK 322
Query: 362 GIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFV 421
I EN+AGPEKHQAVALRV +D++IFY C++DGYQ TL+A + RQFYRDC ISGTID +
Sbjct: 323 DISIENTAGPEKHQAVALRVTADKAIFYNCQIDGYQATLFAESQRQFYRDCSISGTIDMI 382
Query: 422 FGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKN 481
+GD+ AV QNC +VRKPLE QQC V A GR + + +G + Q +P+ + K
Sbjct: 383 YGDAFAVFQNCKLIVRKPLEEQQCFVAADGRTKSDSSSGFVFQSCHFTGEPEVAKIDPK- 441
Query: 482 KAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVK 541
AYL RPWK +S+ + +D+ I D+ PEGYMPW G DTC + EYNN+GPG+D
Sbjct: 442 IAYLGRPWKSYSKVVIMDSNIDDIFDPEGYMPWM---GSAFKDTCTFYEYNNKGPGADTS 498
Query: 542 QRVKWQGVKTITSEGAASFVPIRFFH------GDDWIRVTRVPYSPGA 583
+RVKW GVK+I+S AA+F P +FF D WI + VPYS A
Sbjct: 499 KRVKWPGVKSISSTEAAAFYPGKFFEIANATDRDTWIVKSGVPYSLAA 546
>UniRef100_Q9SC89 Pectin methyl-esterase PEF1 precursor [Medicago truncatula]
Length = 565
Score = 456 bits (1173), Expect = e-127
Identities = 250/587 (42%), Positives = 347/587 (58%), Gaps = 40/587 (6%)
Query: 5 GDAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCA 64
G + K+ A++GVS LLVAMV V V++ G + +++H+S S K V LC
Sbjct: 8 GGQGQGKKHALLGVSCILLVAMVGVVAVSL------TKGGDGEQKAHISNSQKNVDMLCQ 61
Query: 65 PTDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQAL 124
T +K+ C +L + + K IK A T ++ + + + L QE D TKQA+
Sbjct: 62 STKFKETCHKTL--EKASFSNMKNRIKGALGATEEELRKHINNSALYQELATDSMTKQAM 119
Query: 125 DTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTD 184
+ C +V+ ++D +S+ FD + L +KVWL+G +++Q+TCLD F NT T
Sbjct: 120 EICNEVLDYAVDGIHKSVGTLDQFDFHKLSEYAFDIKVWLTGTLSHQQTCLDGFVNTKTH 179
Query: 185 AGKKMKEVLQTSMHMSSNGLS---IINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGG 241
AG+ M +VL+TSM +SSN + +++++ K F + RRLL + DG
Sbjct: 180 AGETMAKVLKTSMELSSNAIDMMDVVSRILKGFHPSQYGVSRRLLSD--DG--------- 228
Query: 242 DFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVI 301
+P WV D G R LL G ++A+ VVA+DGSG F T+T+ALK VP N PFVI
Sbjct: 229 ---IPSWVSD--GHRHLL---AGGNVKANAVVAQDGSGQFKTLTDALKTVPPTNAAPFVI 280
Query: 302 YIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGI 361
Y+K GVYKE V V K M +V IGDG KT+ TG+ N+ DG+ T+KTA+ + G F+
Sbjct: 281 YVKAGVYKETVNVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYKTATFGVNGANFMAK 340
Query: 362 GIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFV 421
IGFEN+AG K QAVALRV +D++IF+ C+MDG+QDTL+ + RQFYRDC ISGTIDFV
Sbjct: 341 DIGFENTAGTSKFQAVALRVTADQAIFHNCQMDGFQDTLFVESQRQFYRDCAISGTIDFV 400
Query: 422 FGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKN 481
FGD+ V QNC + R P + Q+C+VTA GR ++N + L+ +P V K
Sbjct: 401 FGDAFGVFQNCKLICRVPAKGQKCLVTAGGRDKQNSASALVFLSSHFTGEPALTSVTPK- 459
Query: 482 KAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVK 541
+YL RPWK +S+ + +D+ I M PEGYMP G DTC + EYNN+GPG+D
Sbjct: 460 LSYLGRPWKLYSKVVIMDSTIDAMFAPEGYMPM---VGGAFKDTCTFYEYNNKGPGADTN 516
Query: 542 QRVKWQGVKTITSEGAASFVPIRFFH------GDDWIRVTRVPYSPG 582
RVKW GVK +TS AA + P +FF D WI + VPYS G
Sbjct: 517 LRVKWHGVKVLTSNVAAEYYPGKFFEIVNATARDTWIVKSGVPYSLG 563
>UniRef100_Q7F0H7 Putative pectin methylesterase [Oryza sativa]
Length = 566
Score = 455 bits (1170), Expect = e-126
Identities = 250/571 (43%), Positives = 343/571 (59%), Gaps = 22/571 (3%)
Query: 14 AIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTDYKKECE 73
AIIG ST L+VA+V AV V V F N G ++ +S SVK+VK C PTDY++ CE
Sbjct: 5 AIIGASTVLVVAVVAAVCV-VSFKN----GGGGKEDGELSTSVKSVKAFCQPTDYQQTCE 59
Query: 74 DSLIAHAGN-ITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALDTCKQVMQ 132
+ L AGN + P +L K F +T KIS+ + ++ L+E + D+RT AL CK++++
Sbjct: 60 EELGKAAGNGASSPTDLAKAMFAVTSEKISKAISESSTLEELKNDKRTSGALQNCKELLE 119
Query: 133 LSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKMKEV 192
++D+ + S E+ F++ + + + L+ WLS A+TYQ TCLD F NTTTDA KMK
Sbjct: 120 YAVDDLKTSFEKLGGFEMTNFHKAVDDLRTWLSAALTYQGTCLDGFLNTTTDAADKMKSA 179
Query: 193 LQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFELPEWVDDR 252
L +S ++ + L++++Q S T + RRLL + DG + GG +L E
Sbjct: 180 LNSSQELTEDILAVVDQFSATLGSLNI-GRRRLLAD--DGMPVWMSEGGRRQLLEAAGPE 236
Query: 253 AGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYV 312
AG + + V VA DGSG+ TI EA+ VP KN + + IY+K G Y EYV
Sbjct: 237 AGPVEF---------KPDVTVAADGSGDVKTIGEAVAKVPPKNKERYTIYVKAGTYNEYV 287
Query: 313 EVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPE 372
V + T+V IGDG KT ITGNKNF + T TA++ G+ F GI EN+AGPE
Sbjct: 288 SVGRPATNVNMIGDGIGKTIITGNKNFKMNLTTKDTATMEAIGNGFFMRGITVENTAGPE 347
Query: 373 KHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNC 432
HQAVALR QSD ++FY+C DGYQDTLY H RQF+RDC +SGTIDF+FG+S VLQNC
Sbjct: 348 NHQAVALRAQSDMAVFYQCEFDGYQDTLYPHAQRQFFRDCTVSGTIDFIFGNSQVVLQNC 407
Query: 433 TFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDF 492
RKP++NQ I+TAQGR+EK G +I ++ P K K YLARPWK++
Sbjct: 408 LLQPRKPMDNQVNIITAQGRREKRSAGGTVIHNCTVAPHPDLEKFTDKVKTYLARPWKEY 467
Query: 493 SRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTI 552
SRTIF+ IG ++ P G++ W G DT YY E +N GPG+D+ +R KW+GV+++
Sbjct: 468 SRTIFVQNEIGAVVDPVGWLEWN---GNFALDTLYYAEVDNHGPGADMSKRAKWKGVQSL 524
Query: 553 TSEGA-ASFVPIRFFHGDDWIRVTRVPYSPG 582
T + F F G ++I VPY PG
Sbjct: 525 TYQDVQKEFTVEAFIQGQEFIPKFGVPYIPG 555
>UniRef100_Q42920 Pectinesterase precursor [Medicago sativa]
Length = 447
Score = 420 bits (1079), Expect = e-116
Identities = 218/468 (46%), Positives = 293/468 (62%), Gaps = 32/468 (6%)
Query: 124 LDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTT 183
++ C +V+ ++D +S+ FD + L LKVWL+G +++Q+TCLD F NTTT
Sbjct: 1 MEICNEVLDYAVDGIHKSVGTLDQFDFHKLSEYAFDLKVWLTGTLSHQQTCLDGFANTTT 60
Query: 184 DAGKKMKEVLQTSMHMSSNGLSIINQLS---KTFEEMKQPAGRRLLKESVDGEEDVLGHG 240
AG+ M +VL+TSM +SSN + +++ +S K F+ + RRLL + DG
Sbjct: 61 KAGETMTKVLKTSMELSSNAIDMMDAVSRILKGFDTSQYSVSRRLLSD--DG-------- 110
Query: 241 GDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFV 300
+P WV+D G R+LL G +Q + VVA+DGSG F T+T+ALK VP KN PFV
Sbjct: 111 ----IPSWVND--GHRRLL---AGGNVQPNAVVAQDGSGQFKTLTDALKTVPPKNAVPFV 161
Query: 301 IYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVG 360
I++K GVYKE V V K M +V IGDG KT+ TG+ N+ DG+ T+ TA+ + G F+
Sbjct: 162 IHVKAGVYKETVNVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYNTATFGVNGANFMA 221
Query: 361 IGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDF 420
IGFEN+AG KHQAVALRV +D++IFY C+MDG+QDTLY + RQFYRDC ISGTIDF
Sbjct: 222 KDIGFENTAGTGKHQAVALRVTADQAIFYNCQMDGFQDTLYVQSQRQFYRDCSISGTIDF 281
Query: 421 VFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLK 480
VFG+ V QNC V R P + QQC+VTA GR+++N + L+ Q +P V K
Sbjct: 282 VFGERFGVFQNCKLVCRLPAKGQQCLVTAGGREKQNSASALVFQSSHFTGEPALTSVTPK 341
Query: 481 NKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDV 540
+YL RPWK +S+ + +D+ I + PEGYMPW G +TC Y EYNN+GPG+D
Sbjct: 342 -VSYLGRPWKQYSKVVIMDSTIDAIFVPEGYMPWM---GSAFKETCTYYEYNNKGPGADT 397
Query: 541 KQRVKWQGVKTITSEGAASFVPIRFFH------GDDWIRVTRVPYSPG 582
RVKW GVK +TS AA + P +FF D WI + VPYS G
Sbjct: 398 NLRVKWHGVKVLTSNVAAEYYPGKFFEIVNATARDTWIVKSGVPYSLG 445
>UniRef100_Q7XUZ9 OSJNBa0036B21.20 protein [Oryza sativa]
Length = 568
Score = 414 bits (1064), Expect = e-114
Identities = 222/570 (38%), Positives = 328/570 (56%), Gaps = 17/570 (2%)
Query: 17 GVSTFLLVAMVVAVTVNVGFNNKKEPGEETSK-ESHVSQSVKAVKTLCAPTDYKKECEDS 75
GV L+VA+VV V V + K T E++++ S K+V++LCAPT YK+ CE +
Sbjct: 9 GVGAILVVAVVVGVVATVTRSGNKAGDNFTVPGEANLATSGKSVESLCAPTLYKESCEKT 68
Query: 76 LIAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAE-KDERTKQALDTCKQVMQLS 134
L PKE+ + I ++K+ + EA+ D T+ A + CK +++ S
Sbjct: 69 LTTATSGTENPKEVFSTVAKSALESIKSAVEKSKAIGEAKTSDSMTESAREDCKALLEDS 128
Query: 135 IDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKMKEVLQ 194
+D+ R + + D+ L L+ WL+G +T+ +TC D F + A M VL+
Sbjct: 129 VDDL-RGMVEMAGGDVKVLFSRSDDLEHWLTGVMTFMDTCADGFADEKLKAD--MHSVLR 185
Query: 195 TSMHMSSNGLSIINQLSKTFEEMKQPA--GRRLLKESVDGEEDVLGHGGDFELPEWVDDR 252
+ +SSN L+I N L F+++ G + S+ E++ +G P W+ +
Sbjct: 186 NASELSSNALAITNTLGAIFKKLDLDMFKGENPIHRSLIAEQETVGG-----FPSWM--K 238
Query: 253 AGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYV 312
A RKLL + Q + VVA+DGSG F TI EA+ +PK + +VIY+K G+Y E V
Sbjct: 239 APDRKLLASGDRNRPQPNAVVAQDGSGQFKTIQEAVNSMPKGHQGRYVIYVKAGLYDEIV 298
Query: 313 EVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPE 372
V K ++ GDG +++R+TG K+F DG+ T KTA+ ++ F+ +GF N+AG E
Sbjct: 299 MVPKDKVNIFMYGDGPKRSRVTGRKSFADGITTMKTATFSVEAAGFICKNMGFHNTAGAE 358
Query: 373 KHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNC 432
+HQAVALR+ D FY CR D +QDTLY H RQF+R+C+ISGTIDF+FG+S AV QNC
Sbjct: 359 RHQAVALRINGDLGAFYNCRFDAFQDTLYVHARRQFFRNCVISGTIDFIFGNSAAVFQNC 418
Query: 433 TFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDF 492
+ R+P++NQQ VTA GR + N +GL+IQ +V D K +P R K +YL RPWK++
Sbjct: 419 LIITRRPMDNQQNSVTAHGRTDPNMKSGLVIQNCRLVPDQKLFPDRFKIPSYLGRPWKEY 478
Query: 493 SRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTI 552
SR + +++ I D I PEGYMPW G +T YY E+NNRGPG+ +RV W+G + I
Sbjct: 479 SRLVIMESTIADFIKPEGYMPWN---GEFALNTLYYAEFNNRGPGAGTSKRVNWKGFRVI 535
Query: 553 TSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
+ A F F G W++ T P+ G
Sbjct: 536 GQKEAEQFTAGPFVDGGTWLKFTGTPHFLG 565
>UniRef100_Q9MBB6 Pectin methylesterase [Salix gilgiana]
Length = 596
Score = 408 bits (1048), Expect = e-112
Identities = 232/577 (40%), Positives = 335/577 (57%), Gaps = 28/577 (4%)
Query: 15 IIGVSTFLLVAMVVAVTVNVGFNNKKEPGEET---SKESHVSQSVKAVKTLCAPTDYKKE 71
I+G F LV+ ++ NN P T K +S + +KT+C T Y+
Sbjct: 38 IVGAGVFSLVSNH---DISSPGNNGGSPSAATRPAEKAKPISHVARVIKTVCNATTYQDT 94
Query: 72 CEDSL----IAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALDTC 127
C+++L + + +PK+L+KIA +I + +KK + + R K A D C
Sbjct: 95 CQNTLEKGVLGKDPSSVQPKDLLKIAIKAADEEIDKVIKKASSFKFDKP--REKAAFDDC 152
Query: 128 KQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGK 187
++++ + +E + S++ N D+ L L WLS ++YQ+TC+D F +
Sbjct: 153 LELIEDAKEELKNSVDCIGN-DIGKLASNAPDLSNWLSAVMSYQQTCIDGFPEGKLKSD- 210
Query: 188 KMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPA--GRRLLKESVDGEEDVLGHGGDFEL 245
M++ + + ++SN L++++ L + RRLL E + L G +
Sbjct: 211 -MEKTFKATRELTSNSLAMVSSLVSFLKNFSFSGTLNRRLLAEEQNSPS--LDKDG---V 264
Query: 246 PEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKE 305
P W+ R++L K + +V VAKDGSG+F TI+EAL +P K +VI++K+
Sbjct: 265 PGWMSHED--RRILKGADKDKPKPNVSVAKDGSGDFKTISEALAAMPAKYEGRYVIFVKQ 322
Query: 306 GVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGF 365
GVY E V VTK M ++ GDG +KT +TGNKNF DGV TF+TA+ A+ GD F+ +GF
Sbjct: 323 GVYDETVTVTKKMANITMYGDGSQKTIVTGNKNFADGVQTFRTATFAVLGDGFLCKFMGF 382
Query: 366 ENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDS 425
N+AGPEKHQAVA+RVQ+DR+IF CR +GYQDTLYA T RQFYR C+I+GT+DF+FGD+
Sbjct: 383 RNTAGPEKHQAVAIRVQADRAIFLNCRFEGYQDTLYAQTHRQFYRSCVITGTVDFIFGDA 442
Query: 426 IAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYL 485
+V QNC VRKPLENQQ IVTAQGR + ++ TG+++Q I D PV+ K ++YL
Sbjct: 443 TSVFQNCLITVRKPLENQQNIVTAQGRIDGHETTGIVLQSCRIEPDKDLVPVKNKIRSYL 502
Query: 486 ARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVK 545
RPWK+FSRT+ +D+ IGD I P G++PWQ G G T YY EY+N+G G+ R+K
Sbjct: 503 GRPWKEFSRTVIMDSTIGDFIHPGGWLPWQ---GDFGLKTLYYAEYSNKGGGAQTNARIK 559
Query: 546 WQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
W G I E A F F+ G DWI + P G
Sbjct: 560 WPGYHIIKKEEAMKFTIENFYQG-DWISASGSPVHLG 595
>UniRef100_Q69PI0 Putative pectinesterase [Oryza sativa]
Length = 617
Score = 393 bits (1009), Expect = e-108
Identities = 238/606 (39%), Positives = 332/606 (54%), Gaps = 53/606 (8%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSV----------- 56
+R + + +G + +++ +V+ K E E SK S +S
Sbjct: 23 QRRRIMIALGTVSIIIILIVMGAAAITYSGKKSEEDEGGSKGSSKGKSKGGGGGDDEDGG 82
Query: 57 ------------KAVKTLCAPTDYKKECEDSLIAHAG-NITEPKELIKIAFNITIAKISE 103
K++K +CA TD+ C S+ A +++ PK++I+ A ++ + +
Sbjct: 83 GGGGKADLRAVSKSIKMMCAQTDFADSCATSIGKAANASVSSPKDIIRTAVDVIGGAVDQ 142
Query: 104 GLKKTHLLQEAEKDERTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVW 163
+ L+ D R K A+ CK++ + D+ +L+ D L + L+VW
Sbjct: 143 AFDRADLIMS--NDPRVKAAVADCKELFDDAKDDLNCTLKGIDGKD--GLKQGF-QLRVW 197
Query: 164 LSGAITYQETCLDAFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGR 223
LS I ETC+D F + + K+KE +SN L++I + S +K + R
Sbjct: 198 LSAVIANMETCIDGFPDG--EFRDKVKESFNNGREFTSNALALIEKASSFLSALKG-SQR 254
Query: 224 RLLKESVDGEEDVLGHGGDFEL-------PEWVDDRAGVRKLLNKMTGRK--LQAHVVVA 274
RLL GEED G D L PEWV D G R++L K G K L +V+VA
Sbjct: 255 RLLA----GEEDNGGGAADPHLALAEDGIPEWVPD--GDRRVL-KGGGFKNNLTPNVIVA 307
Query: 275 KDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRIT 334
KDGSG F TI EAL +PK +VIY+KEGVY EYV +TK M V GDG RK+ +T
Sbjct: 308 KDGSGKFKTINEALAAMPKTYSGRYVIYVKEGVYAEYVTITKKMASVTMYGDGSRKSIVT 367
Query: 335 GNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMD 394
G+KNF DG+ TFKTA+ A GD F+ IG+GF+N+AG KHQAVAL VQSD+S+F C MD
Sbjct: 368 GSKNFADGLTTFKTATFAAQGDGFMAIGMGFQNTAGAAKHQAVALLVQSDKSVFLNCWMD 427
Query: 395 GYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKE 454
G+QDTLYAH+ QFYR+C+I+GTIDFVFGD+ AV QNC +R+P++NQQ I TAQGR +
Sbjct: 428 GFQDTLYAHSKAQFYRNCVITGTIDFVFGDAAAVFQNCVLTLRRPMDNQQNIATAQGRAD 487
Query: 455 KNQPTGLIIQGGSIVADPKYYPVRLKN-KAYLARPWKDFSRTIFLDTYIGDMITPEGYMP 513
+ TG ++Q A+P +L + YL RPW++FSRT+ +++ I +I GYMP
Sbjct: 488 GREATGFVLQKCEFNAEPALTDAKLPPIRNYLGRPWREFSRTVIMESDIPAIIDKAGYMP 547
Query: 514 WQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEG-AASFVPIRFFHGDDWI 572
W G T YY EY N+GPG+D RV W G K + S+ A F F H WI
Sbjct: 548 WN---GEFALKTLYYAEYANKGPGADTAGRVAWPGYKKVISKADATKFTVDNFLHAKPWI 604
Query: 573 RVTRVP 578
T P
Sbjct: 605 DPTGTP 610
>UniRef100_Q9FF78 Pectinesterase [Arabidopsis thaliana]
Length = 564
Score = 391 bits (1005), Expect = e-107
Identities = 234/579 (40%), Positives = 332/579 (56%), Gaps = 43/579 (7%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGF--NNKKEPGEETSKESHVSQSVKAVKTLCAP 65
K KR+AII +S+ +LV +VV V N+KK P E + VS VK LC
Sbjct: 20 KTKKRIAIIAISSIVLVCIVVGAVVGTTARDNSKKPPTENNGEPISVS-----VKALCDV 74
Query: 66 TDYKKECEDSL-IAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQAL 124
T +K++C ++L A + + P+EL K A +TI ++S+ L D T A+
Sbjct: 75 TLHKEKCFETLGSAPNASRSSPEELFKYAVKVTITELSKVLDG--FSNGEHMDNATSAAM 132
Query: 125 DTCKQVMQLSIDEFQRSL-ERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTT 183
C +++ L++D+ ++ NFD L+ WLS TYQETC+DA
Sbjct: 133 GACVELIGLAVDQLNETMTSSLKNFD---------DLRTWLSSVGTYQETCMDALVEANK 183
Query: 184 DAGKKMKEV-LQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGD 242
+ E L+ S M+SN L+II L K + +K RR L E+ + + V D
Sbjct: 184 PSLTTFGENHLKNSTEMTSNALAIITWLGKIADTVK--FRRRRLLETGNAKVVV----AD 237
Query: 243 FELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIY 302
+ E R+LL +K +A +VVAKDGSG + TI EAL V +KN KP +IY
Sbjct: 238 LPMMEG-------RRLLESGDLKK-KATIVVAKDGSGKYRTIGEALAEVEEKNEKPTIIY 289
Query: 303 IKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIG 362
+K+GVY E V V KT +VV +GDG KT ++ NFIDG TF+TA+ A+ G F+
Sbjct: 290 VKKGVYLENVRVEKTKWNVVMVGDGQSKTIVSAGLNFIDGTPTFETATFAVFGKGFMARD 349
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
+GF N+AGP KHQAVAL V +D S+FYKC MD +QDT+YAH RQFYRDC+I GT+DF+F
Sbjct: 350 MGFINTAGPAKHQAVALMVSADLSVFYKCTMDAFQDTMYAHAQRQFYRDCVILGTVDFIF 409
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
G++ V Q C + R+P++ QQ +TAQGRK+ NQ TG+ I +I + +
Sbjct: 410 GNAAVVFQKCEILPRRPMKGQQNTITAQGRKDPNQNTGISIHNCTIKPLDNLTDI----Q 465
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
+L RPWKDFS T+ + +++ I P+G++PW G T DT +Y EY N GPG+ K
Sbjct: 466 TFLGRPWKDFSTTVIMKSFMDKFINPKGWLPW---TGDTAPDTIFYAEYLNSGPGASTKN 522
Query: 543 RVKWQGVKT-ITSEGAASFVPIRFFHGDDWIRVTRVPYS 580
RVKWQG+KT +T + A F F G++W+ T+VP++
Sbjct: 523 RVKWQGLKTSLTKKEANKFTVKPFIDGNNWLPATKVPFN 561
>UniRef100_Q84W31 Hypothetical protein At5g04960 [Arabidopsis thaliana]
Length = 564
Score = 391 bits (1005), Expect = e-107
Identities = 235/579 (40%), Positives = 332/579 (56%), Gaps = 43/579 (7%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGF--NNKKEPGEETSKESHVSQSVKAVKTLCAP 65
K KR+AII +S+ +LV +VV V N+KK P E + VS VK LC
Sbjct: 20 KTKKRIAIIAISSIVLVCIVVGAVVGTTARDNSKKPPTENNGEPISVS-----VKALCDV 74
Query: 66 TDYKKECEDSL-IAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQAL 124
T +K++C ++L A + + P+EL K A +TI ++S+ L D T A+
Sbjct: 75 TLHKEKCFETLGSAPNASRSSPEELFKYAVKVTITELSKVLDG--FSNGEHMDNATSAAM 132
Query: 125 DTCKQVMQLSIDEFQRSL-ERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTT 183
C +++ L++D+ ++ NFD L+ WLS TYQETC+DA
Sbjct: 133 GACVELIGLAVDQLNETMTSSLKNFD---------DLRTWLSSVGTYQETCMDALVEANK 183
Query: 184 DAGKKMKEV-LQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGD 242
+ E L+ S M+SN L+II L K + +K RR L E+ + + V D
Sbjct: 184 PSLTTFGENHLKNSTEMTSNALAIITWLGKIADTVK--FRRRRLLETGNAKVVV----AD 237
Query: 243 FELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIY 302
+ E R+LL +K +A +VVAKDGSG + TI EAL V +KN KP +IY
Sbjct: 238 LPMMEG-------RRLLESGDLKK-KATIVVAKDGSGKYRTIGEALAEVEEKNEKPTIIY 289
Query: 303 IKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIG 362
+K+GVY E V V KT +VV +GDG KT ++ NFIDG TF+TA+ A+ G F+
Sbjct: 290 VKKGVYLENVRVEKTKWNVVMVGDGQSKTIVSAGLNFIDGTPTFETATFAVFGKGFMARD 349
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
+GF N+AGP KHQAVAL V +D S+FYKC MD +QDT+YAH RQFYRDC+I GT+DF+F
Sbjct: 350 MGFINTAGPAKHQAVALMVSADLSVFYKCTMDAFQDTMYAHAQRQFYRDCVILGTVDFIF 409
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
G++ V Q C + R+P++ QQ +TAQGRK+ NQ TG+ I +I K +
Sbjct: 410 GNAAVVFQKCEILPRRPMKGQQNTITAQGRKDPNQNTGISIHNCTI----KPLDNLTDTQ 465
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
+L RPWKDFS T+ + +++ I P+G++PW G T DT +Y EY N GPG+ K
Sbjct: 466 TFLDRPWKDFSTTVIMKSFMDKFINPKGWLPW---TGDTAPDTIFYAEYLNSGPGASTKN 522
Query: 543 RVKWQGVKT-ITSEGAASFVPIRFFHGDDWIRVTRVPYS 580
RVKWQG+KT +T + A F F G++W+ T+VP++
Sbjct: 523 RVKWQGLKTSLTKKEANKFTVKPFIDGNNWLPATKVPFN 561
>UniRef100_Q7Y201 Putative pectin methylesterase [Arabidopsis thaliana]
Length = 614
Score = 382 bits (982), Expect = e-104
Identities = 216/558 (38%), Positives = 323/558 (57%), Gaps = 37/558 (6%)
Query: 37 NNKKEPGEET-SKESHVS--------QSVKAVKTLCAPTDYKKECEDSLIAHAGN---IT 84
++KK P T S+++ VS Q K ++TLC+ T Y + CE +L +
Sbjct: 78 SDKKSPSPPTPSQKAPVSAAQSVKPGQGDKIIQTLCSSTLYMQICEKTLKNRTDKGFALD 137
Query: 85 EPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALDTCKQVMQLSIDEFQRSLER 144
P +K A + L+K L+ +D+ K A++ CK +++ + +E SL +
Sbjct: 138 NPTTFLKSAIEAVNEDLDLVLEKVLSLKTENQDD--KDAIEQCKLLVEDAKEETVASLNK 195
Query: 145 FSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKMKEVLQTSMHMSSNGL 204
+ ++NS ++V+ L+ WLS ++YQETCLD FE + ++K + +S ++SN L
Sbjct: 196 INVTEVNSFEKVVPDLESWLSAVMSYQETCLDGFEEGNLKS--EVKTSVNSSQVLTSNSL 253
Query: 205 SIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTG 264
++I KTF E P + + + +DG +P WV + R++L +
Sbjct: 254 ALI----KTFTENLSPVMKVVERHLLDG------------IPSWVSNDD--RRMLRAVDV 295
Query: 265 RKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFI 324
+ L+ + VAKDGSG+FTTI +AL+ +P+K ++IY+K+G+Y EYV V K ++ +
Sbjct: 296 KALKPNATVAKDGSGDFTTINDALRAMPEKYEGRYIIYVKQGIYDEYVTVDKKKANLTMV 355
Query: 325 GDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSD 384
GDG +KT +TGNK+ + TF TA+ G+ F+ +GF N+AG E HQAVA+RVQSD
Sbjct: 356 GDGSQKTIVTGNKSHAKKIRTFLTATFVAQGEGFMAQSMGFRNTAGSEGHQAVAIRVQSD 415
Query: 385 RSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQ 444
RSIF CR +GYQDTLYA+T RQ+YR C+I GTIDF+FGD+ A+ QNC +RK L Q+
Sbjct: 416 RSIFLNCRFEGYQDTLYAYTHRQYYRSCVIVGTIDFIFGDAAAIFQNCNIFIRKGLPGQK 475
Query: 445 CIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGD 504
VTAQGR +K Q TG ++ I A+ PV+ + K+YL RPWK++SRTI +++ I +
Sbjct: 476 NTVTAQGRVDKFQTTGFVVHNCKIAANEDLKPVKEEYKSYLGRPWKNYSRTIIMESKIEN 535
Query: 505 MITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAASFVPIR 564
+I P G++ WQ DT YY EYNN+G D RVKW G K I E A ++
Sbjct: 536 VIDPVGWLRWQETD--FAIDTLYYAEYNNKGSSGDTTSRVKWPGFKVINKEEALNYTVGP 593
Query: 565 FFHGDDWIRVTRVPYSPG 582
F G DWI + P G
Sbjct: 594 FLQG-DWISASGSPVKLG 610
>UniRef100_O23447 Pectinesterase [Arabidopsis thaliana]
Length = 701
Score = 382 bits (981), Expect = e-104
Identities = 186/338 (55%), Positives = 241/338 (71%), Gaps = 5/338 (1%)
Query: 244 ELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYI 303
E P WV + R L + ++A+VVVAKDGSG TI +AL VP KN K FVI+I
Sbjct: 366 EFPPWVTPHSR-RLLARRPRNNGIKANVVVAKDGSGKCKTIAQALAMVPMKNTKKFVIHI 424
Query: 304 KEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFI-DGVGTFKTASVAITGDFFVGIG 362
KEGVYKE VEVTK M HV+F+GDG KT ITG+ F+ D VGT++TASVA+ GD+F+
Sbjct: 425 KEGVYKEKVEVTKKMLHVMFVGDGPTKTVITGDIAFLPDQVGTYRTASVAVNGDYFMAKD 484
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
IGFEN+AG +HQAVALRV +D ++F+ C M+GYQDTLY HT RQFYR+C +SGTIDFVF
Sbjct: 485 IGFENTAGAARHQAVALRVSADFAVFFNCHMNGYQDTLYVHTHRQFYRNCRVSGTIDFVF 544
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
GD+ AV QNC FV+R+P+E+QQCIVTAQGRK++ + TG++I I D Y PV+ KN+
Sbjct: 545 GDAKAVFQNCEFVIRRPMEHQQCIVTAQGRKDRRETTGIVIHNSRITGDASYLPVKAKNR 604
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
A+L RPWK+FSRTI ++T I D+I PEG++ W + +T +Y EY NRG GS +
Sbjct: 605 AFLGRPWKEFSRTIIMNTEIDDVIDPEGWLKWNETFAL---NTLFYTEYRNRGRGSGQGR 661
Query: 543 RVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYS 580
RV+W+G+K I+ A F P F G+ WI TR+PY+
Sbjct: 662 RVRWRGIKRISDRAAREFAPGNFLRGNTWIPQTRIPYN 699
Score = 135 bits (339), Expect = 4e-30
Identities = 74/211 (35%), Positives = 122/211 (57%), Gaps = 6/211 (2%)
Query: 12 RLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTDYKKE 71
+ ++GV T L++AMV+ V N G N+ EE +ES + + V +CA TDYK++
Sbjct: 3 KYVLLGV-TALIMAMVICVEANDG-NSPSRKMEEVHRESKLMITKTTVSIICASTDYKQD 60
Query: 72 CEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKK--THLLQEAEKDERTKQALDTCKQ 129
C SL +P+ LI+ AF++ I I G+ + L A+ D T++AL+TC++
Sbjct: 61 CTTSLATVRS--PDPRNLIRSAFDLAIISIRSGIDRGMIDLKSRADADMHTREALNTCRE 118
Query: 130 VMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKM 189
+M +ID+ +++ ++F F L + L VWLSG+ITYQ+TC+D FE ++A M
Sbjct: 119 LMDDAIDDLRKTRDKFRGFLFTRLSDFVEDLCVWLSGSITYQQTCIDGFEGIDSEAAVMM 178
Query: 190 KEVLQTSMHMSSNGLSIINQLSKTFEEMKQP 220
+ V++ H++SNGL+I L K + + P
Sbjct: 179 ERVMRKGQHLTSNGLAIAANLDKLLKAFRIP 209
>UniRef100_Q43111 Pectinesterase 3 precursor [Phaseolus vulgaris]
Length = 581
Score = 381 bits (978), Expect = e-104
Identities = 222/577 (38%), Positives = 324/577 (55%), Gaps = 38/577 (6%)
Query: 6 DAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKE---PGEETSKESHVSQSVKAVKTL 62
+ K KRL II VS+ +L+A+++A V +N+ P ++ ++ +S + ++K +
Sbjct: 22 EKKTRKRLIIIAVSSIVLIAVIIAAVAGVVIHNRNSESSPSSDSVPQTELSPAA-SLKAV 80
Query: 63 CAPTDYKKECEDSLIA-HAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTK 121
C T Y C S+ + N T+P+ L K++ + I ++S K L AE+D R +
Sbjct: 81 CDTTRYPSSCFSSISSLPESNTTDPELLFKLSLRVAIDELSSFPSK--LRANAEQDARLQ 138
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRV-LTSLKVWLSGAITYQETCLDAFEN 180
+A+D C V ++D S+ +++++ WLS A+T Q+TCLDA
Sbjct: 139 KAIDVCSSVFGDALDRLNDSISALGTVAGRIASSASVSNVETWLSAALTDQDTCLDAVGE 198
Query: 181 TTTDAGKKMKEVLQTSMHMS----SNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDV 236
+ A + + ++T+M S SN L+I+ ++ + P R L
Sbjct: 199 LNSTAARGALQEIETAMRNSTEFASNSLAIVTKILGLLSRFETPIHHRRL---------- 248
Query: 237 LGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNL 296
LG PEW+ A R+LL + VVAKDGSG F TI EALK V KK+
Sbjct: 249 LG------FPEWLG--AAERRLLEEKNNDSTP-DAVVAKDGSGQFKTIGEALKLVKKKSE 299
Query: 297 KPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGD 356
+ F +Y+KEG Y E +++ K +V+ GDG KT + G++NF+DG TF+TA+ A+ G
Sbjct: 300 ERFSVYVKEGRYVENIDLDKNTWNVMIYGDGKDKTFVVGSRNFMDGTPTFETATFAVKGK 359
Query: 357 FFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISG 416
F+ IGF N+AG KHQAVALR SDRS+F++C DG+QDTLYAH+ RQFYRDC I+G
Sbjct: 360 GFIAKDIGFVNNAGASKHQAVALRSGSDRSVFFRCSFDGFQDTLYAHSNRQFYRDCDITG 419
Query: 417 TIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYP 476
TIDF+FG++ V Q+C + R+PL NQ +TAQG+K+ NQ TG+IIQ +I +
Sbjct: 420 TIDFIFGNAAVVFQSCKIMPRQPLPNQFNTITAQGKKDPNQNTGIIIQKSTITP----FG 475
Query: 477 VRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGP 536
L YL RPWKDFS T+ + + IG ++ P G+M W + T +Y EY N GP
Sbjct: 476 NNLTAPTYLGRPWKDFSTTVIMQSDIGALLNPVGWMSW--VPNVEPPTTIFYAEYQNSGP 533
Query: 537 GSDVKQRVKWQGVK-TITSEGAASFVPIRFFHGDDWI 572
G+DV QRVKW G K TIT A F F G +W+
Sbjct: 534 GADVSQRVKWAGYKPTITDRNAEEFTVQSFIQGPEWL 570
>UniRef100_Q9SG77 Putative pectinesterase [Arabidopsis thaliana]
Length = 561
Score = 377 bits (969), Expect = e-103
Identities = 223/577 (38%), Positives = 329/577 (56%), Gaps = 39/577 (6%)
Query: 8 KRNKRLAIIGVSTFLLVAMVV-AVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPT 66
K K +AII VS +L +V+ AV + E E + +S SVKAV C T
Sbjct: 21 KTRKNIAIIAVSLVILAGIVIGAVFGTMAHKKSPETVETNNNGDSISVSVKAV---CDVT 77
Query: 67 DYKKECEDSL-IAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
+K++C ++L A + P+EL + A ITIA++S+ + + + DE+ ++
Sbjct: 78 LHKEKCFETLGSAPNASSLNPEELFRYAVKITIAEVSKAI---NAFSSSLGDEKNNITMN 134
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDA 185
C +++ L+ID +L SN D+ ++ ++ L+ WLS A TYQ TC++
Sbjct: 135 ACAELLDLTIDNLNNTLTSSSNGDV-TVPELVDDLRTWLSSAGTYQRTCVETLAPDMRPF 193
Query: 186 GKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFEL 245
G+ L+ S ++SN L+II L K + K RR L + D E D H G
Sbjct: 194 GESH---LKNSTELTSNALAIITWLGKIADSFKL---RRRLLTTADVEVDF--HAG---- 241
Query: 246 PEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKE 305
R+LL RK+ A +VVAKDGSG + TI AL+ VP+K+ K +IY+K+
Sbjct: 242 ----------RRLLQSTDLRKV-ADIVVAKDGSGKYRTIKRALQDVPEKSEKRTIIYVKK 290
Query: 306 GVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGF 365
GVY E V+V K M +V+ +GDG K+ ++G N IDG TFKTA+ A+ G F+ +GF
Sbjct: 291 GVYFENVKVEKKMWNVIVVGDGESKSIVSGRLNVIDGTPTFKTATFAVFGKGFMARDMGF 350
Query: 366 ENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDS 425
N+AGP KHQAVAL V +D + FY+C M+ YQDTLY H RQFYR+C I GT+DF+FG+S
Sbjct: 351 INTAGPSKHQAVALMVSADLTAFYRCTMNAYQDTLYVHAQRQFYRECTIIGTVDFIFGNS 410
Query: 426 IAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYL 485
+VLQ+C + R+P++ QQ +TAQGR + N TG+ I +I V +L
Sbjct: 411 ASVLQSCRILPRRPMKGQQNTITAQGRTDPNMNTGISIHRCNISPLGDLTDV----MTFL 466
Query: 486 ARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVK 545
RPWK+FS T+ +D+Y+ I +G++PW G + DT +YGEY N GPG+ K RVK
Sbjct: 467 GRPWKNFSTTVIMDSYLHGFIDRKGWLPW---TGDSAPDTIFYGEYKNTGPGASTKNRVK 523
Query: 546 WQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
W+G++ ++++ A F F G W+ T+VP+ G
Sbjct: 524 WKGLRFLSTKEANRFTVKPFIDGGRWLPATKVPFRSG 560
>UniRef100_Q9FY03 Putative pectin methylesterase precursor [Populus tremula x Populus
tremuloides]
Length = 579
Score = 373 bits (958), Expect = e-102
Identities = 222/584 (38%), Positives = 324/584 (55%), Gaps = 36/584 (6%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTD 67
K+NKRL + + LVA + AV V N + GE + +K+ C+ T
Sbjct: 22 KKNKRLLLASFAALFLVATIAAVVAGV---NSHKNGENEGAHA-------VLKSACSSTL 71
Query: 68 YKKECEDSLIAH---AGNITEPKELIKIAFNITIAKISEGLKKTH-LLQEAEKDERTKQA 123
Y + C ++ GN+ K++I+++ N+T + + L+ + + +R K A
Sbjct: 72 YPELCYSAIATVPGVTGNLASLKDVIELSINLTTKTVQQNYFTVEKLIAKTKLTKREKTA 131
Query: 124 LDTCKQVMQLSIDEFQRSLERFSNF-DLNSLDRVLTSLKVWLSGAITYQETCLDAFENTT 182
L C + + ++DE + S + + SL +L LS AIT QETCLD F +
Sbjct: 132 LHDCLETIDETLDELHEAQVDISGYPNKKSLKEQADNLITLLSSAITNQETCLDGFSHDG 191
Query: 183 TDAGKKMKEVLQTSMH---MSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGH 239
D K K +L+ H M SN L++I ++ T + R LKE +G E V
Sbjct: 192 ADK-KVRKALLKGQTHVEKMCSNALAMIKNMTDTDIANELQNTNRKLKEEKEGNERVW-- 248
Query: 240 GGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPF 299
PEW+ R+LL + + +VVVA DGSG++ T++EA+ PKK+ K +
Sbjct: 249 ------PEWMS--VADRRLLQSSS---VTPNVVVAADGSGDYKTVSEAVAAAPKKSSKRY 297
Query: 300 VIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFV 359
+I IK GVY+E VEV K +++F+GDG + T IT ++N +DG TFK+A+VA G F+
Sbjct: 298 IIQIKAGVYRENVEVPKDKHNIMFLGDGRKTTIITASRNVVDGSTTFKSATVAAVGQGFL 357
Query: 360 GIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTID 419
G+ FEN+AGP KHQAVALRV SD S FY+C M YQDTLY H+ RQF+ +C ++GT+D
Sbjct: 358 ARGVTFENTAGPSKHQAVALRVGSDLSAFYECDMLAYQDTLYVHSNRQFFINCFVAGTVD 417
Query: 420 FVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRL 479
F+FG++ AV Q+C + R+P Q+ +VTAQGR + NQ TG++IQ I A PV+
Sbjct: 418 FIFGNAAAVFQDCDYHARRPDSGQKNMVTAQGRTDPNQNTGIVIQKSRIGATSDLLPVQS 477
Query: 480 KNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSD 539
YL RPWK++SRT+ + + I D+I P G+ W +G T +Y EY N G G+
Sbjct: 478 SFPTYLGRPWKEYSRTVIMQSSITDVIQPAGWHEW---SGSFALSTLFYAEYQNSGAGAG 534
Query: 540 VKQRVKWQGVKTITS-EGAASFVPIRFFHGDDWIRVTRVPYSPG 582
RVKW+G K ITS A +F P F G W+ T P+S G
Sbjct: 535 TSSRVKWEGYKVITSATEAQAFAPGNFIAGSSWLGSTSFPFSLG 578
>UniRef100_P83948 Pectinesterase 3 precursor [Citrus sinensis]
Length = 584
Score = 372 bits (955), Expect = e-101
Identities = 224/590 (37%), Positives = 336/590 (55%), Gaps = 46/590 (7%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTD 67
K+ K+L + +T L+VA V+ + G N++K G+ ++ H +K+ C+ T
Sbjct: 25 KKKKKLFLALFATLLVVAAVIGIVA--GVNSRKNSGDNGNEPHHA-----ILKSSCSSTR 77
Query: 68 YKKECEDSLIA---HAGNITEPKELIKIAFNITIAKISE---GLKKTHLLQEAEKDERTK 121
Y C ++ A + +T K++I+++ NIT + G++K LL+ +R K
Sbjct: 78 YPDLCFSAIAAVPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQK--LLKRTNLTKREK 135
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNF-DLNSLDRVLTSLKVWLSGAITYQETCLDAFEN 180
AL C + + ++DE +++E + + SL + LK +S A+T Q TCLD F +
Sbjct: 136 VALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGTCLDGFSH 195
Query: 181 TTTDAGKKMKEVLQTSM----HMSSNGLSIINQLSKT-FEEMKQPAGRRLLKES--VDGE 233
DA K +++ L M SN L++I ++ T M+ R+L++E+ VDG
Sbjct: 196 D--DANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTDMMIMRTSNNRKLIEETSTVDG- 252
Query: 234 EDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPK 293
P W+ G R+LL + + +VVVA DGSGNF T+ ++ P+
Sbjct: 253 -----------WPAWLS--TGDRRLLQSSS---VTPNVVVAADGSGNFKTVAASVAAAPQ 296
Query: 294 KNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAI 353
K ++I IK GVY+E VEVTK +++FIGDG +T ITG++N +DG TFK+A+VA+
Sbjct: 297 GGTKRYIIRIKAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATVAV 356
Query: 354 TGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCI 413
G+ F+ I F+N+AGP KHQAVALRV +D S FY C M YQDTLY H+ RQF+ +C+
Sbjct: 357 VGEGFLARDITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCL 416
Query: 414 ISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPK 473
I+GT+DF+FG++ AVLQNC RKP Q+ +VTAQGR + NQ TG++IQ I A
Sbjct: 417 IAGTVDFIFGNAAAVLQNCDIHARKPNSGQKNMVTAQGRADPNQNTGIVIQKSRIGATSD 476
Query: 474 YYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNN 533
PV+ YL RPWK++SRT+ + + I D+I P G+ W G +T +YGE+ N
Sbjct: 477 LKPVQGSFPTYLGRPWKEYSRTVIMQSSITDVIHPAGWHEWD---GNFALNTLFYGEHQN 533
Query: 534 RGPGSDVKQRVKWQGVKTITS-EGAASFVPIRFFHGDDWIRVTRVPYSPG 582
G G+ RVKW+G + ITS A +F P F G W+ T P+S G
Sbjct: 534 AGAGAGTSGRVKWKGFRVITSATEAQAFTPGSFIAGSSWLGSTGFPFSLG 583
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,008,041,386
Number of Sequences: 2790947
Number of extensions: 43395448
Number of successful extensions: 107002
Number of sequences better than 10.0: 349
Number of HSP's better than 10.0 without gapping: 254
Number of HSP's successfully gapped in prelim test: 95
Number of HSP's that attempted gapping in prelim test: 105967
Number of HSP's gapped (non-prelim): 401
length of query: 593
length of database: 848,049,833
effective HSP length: 133
effective length of query: 460
effective length of database: 476,853,882
effective search space: 219352785720
effective search space used: 219352785720
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC144541.15


