

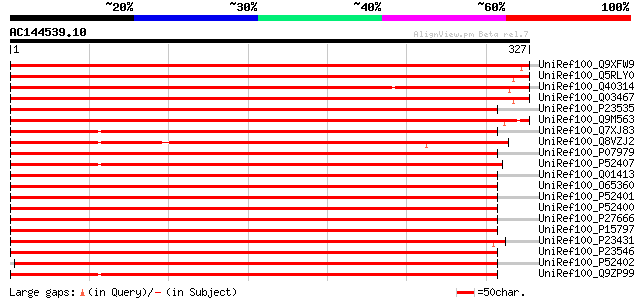
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144539.10 - phase: 0
(327 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9XFW9 Glucan-endo-1,3-beta-glucosidase precursor [Cic... 526 e-148
UniRef100_Q5RLY0 Acidic glucanase [Medicago sativa] 518 e-146
UniRef100_Q40314 Acidic glucanase [Medicago sativa] 504 e-141
UniRef100_Q03467 Glucan endo-1,3-beta-glucosidase precursor (EC ... 503 e-141
UniRef100_P23535 Glucan endo-1,3-beta-glucosidase, basic isoform... 498 e-140
UniRef100_Q9M563 Beta-1,3-glucanase [Vitis vinifera] 418 e-116
UniRef100_Q7XJ83 Beta-1,3-glucanase [Hevea brasiliensis] 405 e-112
UniRef100_Q8VZJ2 AT4g16260/dl4170c [Arabidopsis thaliana] 404 e-111
UniRef100_P07979 Lichenase precursor [Nicotiana plumbaginifolia] 403 e-111
UniRef100_P52407 Glucan endo-1,3-beta-glucosidase, basic vacuola... 402 e-111
UniRef100_Q01413 Glucan endo-1,3-beta-glucosidase B precursor (E... 400 e-110
UniRef100_O65360 1,3-beta-glucan glucanohydrolase [Solanum tuber... 397 e-109
UniRef100_P52401 Glucan endo-1,3-beta-glucosidase, basic isoform... 397 e-109
UniRef100_P52400 Glucan endo-1,3-beta-glucosidase, basic isoform... 395 e-109
UniRef100_P27666 Glucan endo-1,3-beta-glucosidase, basic vacuola... 394 e-108
UniRef100_P15797 Glucan endo-1,3-beta-glucosidase, basic vacuola... 394 e-108
UniRef100_P23431 Glucan endo-1,3-beta-glucosidase, basic vacuola... 393 e-108
UniRef100_P23546 Glucan endo-1,3-beta-glucosidase, basic vacuola... 392 e-108
UniRef100_P52402 Glucan endo-1,3-beta-glucosidase, basic isoform... 391 e-107
UniRef100_Q9ZP99 Beta-1,3-glucanase [Hevea brasiliensis] 388 e-106
>UniRef100_Q9XFW9 Glucan-endo-1,3-beta-glucosidase precursor [Cicer arietinum]
Length = 372
Score = 526 bits (1354), Expect = e-148
Identities = 260/332 (78%), Positives = 286/332 (85%), Gaps = 5/332 (1%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLP E IDL K+NNIKRMRLYDPNQAAL+ALRNSGIEL+LGVPNSDLQ++ATNN
Sbjct: 41 MMGNNLPPANEVIDLYKANNIKRMRLYDPNQAALQALRNSGIELILGVPNSDLQSLATNN 100
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
DIAIQWVQKNVLNFYPSVKIKYIAVGNEV+P+GGSS A++VLPA QNIYQAIRAKNLHD
Sbjct: 101 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVSPIGGSSWLAQYVLPATQNIYQAIRAKNLHD 160
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKVST+IDMT+IG S+PPSKGSFRSDVRSYLDP IGYLVYA APL N+Y YFSY NP
Sbjct: 161 QIKVSTSIDMTLIGNSFPPSKGSFRSDVRSYLDPFIGYLVYAGAPLLVNVYPYFSYVGNP 220
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALFTSPNV+V DG GYQNLFDA+LDS+HAA+DNTGIG+V VVVSESGWPSDG
Sbjct: 221 RDISLPYALFTSPNVMVQDGQYGYQNLFDAMLDSVHAALDNTGIGWVNVVVSESGWPSDG 280
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
G AT+YDNAR+YLDNLIRHV GTP R ETYIF +FDENQK+PELEKHFGVF PNKQ
Sbjct: 281 GSATSYDNARIYLDNLIRHVGKGTPRRPWATETYIFAMFDENQKSPELEKHFGVFNPNKQ 340
Query: 301 KKYPFGFQGKIDGTNLFNVSF-----PLKSDI 327
KKYPFGF G+ + N F LKSD+
Sbjct: 341 KKYPFGFGGERRNGEIVNDDFNATTVSLKSDM 372
>UniRef100_Q5RLY0 Acidic glucanase [Medicago sativa]
Length = 370
Score = 518 bits (1335), Expect = e-146
Identities = 255/330 (77%), Positives = 286/330 (86%), Gaps = 3/330 (0%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLP E IDL K+NNIKRMRLYDPNQAAL ALRNSGIEL+LGVPNSDLQ++ATN+
Sbjct: 41 MMGNNLPPANEVIDLYKANNIKRMRLYDPNQAALNALRNSGIELILGVPNSDLQSLATNS 100
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
D A QWVQ+NVLNF+PSVKIKYIAVGNEV+PVGGSS A++VLPA QNIYQAIRAKNLHD
Sbjct: 101 DNARQWVQRNVLNFWPSVKIKYIAVGNEVSPVGGSSWLAQYVLPATQNIYQAIRAKNLHD 160
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKVSTAIDMT+IG S+PPSKGSFR+DVR+YLDP IGYLVYA APL N+Y YFS+ NP
Sbjct: 161 QIKVSTAIDMTLIGNSFPPSKGSFRNDVRAYLDPFIGYLVYAGAPLLVNVYPYFSHVGNP 220
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALFTSP V+V DG GYQNLFDA+LDS+HAA+DNTGIG+V VVVSESGWPSDG
Sbjct: 221 RDISLPYALFTSPGVMVQDGPNGYQNLFDAMLDSVHAALDNTGIGWVNVVVSESGWPSDG 280
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
G AT+YDNAR+YLDNLIRHV GTP R ETYIF +FDENQK+PELEKHFGVFYPNKQ
Sbjct: 281 GAATSYDNARIYLDNLIRHVGKGTPRRPWATETYIFAMFDENQKSPELEKHFGVFYPNKQ 340
Query: 301 KKYPFGFQGKIDGTNL---FNVSFPLKSDI 327
KKYPFGF G+ +G + FN + LKSD+
Sbjct: 341 KKYPFGFGGERNGEIVNGDFNATISLKSDM 370
>UniRef100_Q40314 Acidic glucanase [Medicago sativa]
Length = 368
Score = 504 bits (1298), Expect = e-141
Identities = 251/329 (76%), Positives = 281/329 (85%), Gaps = 3/329 (0%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLP E IDL K+NNIKRMRLYDPNQAAL ALRNSGIEL+LGVPNSDLQ++ATN+
Sbjct: 41 MMGNNLPPANEVIDLYKANNIKRMRLYDPNQAALNALRNSGIELILGVPNSDLQSLATNS 100
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
D A QWVQ+NVLNF+PSVKIKYIAVGNEV+PVGGSS ++VLPA QNIYQAIRAKNLHD
Sbjct: 101 DNARQWVQRNVLNFWPSVKIKYIAVGNEVSPVGGSSWLGQYVLPATQNIYQAIRAKNLHD 160
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QI VSTAIDMT+IG S+PPSKGSFR+DVR+YLDP IGYLVYA APL N+Y YFS+ NP
Sbjct: 161 QILVSTAIDMTLIGNSFPPSKGSFRNDVRAYLDPFIGYLVYAGAPLLVNVYPYFSHVGNP 220
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALFTSP V+V DG GYQNLFDA+LDS+HAA+DNTGIG+V VVVSESGWPSDG
Sbjct: 221 RDISLPYALFTSPGVMVQDGPNGYQNLFDAMLDSVHAALDNTGIGWVNVVVSESGWPSDG 280
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
G AT+YDNAR+YLDNLIR+ GTP R ETYIF +FDENQK+PELEKHFGVFYPNKQ
Sbjct: 281 G-ATSYDNARIYLDNLIRYEGKGTPRRPWATETYIFAMFDENQKSPELEKHFGVFYPNKQ 339
Query: 301 KKYPFGFQGKIDG--TNLFNVSFPLKSDI 327
KKYPFGF G+ G FN + LKSD+
Sbjct: 340 KKYPFGFGGERMGIVNGDFNATISLKSDM 368
>UniRef100_Q03467 Glucan endo-1,3-beta-glucosidase precursor (EC 3.2.1.39)
((1->3)-beta- glucan endohydrolase)
((1->3)-beta-glucanase) [Pisum sativum]
Length = 370
Score = 503 bits (1294), Expect = e-141
Identities = 249/331 (75%), Positives = 280/331 (84%), Gaps = 4/331 (1%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLP E I L K+NNIKRMRLYDPNQ AL ALR+SGIEL+LG+PNSDLQ +ATN
Sbjct: 40 MMGNNLPPANEVIALYKANNIKRMRLYDPNQPALNALRDSGIELILGIPNSDLQTLATNQ 99
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
D A QWVQ+NVLNFYPSVKIKYIAVGNEV+PVGGSS A++VLPA QN+YQAIRA+ LHD
Sbjct: 100 DSARQWVQRNVLNFYPSVKIKYIAVGNEVSPVGGSSWLAQYVLPATQNVYQAIRAQGLHD 159
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKV+TAIDMT+IG S+PPSKGSFRSDVRSYLDP IGYLVYA APL N+Y YFS+ NP
Sbjct: 160 QIKVTTAIDMTLIGNSFPPSKGSFRSDVRSYLDPFIGYLVYAGAPLLVNVYPYFSHIGNP 219
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALFTSP V+V DG GYQNLFDA+LDS+HAA+DNTGIG+V VVVSESGWPSDG
Sbjct: 220 RDISLPYALFTSPGVMVQDGPNGYQNLFDAMLDSVHAALDNTGIGWVNVVVSESGWPSDG 279
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
G AT+YDNAR+YLDNLIRHV GTP R E Y+F +FDENQK+PELEKHFGVFYPNKQ
Sbjct: 280 GSATSYDNARIYLDNLIRHVGKGTPRRPWATEAYLFAMFDENQKSPELEKHFGVFYPNKQ 339
Query: 301 KKYPFGFQG-KIDGTNL---FNVSFPLKSDI 327
KKYPFGF G + DG + FN + LKSD+
Sbjct: 340 KKYPFGFGGERRDGEIVEGDFNGTVSLKSDM 370
>UniRef100_P23535 Glucan endo-1,3-beta-glucosidase, basic isoform precursor (EC
3.2.1.39) ((1->3)-beta-glucan endohydrolase)
((1->3)-beta- glucanase) [Phaseolus vulgaris]
Length = 348
Score = 498 bits (1283), Expect = e-140
Identities = 240/308 (77%), Positives = 274/308 (88%), Gaps = 1/308 (0%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLPS E I+L +SNNI+RMRLYDPNQAAL+ALRNSGIEL+LGVPNSDLQ +ATN
Sbjct: 8 MMGNNLPSANEVINLYRSNNIRRMRLYDPNQAALQALRNSGIELILGVPNSDLQGLATNA 67
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
D A QWVQ+NVLNF+PSVKIKYIAVGNEV+PVGGSS +A++VLPA+QN+YQA+RA+ LHD
Sbjct: 68 DTARQWVQRNVLNFWPSVKIKYIAVGNEVSPVGGSSWYAQYVLPAVQNVYQAVRAQGLHD 127
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
QIKVSTAIDMT+IG SYPPS+GSFR DVRSYLDPIIGYL+YA+APL N+Y YFSY NP
Sbjct: 128 QIKVSTAIDMTLIGNSYPPSQGSFRGDVRSYLDPIIGYLLYASAPLHVNVYPYFSYSGNP 187
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALFTSPNVVV DG GYQNLFDA+LDS+HAAIDNT IG+V+VVVSESGWPSDG
Sbjct: 188 RDISLPYALFTSPNVVVRDGQYGYQNLFDAMLDSVHAAIDNTRIGYVEVVVSESGWPSDG 247
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMR-SGPIETYIFGLFDENQKNPELEKHFGVFYPNK 299
GF TYDNARVYLDNL+R G+P R S P ETYIF +FDENQK+PE+EKHFG+F P+K
Sbjct: 248 GFGATYDNARVYLDNLVRRAGRGSPRRPSKPTETYIFAMFDENQKSPEIEKHFGLFKPSK 307
Query: 300 QKKYPFGF 307
+KKYPFGF
Sbjct: 308 EKKYPFGF 315
>UniRef100_Q9M563 Beta-1,3-glucanase [Vitis vinifera]
Length = 360
Score = 418 bits (1075), Expect = e-116
Identities = 209/333 (62%), Positives = 257/333 (76%), Gaps = 7/333 (2%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M+GNNLP + + L KS NI RMR+YDPNQAAL+ALR S I+LMLGVPNSDLQ +ATN
Sbjct: 29 MLGNNLPPASQVVALYKSRNIDRMRIYDPNQAALQALRGSNIQLMLGVPNSDLQGLATNP 88
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPV-GGSSQFAKFVLPAIQNIYQAIRAKNLH 119
A WVQ+NV N++P V +YIAVGNEV+PV GG+S+FA+FVLPA++NI A+ + L
Sbjct: 89 SQAQSWVQRNVRNYWPGVSFRYIAVGNEVSPVNGGTSRFAQFVLPAMRNIRAALASAGLQ 148
Query: 120 DQIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDN 179
D++KVSTAID+T++G SYPPS+G+FR DVR YLDPII +LV +PL ANIY YFSY N
Sbjct: 149 DRVKVSTAIDLTLLGNSYPPSQGAFRGDVRGYLDPIIRFLVDNKSPLLANIYPYFSYSGN 208
Query: 180 PKDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSD 239
PKDISL YALFT+ +VVVWDG RGY+NLFDA+LD+L++A++ G ++VV+SESGWPS
Sbjct: 209 PKDISLPYALFTANSVVVWDGQRGYKNLFDAMLDALYSALERAGGASLEVVLSESGWPSA 268
Query: 240 GGFATTYDNARVYLDNLIRHVKGGTPMRSG-PIETYIFGLFDENQKNPELEKHFGVFYPN 298
GGF TT DNAR Y NLIRHVKGGTP R G IETY+F +FDEN+K P+LEKHFG+F+PN
Sbjct: 269 GGFGTTVDNARTYNSNLIRHVKGGTPKRPGRAIETYLFAMFDENKKEPQLEKHFGLFFPN 328
Query: 299 KQKKYPFGFQGK----IDGTNLFNVSFPLKSDI 327
KQ KY F G+ + N N LKSDI
Sbjct: 329 KQPKYSINFSGEKPWDVSSENDTNEE-SLKSDI 360
>UniRef100_Q7XJ83 Beta-1,3-glucanase [Hevea brasiliensis]
Length = 374
Score = 405 bits (1041), Expect = e-112
Identities = 200/309 (64%), Positives = 245/309 (78%), Gaps = 3/309 (0%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M GNNLP E I L K +NIKRMR+YDPN+A LEALR S IEL+LGVPNSDLQ++ TN
Sbjct: 44 MQGNNLPPVSEVIALYKQSNIKRMRIYDPNRAVLEALRGSNIELILGVPNSDLQSL-TNP 102
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPV-GGSSQFAKFVLPAIQNIYQAIRAKNLH 119
A WVQKNV F+ SV +YIAVGNE++PV GG++ A+FVLPA++NI+ AIR+ L
Sbjct: 103 SNANSWVQKNVRGFWSSVLFRYIAVGNEISPVNGGTAWLAQFVLPAMRNIHDAIRSAGLQ 162
Query: 120 DQIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDN 179
DQIKVSTAID+T++G SYPPS G+FR DVRSYLDPIIG+L +PL ANIY YF+Y N
Sbjct: 163 DQIKVSTAIDLTLVGNSYPPSAGAFRDDVRSYLDPIIGFLSSIRSPLLANIYPYFTYAGN 222
Query: 180 PKDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSD 239
P+DISL YALFTSP+VVVWDG RGY+NLFDA LD+L++A++ G ++VVVSESGWPS
Sbjct: 223 PRDISLPYALFTSPSVVVWDGQRGYKNLFDATLDALYSALERASGGSLEVVVSESGWPSA 282
Query: 240 GGFATTYDNARVYLDNLIRHVKGGTPMR-SGPIETYIFGLFDENQKNPELEKHFGVFYPN 298
G FA T+DN R YL NLI+HVKGGTP R + IETY+F +FDEN+K PE+EKHFG+F+P+
Sbjct: 283 GAFAATFDNGRTYLSNLIQHVKGGTPKRPNRAIETYLFAMFDENKKQPEVEKHFGLFFPD 342
Query: 299 KQKKYPFGF 307
K+ KY F
Sbjct: 343 KRPKYNLNF 351
>UniRef100_Q8VZJ2 AT4g16260/dl4170c [Arabidopsis thaliana]
Length = 344
Score = 404 bits (1039), Expect = e-111
Identities = 198/317 (62%), Positives = 248/317 (77%), Gaps = 8/317 (2%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLPSQ + I L + NNI+R+RLYDPNQAAL ALRN+GIE+++GVPN+DL+++ TN
Sbjct: 30 MMGNNLPSQSDTIALFRQNNIRRVRLYDPNQAALNALRNTGIEVIIGVPNTDLRSL-TNP 88
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
A W+Q NVLN+YP+V KYIAVGNEV+P G VLPA++N+Y A+R NL D
Sbjct: 89 SSARSWLQNNVLNYYPAVSFKYIAVGNEVSPSNGGD----VVLPAMRNVYDALRGANLQD 144
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
+IKVSTAIDMT+IG S+PPS G FR DVR Y+DP+IG+L N+ L ANIY YFSY DNP
Sbjct: 145 RIKVSTAIDMTLIGNSFPPSSGEFRGDVRWYIDPVIGFLTSTNSALLANIYPYFSYVDNP 204
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALFTSP+VVVWDGSRGYQNLFDALLD +++A++ +G G + VVVSESGWPS+G
Sbjct: 205 RDISLSYALFTSPSVVVWDGSRGYQNLFDALLDVVYSAVERSGGGSLPVVVSESGWPSNG 264
Query: 241 GFATTYDNARVYLDNLIRHVK--GGTPMRSG-PIETYIFGLFDENQKNPELEKHFGVFYP 297
G A ++DNAR + NL V+ GTP R G +ETY+F +FDENQK+PE+EK+FG+F+P
Sbjct: 265 GNAASFDNARAFYTNLASRVRENRGTPKRPGRGVETYLFAMFDENQKSPEIEKNFGLFFP 324
Query: 298 NKQKKYPFGFQGKIDGT 314
NKQ K+P F DGT
Sbjct: 325 NKQPKFPITFSAARDGT 341
>UniRef100_P07979 Lichenase precursor [Nicotiana plumbaginifolia]
Length = 370
Score = 403 bits (1036), Expect = e-111
Identities = 194/308 (62%), Positives = 242/308 (77%), Gaps = 1/308 (0%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M+GNNLP + + L KS NI+RMRLYDPNQAAL+ALR S IE+MLGVPNSDLQNIA N
Sbjct: 39 MLGNNLPPASQVVQLYKSKNIRRMRLYDPNQAALQALRGSNIEVMLGVPNSDLQNIAANP 98
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
A WVQ+NV NF+P+VK +YIAVGNEV+PV G+S +++LPA++NI AI + L +
Sbjct: 99 SNANNWVQRNVRNFWPAVKFRYIAVGNEVSPVTGTSSLTRYLLPAMRNIRNAISSAGLQN 158
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVS+++DMT+IG S+PPS+GSFR+DVRS++DPIIG++ N+PL NIY YFSY NP
Sbjct: 159 NIKVSSSVDMTLIGNSFPPSQGSFRNDVRSFIDPIIGFVRRINSPLLVNIYPYFSYAGNP 218
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
+DISL YALFT+PNVVV DGS GY+NLFDA+ D+++AA+ G G +++VVSESGWPS G
Sbjct: 219 RDISLPYALFTAPNVVVQDGSLGYRNLFDAMSDAVYAALSRAGGGSIEIVVSESGWPSAG 278
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGP-IETYIFGLFDENQKNPELEKHFGVFYPNK 299
FA T +NA Y NLI+HVK G+P R IETY+F +FDEN KNPELEKHFG+F PNK
Sbjct: 279 AFAATTNNAATYYKNLIQHVKRGSPRRPNKVIETYLFAMFDENNKNPELEKHFGLFSPNK 338
Query: 300 QKKYPFGF 307
Q KYP F
Sbjct: 339 QPKYPLSF 346
>UniRef100_P52407 Glucan endo-1,3-beta-glucosidase, basic vacuolar isoform precursor
(EC 3.2.1.39) ((1->3)-beta-glucan endohydrolase)
((1->3)-beta- glucanase) [Hevea brasiliensis]
Length = 374
Score = 402 bits (1032), Expect = e-111
Identities = 199/312 (63%), Positives = 244/312 (77%), Gaps = 3/312 (0%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M GNNLP E I L K +NI RMR+YDPN+A LEALR S IEL+LGVPNSDLQ++ TN
Sbjct: 44 MQGNNLPPVSEVIALYKKSNITRMRIYDPNRAVLEALRGSNIELILGVPNSDLQSL-TNP 102
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVG-GSSQFAKFVLPAIQNIYQAIRAKNLH 119
A WVQKNV F+ SV +YIAVGNE++PV G++ A+FVLPA++NI+ AIR+ L
Sbjct: 103 SNAKSWVQKNVRGFWSSVLFRYIAVGNEISPVNRGTAWLAQFVLPAMRNIHDAIRSAGLQ 162
Query: 120 DQIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDN 179
DQIKVSTAID+T++G SYPPS G+FR DVRSYLDPIIG+L +PL ANIY YF+Y N
Sbjct: 163 DQIKVSTAIDLTLVGNSYPPSAGAFRDDVRSYLDPIIGFLSSIRSPLLANIYPYFTYAYN 222
Query: 180 PKDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSD 239
P+DISL YALFTSP+VVVWDG RGY+NLFDA LD+L++A++ G ++VVVSESGWPS
Sbjct: 223 PRDISLPYALFTSPSVVVWDGQRGYKNLFDATLDALYSALERASGGSLEVVVSESGWPSA 282
Query: 240 GGFATTYDNARVYLDNLIRHVKGGTPMR-SGPIETYIFGLFDENQKNPELEKHFGVFYPN 298
G FA T+DN R YL NLI+HVKGGTP R + IETY+F +FDEN+K PE+EKHFG+F+PN
Sbjct: 283 GAFAATFDNGRTYLSNLIQHVKGGTPKRPNRAIETYLFAMFDENKKQPEVEKHFGLFFPN 342
Query: 299 KQKKYPFGFQGK 310
K +KY F +
Sbjct: 343 KWQKYNLNFSAE 354
>UniRef100_Q01413 Glucan endo-1,3-beta-glucosidase B precursor (EC 3.2.1.39) ((1->3)-
beta-glucan endohydrolase B) ((1->3)-beta-glucanase B)
[Lycopersicon esculentum]
Length = 360
Score = 400 bits (1028), Expect = e-110
Identities = 195/307 (63%), Positives = 238/307 (77%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLPS E I L KS NI+R+RLYDPN AL ALR S IE++LG+PN D+++I++
Sbjct: 33 MMGNNLPSHSEVIQLYKSRNIRRLRLYDPNHGALNALRGSNIEVILGLPNVDVKHISSGM 92
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WVQKNV +F+P VKIKYIAVGNE++PV G+S A F +PA+ NIY+AI L +
Sbjct: 93 EHARWWVQKNVRDFWPHVKIKYIAVGNEISPVTGTSNLAPFQVPALVNIYKAIGEAGLGN 152
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVST++DMT+IG SYPPS+GSFR+DVR + DPI+G+L APL NIY YFSY NP
Sbjct: 153 DIKVSTSVDMTLIGNSYPPSQGSFRNDVRWFTDPIVGFLRDTRAPLLVNIYPYFSYSGNP 212
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
ISL YALFT+PNVVV DGSR Y+NLFDA+LDS++AA+D TG G V +VVSESGWPS G
Sbjct: 213 GQISLPYALFTAPNVVVQDGSRQYRNLFDAMLDSVYAAMDRTGGGSVGIVVSESGWPSAG 272
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
F T++NA+ YL NLI+H K G+P + GPIETYIF +FDEN KNPELEKHFG+F PNKQ
Sbjct: 273 AFGATHENAQTYLRNLIQHAKEGSPRKPGPIETYIFAMFDENNKNPELEKHFGMFSPNKQ 332
Query: 301 KKYPFGF 307
KY F
Sbjct: 333 PKYNLNF 339
>UniRef100_O65360 1,3-beta-glucan glucanohydrolase [Solanum tuberosum]
Length = 363
Score = 397 bits (1020), Expect = e-109
Identities = 195/307 (63%), Positives = 234/307 (75%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLPS E I L KS NI R+RLYDPN AL ALR S IE++LG+PN D+++IA+
Sbjct: 33 MMGNNLPSHSEVIQLYKSRNIGRLRLYDPNHGALNALRRSNIEVILGLPNVDVKHIASGM 92
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WVQKNV +F+P VKIKYIAVGNE++PV G+S F +PA+ NIY+AI L +
Sbjct: 93 EHARWWVQKNVKDFWPDVKIKYIAVGNEISPVTGTSSLTSFQVPALVNIYKAIGEAGLGN 152
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVST++DMT+IG SYPPS+GSFR+DVR + DPI+G+L APL NIY YFSY NP
Sbjct: 153 DIKVSTSVDMTLIGNSYPPSQGSFRNDVRWFTDPIVGFLRDTRAPLLVNIYPYFSYSGNP 212
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
ISL YALFT+PNVVV DGSR Y+NLFDA+LDS++AA++ TG G V +VVSESGWPS G
Sbjct: 213 GQISLPYALFTAPNVVVQDGSRQYRNLFDAMLDSVYAAMERTGGGSVGIVVSESGWPSAG 272
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
F T DNA YL NLI+H K G+P + GPIETYIF +FDEN KNPELEKHFG+F PNKQ
Sbjct: 273 AFGATQDNAATYLRNLIQHAKEGSPRKPGPIETYIFAMFDENNKNPELEKHFGLFSPNKQ 332
Query: 301 KKYPFGF 307
KY F
Sbjct: 333 PKYNLNF 339
>UniRef100_P52401 Glucan endo-1,3-beta-glucosidase, basic isoform 2 precursor (EC
3.2.1.39) ((1->3)-beta-glucan endohydrolase)
((1->3)-beta- glucanase) [Solanum tuberosum]
Length = 363
Score = 397 bits (1019), Expect = e-109
Identities = 194/307 (63%), Positives = 234/307 (76%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLPS E I L KS NI R+RLYDPNQ AL ALR S IE++LG+PN D+++IA+
Sbjct: 33 MMGNNLPSHSEVIQLYKSRNIGRLRLYDPNQGALNALRGSNIEVILGLPNVDVKHIASGM 92
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WVQKNV +F+P VKIKYIAVGNE++PV G+S F +PA+ NIY+A+ L +
Sbjct: 93 EHARWWVQKNVKDFWPDVKIKYIAVGNEISPVTGTSSLTSFQVPALVNIYKAVGEAGLGN 152
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVST++DMT+IG SYPPS+GSFR+DVR + DPI+G+L APL NIY YFSY NP
Sbjct: 153 DIKVSTSVDMTLIGNSYPPSQGSFRNDVRWFTDPIVGFLRDTRAPLLVNIYPYFSYSGNP 212
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
ISL YALFT+PNVVV DGSR Y+NLFDA+LDS++AA++ TG G V +VVSE GWPS G
Sbjct: 213 GQISLPYALFTAPNVVVQDGSRQYRNLFDAMLDSVYAAMERTGGGSVGIVVSECGWPSAG 272
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
F T DNA YL NLI+H K G+P + GPIETYIF +FDEN KNPELEKHFG+F PNKQ
Sbjct: 273 AFGATQDNAATYLRNLIQHAKEGSPRKPGPIETYIFAMFDENNKNPELEKHFGLFSPNKQ 332
Query: 301 KKYPFGF 307
KY F
Sbjct: 333 PKYNLNF 339
>UniRef100_P52400 Glucan endo-1,3-beta-glucosidase, basic isoform 1 precursor (EC
3.2.1.39) ((1->3)-beta-glucan endohydrolase)
((1->3)-beta- glucanase) [Solanum tuberosum]
Length = 337
Score = 395 bits (1015), Expect = e-109
Identities = 193/307 (62%), Positives = 233/307 (75%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
MMGNNLPS E I L KS NI R+RLYDPN AL ALR S IE++LG+PN D+++IA+
Sbjct: 7 MMGNNLPSHSEVIQLYKSRNIGRLRLYDPNHGALNALRGSNIEVILGLPNVDVKHIASGM 66
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WVQKNV +F+P VKIKYIAVGNE++PV G+S F +PA+ NIY+A+ L +
Sbjct: 67 EHARWWVQKNVKDFWPDVKIKYIAVGNEISPVTGTSSLTSFQVPALVNIYKAVGEAGLGN 126
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVST++DMT+IG SYPPS+GSFR+DVR + DPI+G+L APL NIY YFSY NP
Sbjct: 127 DIKVSTSVDMTLIGNSYPPSQGSFRNDVRWFTDPIVGFLRDTRAPLLVNIYPYFSYSGNP 186
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
ISL YALFT+PN VV DGSR Y+NLFDA+LDS++AA++ TG G V +VVSESGWPS G
Sbjct: 187 GQISLPYALFTAPNAVVQDGSRQYRNLFDAMLDSVYAAMERTGGGSVGIVVSESGWPSAG 246
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
F T DNA YL NLI+H K G+P + GPIETYIF +FDEN KNPELEKHFG+F PNKQ
Sbjct: 247 AFGATQDNAATYLRNLIQHAKEGSPRKPGPIETYIFAMFDENNKNPELEKHFGLFSPNKQ 306
Query: 301 KKYPFGF 307
KY F
Sbjct: 307 PKYNLNF 313
>UniRef100_P27666 Glucan endo-1,3-beta-glucosidase, basic vacuolar isoform GLB
precursor (EC 3.2.1.39) ((1->3)-beta-glucan
endohydrolase) ((1->3)-beta- glucanase) [Nicotiana
tabacum]
Length = 370
Score = 394 bits (1012), Expect = e-108
Identities = 191/307 (62%), Positives = 235/307 (76%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M+GNNLP+ E I L KS NI R+RLYDPN AL+AL+ S IE+MLG+PNSD+++IA+
Sbjct: 41 MLGNNLPNHWEVIQLYKSRNIGRLRLYDPNHGALQALKGSNIEVMLGLPNSDVKHIASGM 100
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WVQKNV +F+P VKIKYIAVGNE++PV G+S F+ PA+ NIY+AI L +
Sbjct: 101 EHARWWVQKNVKDFWPDVKIKYIAVGNEISPVTGTSYLTSFLTPAMVNIYKAIGEAGLGN 160
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVST++DMT+IG SYPPS+GSFR+D R + DPI+G+L APL NIY YFSY NP
Sbjct: 161 NIKVSTSVDMTLIGNSYPPSQGSFRNDARWFTDPIVGFLRDTRAPLLVNIYPYFSYSGNP 220
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
ISL Y+LFT+PNVVV DGSR Y+NLFDA+LDS++AA++ +G V +VVSESGWPS G
Sbjct: 221 GQISLPYSLFTAPNVVVQDGSRQYRNLFDAMLDSVYAALERSGGASVGIVVSESGWPSAG 280
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
F TYDNA YL NLI+H K G+P + GPIETYIF +FDEN KNPELEKHFG+F PNKQ
Sbjct: 281 AFGATYDNAATYLRNLIQHAKEGSPRKPGPIETYIFAMFDENNKNPELEKHFGLFSPNKQ 340
Query: 301 KKYPFGF 307
KY F
Sbjct: 341 PKYNLNF 347
>UniRef100_P15797 Glucan endo-1,3-beta-glucosidase, basic vacuolar isoform precursor
(EC 3.2.1.39) ((1->3)-beta-glucan endohydrolase)
((1->3)-beta- glucanase) [Nicotiana tabacum]
Length = 371
Score = 394 bits (1012), Expect = e-108
Identities = 191/307 (62%), Positives = 235/307 (76%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M+GNNLP+ E I L KS NI R+RLYDPN AL+AL+ S IE+MLG+PNSD+++IA+
Sbjct: 42 MLGNNLPNHWEVIQLYKSRNIGRLRLYDPNHGALQALKGSNIEVMLGLPNSDVKHIASGM 101
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WVQKNV +F+P VKIKYIAVGNE++PV G+S F+ PA+ NIY+AI L +
Sbjct: 102 EHARWWVQKNVKDFWPDVKIKYIAVGNEISPVTGTSYLTSFLTPAMVNIYKAIGEAGLGN 161
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVST++DMT+IG SYPPS+GSFR+D R + DPI+G+L APL NIY YFSY NP
Sbjct: 162 NIKVSTSVDMTLIGNSYPPSQGSFRNDARWFTDPIVGFLRDTRAPLLVNIYPYFSYSGNP 221
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
ISL Y+LFT+PNVVV DGSR Y+NLFDA+LDS++AA++ +G V +VVSESGWPS G
Sbjct: 222 GQISLPYSLFTAPNVVVQDGSRQYRNLFDAMLDSVYAALERSGGASVGIVVSESGWPSAG 281
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
F TYDNA YL NLI+H K G+P + GPIETYIF +FDEN KNPELEKHFG+F PNKQ
Sbjct: 282 AFGATYDNAATYLRNLIQHAKEGSPRKPGPIETYIFAMFDENNKNPELEKHFGLFSPNKQ 341
Query: 301 KKYPFGF 307
KY F
Sbjct: 342 PKYNINF 348
>UniRef100_P23431 Glucan endo-1,3-beta-glucosidase, basic vacuolar isoform precursor
(EC 3.2.1.39) ((1->3)-beta-glucan endohydrolase)
((1->3)-beta- glucanase) [Nicotiana plumbaginifolia]
Length = 365
Score = 393 bits (1010), Expect = e-108
Identities = 192/314 (61%), Positives = 239/314 (75%), Gaps = 2/314 (0%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M+GNNLP+ E I L KS NI R+RLYDPN AL+AL+ S IE+MLG+PNSD+++IA+
Sbjct: 41 MLGNNLPNHWEVIQLYKSRNIGRLRLYDPNHGALQALKGSNIEVMLGLPNSDVKHIASGM 100
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WVQKNV +F+P VKIKYIAVGNE++PV G+S F+ PA+ NIY+AI L +
Sbjct: 101 EHARWWVQKNVKDFWPDVKIKYIAVGNEISPVTGTSYLTSFLTPAMVNIYKAIGEAGLGN 160
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVST++DMT+IG SYPPS+GSFR+D R ++DPI+G+L APL NIY YFSY NP
Sbjct: 161 NIKVSTSVDMTLIGNSYPPSQGSFRNDARWFVDPIVGFLRDTRAPLLVNIYPYFSYSGNP 220
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
ISL Y+LFT+PNVVV DGSR Y+NLFDA+LDS++AA++ +G V +VVSESGWPS G
Sbjct: 221 GQISLPYSLFTAPNVVVQDGSRQYRNLFDAMLDSVYAALERSGGASVGIVVSESGWPSAG 280
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
F TYDNA YL NLI+H K G+P + PIETYIF +FDEN KNPELEKHFG+F PNKQ
Sbjct: 281 AFGATYDNAATYLKNLIQHAKEGSPRKPRPIETYIFAMFDENNKNPELEKHFGLFSPNKQ 340
Query: 301 KKY--PFGFQGKID 312
KY FG G ++
Sbjct: 341 PKYNLNFGVSGSVE 354
>UniRef100_P23546 Glucan endo-1,3-beta-glucosidase, basic vacuolar isoform GGIB50
precursor (EC 3.2.1.39) ((1->3)-beta-glucan
endohydrolase) ((1->3)- beta-glucanase) [Nicotiana
tabacum]
Length = 370
Score = 392 bits (1006), Expect = e-108
Identities = 190/307 (61%), Positives = 235/307 (75%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M+GNNLP+ E I L KS NI R+RLYDPN AL+AL+ S IE+MLG+PNSD+++IA+
Sbjct: 41 MLGNNLPNHWEVIQLYKSRNIGRLRLYDPNHGALQALKGSNIEVMLGLPNSDVKHIASGM 100
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHD 120
+ A WVQKNV +F+P VKIKYIAVGNE++PV G+S F+ PA+ NIY+AI L +
Sbjct: 101 EHARWWVQKNVKDFWPDVKIKYIAVGNEISPVTGTSYLTSFLTPAMVNIYKAIGEAGLGN 160
Query: 121 QIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNP 180
IKVST++DMT+IG SYPPS+GSFR+D R ++D I+G+L APL NIY YFSY NP
Sbjct: 161 NIKVSTSVDMTLIGNSYPPSQGSFRNDARWFVDAIVGFLRDTRAPLLVNIYPYFSYSGNP 220
Query: 181 KDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDG 240
ISL Y+LFT+PNVVV DGSR Y+NLFDA+LDS++AA++ +G V +VVSESGWPS G
Sbjct: 221 GQISLPYSLFTAPNVVVQDGSRQYRNLFDAMLDSVYAALERSGGASVGIVVSESGWPSAG 280
Query: 241 GFATTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQ 300
F TYDNA YL NLI+H K G+P + GPIETYIF +FDEN KNPELEKHFG+F PNKQ
Sbjct: 281 AFGATYDNAATYLRNLIQHAKEGSPRKPGPIETYIFAMFDENNKNPELEKHFGLFSPNKQ 340
Query: 301 KKYPFGF 307
KY F
Sbjct: 341 PKYNINF 347
>UniRef100_P52402 Glucan endo-1,3-beta-glucosidase, basic isoform 3 precursor (EC
3.2.1.39) ((1->3)-beta-glucan endohydrolase)
((1->3)-beta- glucanase) [Solanum tuberosum]
Length = 328
Score = 391 bits (1004), Expect = e-107
Identities = 192/304 (63%), Positives = 231/304 (75%)
Query: 4 NNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNNDIA 63
NNLPS E I L KS NI R+RLYDPN AL ALR S IE++LG+PN D+++IA+ + A
Sbjct: 1 NNLPSHSEVIQLYKSRNIGRLRLYDPNHGALNALRGSNIEVILGLPNVDVKHIASGMEHA 60
Query: 64 IQWVQKNVLNFYPSVKIKYIAVGNEVNPVGGSSQFAKFVLPAIQNIYQAIRAKNLHDQIK 123
WVQKNV +F+P VKIKYIAVGNE++PV G+S F +PA+ NIY+AI L + IK
Sbjct: 61 RWWVQKNVKDFWPDVKIKYIAVGNEISPVTGTSSLTSFQVPALVNIYKAIGEAGLGNDIK 120
Query: 124 VSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDNPKDI 183
VST++DMT+IG SYPPS+GSFR+DVR + DPI+G+L APL NIY YFSY NP I
Sbjct: 121 VSTSVDMTLIGNSYPPSQGSFRNDVRWFTDPIVGFLRDTRAPLLVNIYPYFSYSGNPGQI 180
Query: 184 SLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSDGGFA 243
SL YALFT+PNVVV DGSR Y+NLFDA+LDS++AA++ TG G V +VVSESGWPS G F
Sbjct: 181 SLPYALFTAPNVVVQDGSRQYRNLFDAMLDSVYAAMERTGGGSVGIVVSESGWPSAGAFG 240
Query: 244 TTYDNARVYLDNLIRHVKGGTPMRSGPIETYIFGLFDENQKNPELEKHFGVFYPNKQKKY 303
T DNA YL NLI+H K G+P + GPIETYIF +FDEN KNPELEKHFG+F PNKQ KY
Sbjct: 241 ATQDNAATYLRNLIQHAKEGSPRKPGPIETYIFAMFDENNKNPELEKHFGLFSPNKQPKY 300
Query: 304 PFGF 307
F
Sbjct: 301 NLNF 304
>UniRef100_Q9ZP99 Beta-1,3-glucanase [Hevea brasiliensis]
Length = 352
Score = 388 bits (996), Expect = e-106
Identities = 194/309 (62%), Positives = 239/309 (76%), Gaps = 3/309 (0%)
Query: 1 MMGNNLPSQREAIDLCKSNNIKRMRLYDPNQAALEALRNSGIELMLGVPNSDLQNIATNN 60
M GNNLP E I L K +NI RMR+YDPN+A LEALR S IEL+LGVPNSDLQ++ TN
Sbjct: 44 MQGNNLPPVSEVIALYKKSNITRMRIYDPNRAVLEALRGSNIELILGVPNSDLQSL-TNP 102
Query: 61 DIAIQWVQKNVLNFYPSVKIKYIAVGNEVNPVG-GSSQFAKFVLPAIQNIYQAIRAKNLH 119
A WVQKNV F+ SV +YIAVGNE++PV G++ A+FVLPA++NI+ AIR+ L
Sbjct: 103 SNAKSWVQKNVRGFWSSVLFRYIAVGNEISPVNRGTAWLAQFVLPAMRNIHDAIRSAGLQ 162
Query: 120 DQIKVSTAIDMTMIGTSYPPSKGSFRSDVRSYLDPIIGYLVYANAPLFANIYSYFSYKDN 179
DQIKVSTAID+T++G SYPPS G+FR DVRSYL+PII +L +PL ANIY YF+Y N
Sbjct: 163 DQIKVSTAIDLTLVGNSYPPSAGAFRDDVRSYLNPIIRFLSSIRSPLLANIYPYFTYAGN 222
Query: 180 PKDISLQYALFTSPNVVVWDGSRGYQNLFDALLDSLHAAIDNTGIGFVKVVVSESGWPSD 239
P+DISL YALFTSP+VVVWDG RGY+NLFDA LD L++A++ G ++VVVSESGWPS
Sbjct: 223 PRDISLPYALFTSPSVVVWDGQRGYKNLFDATLDVLYSALERASGGSLEVVVSESGWPSA 282
Query: 240 GGFATTYDNARVYLDNLIRHVKGGTPMR-SGPIETYIFGLFDENQKNPELEKHFGVFYPN 298
G FA T+DN R YL NLI+HVK GTP R + IETY+F +FDEN+K PE+EK FG+F+P+
Sbjct: 283 GAFAATFDNGRTYLSNLIQHVKRGTPKRPNRAIETYLFAMFDENKKQPEVEKQFGLFFPD 342
Query: 299 KQKKYPFGF 307
K +KY F
Sbjct: 343 KWQKYNLNF 351
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 591,045,049
Number of Sequences: 2790947
Number of extensions: 26620298
Number of successful extensions: 58182
Number of sequences better than 10.0: 378
Number of HSP's better than 10.0 without gapping: 343
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 56614
Number of HSP's gapped (non-prelim): 399
length of query: 327
length of database: 848,049,833
effective HSP length: 127
effective length of query: 200
effective length of database: 493,599,564
effective search space: 98719912800
effective search space used: 98719912800
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144539.10


