

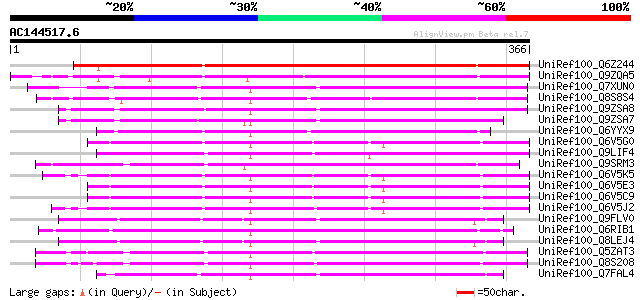
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144517.6 + phase: 0
(366 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6Z244 Putative flavonol synthase [Oryza sativa] 263 6e-69
UniRef100_Q9ZQA5 Putative giberellin beta-hydroxylase [Arabidops... 207 3e-52
UniRef100_Q7XUN0 OSJNBa0064M23.15 protein [Oryza sativa] 203 5e-51
UniRef100_Q8S8S4 Putative flavonol synthase [Arabidopsis thaliana] 203 7e-51
UniRef100_Q9ZSA8 F3H7.16 protein (Putative Fe(II)/ascorbate oxid... 202 9e-51
UniRef100_Q9ZSA7 F3H7.17 protein [Arabidopsis thaliana] 201 2e-50
UniRef100_Q6YYX9 Putative iron deficiency protein Ids3 [Oryza sa... 197 5e-49
UniRef100_Q6V5G0 Fe2+ dioxygenase-like [Capsella rubella] 197 5e-49
UniRef100_Q9LIF4 Arabidopsis thaliana genomic DNA, chromosome 3,... 197 5e-49
UniRef100_Q9SRM3 Leucoanthocyanidin dioxygenase, putative; 41415... 193 7e-48
UniRef100_Q6V5K5 Fe2+ dioxygenase-like protein [Brassica oleracea] 192 1e-47
UniRef100_Q6V5E3 Fe2+ dioxygenase-like [Olimarabidopsis pumila] 189 1e-46
UniRef100_Q6V5C9 Fe2+ dioxygenase-like [Arabidopsis arenosa] 189 1e-46
UniRef100_Q6V5J2 Fe2+ dioxygenase-like [Sisymbrium irio] 188 2e-46
UniRef100_Q9FLV0 Flavanone 3-hydroxylase-like protein [Arabidops... 183 7e-45
UniRef100_Q6RIB1 Flavonol synthase [Ginkgo biloba] 180 5e-44
UniRef100_Q8LEJ4 Flavanone 3-hydroxylase-like protein [Arabidops... 180 6e-44
UniRef100_Q5ZAT3 Putative ethylene-forming enzyme [Oryza sativa] 179 8e-44
UniRef100_Q8S208 Putative iron/ascorbate-dependent oxidoreductas... 179 1e-43
UniRef100_Q7FAL4 OSJNBa0064M23.14 protein [Oryza sativa] 178 2e-43
>UniRef100_Q6Z244 Putative flavonol synthase [Oryza sativa]
Length = 337
Score = 263 bits (672), Expect = 6e-69
Identities = 139/325 (42%), Positives = 200/325 (60%), Gaps = 6/325 (1%)
Query: 46 HDENEIVADQDEVNDP---IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSV 102
H+E A D P IPV+D +LING D+R++ I D+G+ACE+WGFF++TNH V
Sbjct: 10 HEEQSAAAAADGSATPSQGIPVVDLGVLINGAADERSRAIRDLGRACEDWGFFMVTNHGV 69
Query: 103 SKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIV 162
++L E ++D F L E+K+ Y + D I+ GT F D RD++K+
Sbjct: 70 PEALREAIMDACKELFRLPLEEKKEYMRAKPMDP-IRIGTGFYSVVDAVPCRRDYLKMFS 128
Query: 163 HPKFHSPDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLA 222
HP+FH P+KP+ RE + EY+ T L EL K ISESLGL + + +NL+S Q+L
Sbjct: 129 HPEFHCPEKPAKLREIATEYATCTRALLLELTKAISESLGLAGGRLSEALNLESCFQILV 188
Query: 223 ANLYPPCPQPD-LAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVS 281
N YP C +PD AMG+ HSDHGLL LL QNGV GLQV H+G+W+ F V+
Sbjct: 189 GNHYPACSRPDEQAMGLSAHSDHGLLTLLFQNGVDGLQVKHDGEWLLAKPLPGSFFVIAG 248
Query: 282 DHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGM 341
D LEI++NG+YK V+HRA V +RMS ++I P +DTVVEP E+ + + G+
Sbjct: 249 DQLEIVTNGRYKGVLHRAVVGGEQSRMSFVSLIGPCMDTVVEPLPEMA-ADGRGLEFRGI 307
Query: 342 KHIDYMKLQRNNQLYGKSVLNKVKI 366
++ DYM++Q++N + K+ L+ V++
Sbjct: 308 RYRDYMEMQQSNSINEKTALDIVRV 332
>UniRef100_Q9ZQA5 Putative giberellin beta-hydroxylase [Arabidopsis thaliana]
Length = 392
Score = 207 bits (528), Expect = 3e-52
Identities = 127/401 (31%), Positives = 212/401 (52%), Gaps = 51/401 (12%)
Query: 1 MASTLPPQVNQKSNNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVND 60
M + + + N+K G VK L E+ +P+ Y + P + I+ D++
Sbjct: 5 MIAPVMEEENKKYQKG------VKHLCEN-GLTKVPTKYIW---PEPDRPILTKSDKLIK 54
Query: 61 P-----IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFIL------------------ 97
P +P+ID++ L+ N R + I +AC+ +GFF +
Sbjct: 55 PNKNLKLPLIDFAELLGPN---RPHVLRTIAEACKTYGFFQVGSLNPKAHAVISRFLAKQ 111
Query: 98 --------TNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGD 149
NH + + + M+D FF L E++ Y +++ ++YGTSFN D
Sbjct: 112 VSYGNGQVVNHGMEGDVSKNMIDVCKRFFELPYEERSKYMSSDMSAP-VRYGTSFNQIKD 170
Query: 150 KNLFWRDFIKIIVHPKF----HSPDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEV 205
WRDF+K+ HP H P PS FR ++A Y+++T ++ ++K I ESL ++
Sbjct: 171 NVFCWRDFLKLYAHPLPDYLPHWPSSPSDFRSSAATYAKETKEMFEMMVKAILESLEIDG 230
Query: 206 NYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGK 265
+ + L+ G Q++ N YPPCP+P+L +GMPPHSD+G L LL+Q+ V GLQ+L+ +
Sbjct: 231 SD-EAAKELEEGSQVVVVNCYPPCPEPELTLGMPPHSDYGFLTLLLQDEVEGLQILYRDE 289
Query: 266 WINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPA 325
W+ V F+V V DHLEI SNG+YKSV+HR V++ R+S+A++ + L +VV+P+
Sbjct: 290 WVTVDPIPGSFVVNVGDHLEIFSNGRYKSVLHRVLVNSTKPRISVASLHSFPLTSVVKPS 349
Query: 326 SELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVKI 366
+L+D + NP+ Y+ +++ + + K+ L KI
Sbjct: 350 PKLVD-KHNPSQYMDTDFTTFLQYITSREPKWKNFLESRKI 389
>UniRef100_Q7XUN0 OSJNBa0064M23.15 protein [Oryza sativa]
Length = 352
Score = 203 bits (517), Expect = 5e-51
Identities = 120/358 (33%), Positives = 188/358 (51%), Gaps = 31/358 (8%)
Query: 13 SNNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLIN 72
+ +G P + ++ + + PD +++ E IPVID L
Sbjct: 15 AQSGQVPSSHIRPVGDRPDLDNV-------------------DHESGAGIPVIDLKQL-- 53
Query: 73 GNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDK-QVYADK 131
+ R K + IG ACE GFF++ NH + + ++E M+ FF++ E ++ + Y+D
Sbjct: 54 -DGPDRRKVVEAIGSACETDGFFMVKNHGIPEEVVEGMLRVAREFFHMPESERLKCYSDD 112
Query: 132 EVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS----PDKPSGFRETSAEYSRKTW 187
+I+ TSFNV +K WRDF+++ +P P P FR+ YSR+
Sbjct: 113 P--KKAIRLSTSFNVRTEKVSNWRDFLRLHCYPLESFIDQWPSNPPSFRQVVGTYSREAR 170
Query: 188 KLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLL 247
L LL+ ISESLGLE ++ M + Q +A N YPPCPQP+L G+P H D +
Sbjct: 171 ALALRLLEAISESLGLERGHMVSAMGRQA--QHMAVNYYPPCPQPELTYGLPGHKDPNAI 228
Query: 248 NLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATR 307
LL+Q+GVSGLQV NG+W+ V+ + ++ + D ++ +SN +YKSV+HR V++ + R
Sbjct: 229 TLLLQDGVSGLQVQRNGRWVAVNPVPDALVINIGDQIQALSNDRYKSVLHRVIVNSESER 288
Query: 308 MSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVK 365
+S+ T PS D V+ PA L+D +P AY K+ Y N L S L++ +
Sbjct: 289 ISVPTFYCPSPDAVIAPAGALVDGALHPLAYRPFKYQAYYDEFWNMGLQSASCLDRFR 346
>UniRef100_Q8S8S4 Putative flavonol synthase [Arabidopsis thaliana]
Length = 352
Score = 203 bits (516), Expect = 7e-51
Identities = 122/353 (34%), Positives = 190/353 (53%), Gaps = 19/353 (5%)
Query: 20 FTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQ-- 77
FTS TL+ S +P Y P + + +PVID SLL H
Sbjct: 8 FTSAMTLTNS-GVPQVPDRYVLP--PSQRPALGSSLGTSETTLPVIDLSLL----HQPFL 60
Query: 78 RTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDS 137
R+ IH+I AC+E+GFF + NH + S++ +D FF+L E+K + V +
Sbjct: 61 RSLAIHEISMACKEFGFFQVINHGIPSSVVNDALDAATQFFDLPVEEKMLLVSANV-HEP 119
Query: 138 IKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS----PDKPSGFRETSAEYSRKTWKLGREL 193
++YGTS N S D+ +WRDFIK HP P P +++ +Y+ T L ++L
Sbjct: 120 VRYGTSLNHSTDRVHYWRDFIKHYSHPLSKWIDMWPSNPPCYKDKVGKYAEATHLLHKQL 179
Query: 194 LKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQN 253
++ ISESLGLE NY+ + ++ G Q++A N YP CP+P++A+GMPPHSD L +L+Q+
Sbjct: 180 IEAISESLGLEKNYLQE--EIEEGSQVMAVNCYPACPEPEMALGMPPHSDFSSLTILLQS 237
Query: 254 GVSGLQVLH-NGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLAT 312
GLQ++ N W+ V +V + D +E+MSNG YKSV+HR V+ R+S A+
Sbjct: 238 S-KGLQIMDCNKNWVCVPYIEGALIVQLGDQVEVMSNGIYKSVIHRVTVNKEVKRLSFAS 296
Query: 313 VIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVK 365
+ + L + PA +L+ N +N AY D++ +N + ++ +K
Sbjct: 297 LHSLPLHKKISPAPKLV-NPNNAPAYGEFSFNDFLNYISSNDFIQERFIDTIK 348
>UniRef100_Q9ZSA8 F3H7.16 protein (Putative Fe(II)/ascorbate oxidase) [Arabidopsis
thaliana]
Length = 349
Score = 202 bits (515), Expect = 9e-51
Identities = 120/335 (35%), Positives = 179/335 (52%), Gaps = 13/335 (3%)
Query: 35 IPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGF 94
IPS+Y P + +++ + D IP+ID L N R + + AC +GF
Sbjct: 20 IPSNYV---RPISDRPNLSEVESSGDSIPLIDLRDLHGPN---RAVIVQQLASACSTYGF 73
Query: 95 FILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFW 154
F + NH V + + KM FF+ E ++ + + T + + TSFNV DK L W
Sbjct: 74 FQIKNHGVPDTTVNKMQTVAREFFHQPESERVKHYSADPTKTT-RLSTSFNVGADKVLNW 132
Query: 155 RDFIKIIVHPKFHS----PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDK 210
RDF+++ P P P FRE +AEY+ L LL+ ISESLGLE ++I
Sbjct: 133 RDFLRLHCFPIEDFIEEWPSSPISFREVTAEYATSVRALVLRLLEAISESLGLESDHISN 192
Query: 211 TMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVS 270
+ + Q +A N YPPCP+P+L G+P H D ++ +L+Q+ VSGLQV + KW+ VS
Sbjct: 193 ILGKHA--QHMAFNYYPPCPEPELTYGLPGHKDPTVITVLLQDQVSGLQVFKDDKWVAVS 250
Query: 271 STSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLD 330
N F+V + D ++++SN KYKSV+HRA V+ R+S+ T PS D V+ PA EL++
Sbjct: 251 PIPNTFIVNIGDQMQVISNDKYKSVLHRAVVNTENERLSIPTFYFPSTDAVIGPAHELVN 310
Query: 331 NESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVK 365
+ + A Y ++Y N L S L+ K
Sbjct: 311 EQDSLAIYRTYPFVEYWDKFWNRSLATASCLDAFK 345
>UniRef100_Q9ZSA7 F3H7.17 protein [Arabidopsis thaliana]
Length = 348
Score = 201 bits (512), Expect = 2e-50
Identities = 118/319 (36%), Positives = 177/319 (54%), Gaps = 14/319 (4%)
Query: 35 IPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGF 94
+PS+Y P + +++ D IP+ID L N R I+ AC GF
Sbjct: 18 VPSNYV---RPVSDRPKMSEVQTSGDSIPLIDLHDLHGPN---RADIINQFAHACSSCGF 71
Query: 95 FILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFW 154
F + NH V + ++KM++ FF E ++ + + T + + TSFNVS +K W
Sbjct: 72 FQIKNHGVPEETIKKMMNAAREFFRQSESERVKHYSAD-TKKTTRLSTSFNVSKEKVSNW 130
Query: 155 RDFIKIIVHP--KFHS--PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDK 210
RDF+++ +P F + P P FRE +AEY+ L LL+ ISESLGL + +
Sbjct: 131 RDFLRLHCYPIEDFINEWPSTPISFREVTAEYATSVRALVLTLLEAISESLGLAKDRVSN 190
Query: 211 TMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVS 270
T+ Q +A N YP CPQP+L G+P H D L+ +L+Q+ VSGLQV +GKWI V+
Sbjct: 191 TIGKHG--QHMAINYYPRCPQPELTYGLPGHKDANLITVLLQDEVSGLQVFKDGKWIAVN 248
Query: 271 STSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELL- 329
N F+V + D ++++SN KYKSV+HRA V++ R+S+ T PS D V+ PA EL+
Sbjct: 249 PVPNTFIVNLGDQMQVISNEKYKSVLHRAVVNSDMERISIPTFYCPSEDAVISPAQELIN 308
Query: 330 DNESNPAAYVGMKHIDYMK 348
+ E +PA Y + +Y +
Sbjct: 309 EEEDSPAIYRNFTYAEYFE 327
>UniRef100_Q6YYX9 Putative iron deficiency protein Ids3 [Oryza sativa]
Length = 383
Score = 197 bits (500), Expect = 5e-49
Identities = 106/282 (37%), Positives = 169/282 (59%), Gaps = 10/282 (3%)
Query: 62 IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLK 121
+PV+D S L + +R + + AC E+GFF + NH V ++ M+D FF L
Sbjct: 80 LPVVDLSRLRVPS--ERGAVLRTLDAACREYGFFQVVNHGVGGEVVGGMLDVARRFFELP 137
Query: 122 EEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS----PDKPSGFRE 177
+ +++ Y +V ++YGTSFN D L WRDF+K+ P P P+ RE
Sbjct: 138 QPERERYMSADVRAP-VRYGTSFNQVRDAVLCWRDFLKLACMPLAAVVESWPTSPADLRE 196
Query: 178 TSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMG 237
++ Y+ ++ E+++ E+LG+ + + +L +G QM+ N YP CPQP+L +G
Sbjct: 197 VASRYAEANQRVFMEVMEAALEALGVGGGGVME--DLAAGTQMMTVNCYPECPQPELTLG 254
Query: 238 MPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVH 297
MPPHSD+G L L++Q+ V+GLQV+H G+W+ V F+V V DHLEI+SNG+Y+SV+H
Sbjct: 255 MPPHSDYGFLTLVLQDEVAGLQVMHAGEWLTVDPLPGSFVVNVGDHLEILSNGRYRSVLH 314
Query: 298 RAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYV 339
R V++ R+S+A+ + + + VV PA EL+D + +P Y+
Sbjct: 315 RVKVNSRRLRVSVASFHSVAPERVVSPAPELID-DRHPRRYM 355
>UniRef100_Q6V5G0 Fe2+ dioxygenase-like [Capsella rubella]
Length = 356
Score = 197 bits (500), Expect = 5e-49
Identities = 108/322 (33%), Positives = 186/322 (57%), Gaps = 16/322 (4%)
Query: 56 DEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVF 115
D ++ +P ID S L NG D+R I +I AC+ GFF + NH +++++++ ++
Sbjct: 38 DSNSEIVPTIDVSRL-NGGDDERRGVIREISLACQHLGFFQIVNHGINQNILDDALEVAK 96
Query: 116 AFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS----PDK 171
FF L ++K+ + +V ++Y TS DK FWR F+K HP P+
Sbjct: 97 GFFELPAKEKKKFMSNDVYAP-VRYSTSLKDGLDKIQFWRIFLKHYAHPLHRWIHLWPEN 155
Query: 172 PSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQ 231
P G+RE ++ + KL E++ I+ESLGL +Y+ M+ ++G+Q++A N YPPCP
Sbjct: 156 PPGYREKMGKFCEEVRKLSIEIMGAITESLGLGRDYLSSRMD-ENGMQVMAVNCYPPCPG 214
Query: 232 PDLAMGMPPHSDHGLLNLLIQNGVSGLQVLH------NGKWINVSSTSNCFLVLVSDHLE 285
PD A+G+PPHSD+ + +L+QN ++GL++ +G+W+ V + V + DH+E
Sbjct: 215 PDAALGLPPHSDYSCITILLQN-LAGLKIFDPMAHGGSGRWVVVPQVTGVLKVHIGDHVE 273
Query: 286 IMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHID 345
++SNG YKSVVH+ ++ TR+SLA++ + ++ + EL++NE NP Y D
Sbjct: 274 VLSNGLYKSVVHKVTLNEENTRISLASLHSLDMNDKMSVPCELVNNE-NPVRYKESSFND 332
Query: 346 YMK-LQRNNQLYGKSVLNKVKI 366
++ L +N+ G ++ ++I
Sbjct: 333 FLDFLVKNDISQGDRFIDTLRI 354
>UniRef100_Q9LIF4 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone:MHC9
[Arabidopsis thaliana]
Length = 364
Score = 197 bits (500), Expect = 5e-49
Identities = 113/312 (36%), Positives = 170/312 (54%), Gaps = 11/312 (3%)
Query: 62 IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLK 121
IPVID S L ++D I + +ACE+WGFF + NH + ++E + + FF++
Sbjct: 55 IPVIDLSKLSKPDNDDFFFEILKLSQACEDWGFFQVINHGIEVEVVEDIEEVASEFFDMP 114
Query: 122 EEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFR 176
E+K+ Y + T YG +F S D+ L W + + VHP P KP+ F
Sbjct: 115 LEEKKKYPMEPGTVQG--YGQAFIFSEDQKLDWCNMFALGVHPPQIRNPKLWPSKPARFS 172
Query: 177 ETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAM 236
E+ YS++ +L + LLK I+ SLGL+ ++ +Q + N YPPC PDL +
Sbjct: 173 ESLEGYSKEIRELCKRLLKYIAISLGLKEERFEEMFG--EAVQAVRMNYYPPCSSPDLVL 230
Query: 237 GMPPHSDHGLLNLLIQ--NGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKS 294
G+ PHSD L +L Q N GLQ+L + W+ V N ++ + D +E++SNGKYKS
Sbjct: 231 GLSPHSDGSALTVLQQSKNSCVGLQILKDNTWVPVKPLPNALVINIGDTIEVLSNGKYKS 290
Query: 295 VVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQ 354
V HRA + R+++ T AP+ + +EP SEL+D+E+NP Y H DY +N+
Sbjct: 291 VEHRAVTNREKERLTIVTFYAPNYEVEIEPMSELVDDETNPCKYRSYNHGDYSYHYVSNK 350
Query: 355 LYGKSVLNKVKI 366
L GK L+ KI
Sbjct: 351 LQGKKSLDFAKI 362
>UniRef100_Q9SRM3 Leucoanthocyanidin dioxygenase, putative; 41415-43854 [Arabidopsis
thaliana]
Length = 400
Score = 193 bits (490), Expect = 7e-48
Identities = 122/348 (35%), Positives = 178/348 (51%), Gaps = 15/348 (4%)
Query: 19 PFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVND-PIPVIDYSLLINGNHDQ 77
P V++L+ES + S+P Y + + I+ Q EV D IP+ID L +GN D
Sbjct: 52 PIVRVQSLAES-NLTSLPDRYIKPPSQRPQTTIIDHQPEVADINIPIIDLDSLFSGNEDD 110
Query: 78 RTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDS 137
+ + I +AC EWGFF + NH V LM+ + +FFNL E K+VY++ T +
Sbjct: 111 KKR----ISEACREWGFFQVINHGVKPELMDAARETWKSFFNLPVEAKEVYSNSPRTYEG 166
Query: 138 IKYGTSFNVSGDKNLFWRDFIKIIVHP----KFHS-PDKPSGFRETSAEYSRKTWKLGRE 192
YG+ V L W D+ + P F+ P PS RE + EY ++ KLG
Sbjct: 167 --YGSRLGVEKGAILDWNDYYYLHFLPLALKDFNKWPSLPSNIREMNDEYGKELVKLGGR 224
Query: 193 LLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ 252
L+ +S +LGL + + + L N YP CPQP+LA+G+ PHSD G + +L+
Sbjct: 225 LMTILSSNLGLRAEQLQEAFGGEDVGACLRVNYYPKCPQPELALGLSPHSDPGGMTILLP 284
Query: 253 NG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLA 311
+ V GLQV H WI V+ + F+V + D ++I+SN KYKSV HR V++ R+SLA
Sbjct: 285 DDQVVGLQVRHGDTWITVNPLRHAFIVNIGDQIQILSNSKYKSVEHRVIVNSEKERVSLA 344
Query: 312 TVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKS 359
P D ++P +L+ + P Y M Y R GKS
Sbjct: 345 FFYNPKSDIPIQPMQQLV-TSTMPPLYPPMTFDQYRLFIRTQGPRGKS 391
>UniRef100_Q6V5K5 Fe2+ dioxygenase-like protein [Brassica oleracea]
Length = 361
Score = 192 bits (488), Expect = 1e-47
Identities = 111/354 (31%), Positives = 194/354 (54%), Gaps = 26/354 (7%)
Query: 24 KTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIH 83
++L PD ++P SY P D D + +P ID S L G+ D+R I
Sbjct: 21 RSLPYVPDCYAVPPSY----EPRDA------LDSNFEIVPTIDISRL-KGSDDERRGVIQ 69
Query: 84 DIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTS 143
++ AC+ +GFF + NH +++++++ ++ FF L ++K+ + +V ++Y TS
Sbjct: 70 ELSSACQLFGFFQVVNHGINQNILDDALEVAHGFFELPAKEKKKFMSNDVYAP-VRYTTS 128
Query: 144 FNVSGDKNLFWRDFIKIIVHPKFHS----PDKPSGFRETSAEYSRKTWKLGRELLKGISE 199
DK FWR F+K HP P P +RE ++ + L E++ I+E
Sbjct: 129 IKDGLDKIQFWRIFLKHYAHPLHRWIHLWPQNPPEYREKMGKFCEEVRSLSLEIMGAITE 188
Query: 200 SLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQ 259
SLGL +Y+ M+ +G+Q++A N YPPCP P A+G+PPHSD+ + +L+QN ++GL+
Sbjct: 189 SLGLGRDYLSSRMD-KNGMQVMAVNCYPPCPDPTTALGLPPHSDYSCITILLQN-LTGLE 246
Query: 260 VLH------NGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATV 313
+ H +G+W+ V V + DH+E++SNG Y SV+H+ ++ TR+SLA++
Sbjct: 247 IFHLSAHGGSGRWVLVPEVKGVLKVHIGDHVEVLSNGLYNSVIHKVTLNEEKTRISLASL 306
Query: 314 IAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMK-LQRNNQLYGKSVLNKVKI 366
+ +D + +L++NE NPA Y D++ L +N+ G+ ++ ++I
Sbjct: 307 HSLGMDDKMSVPYQLVNNE-NPARYRESSFNDFLAFLVKNDISQGERFIDTLRI 359
>UniRef100_Q6V5E3 Fe2+ dioxygenase-like [Olimarabidopsis pumila]
Length = 356
Score = 189 bits (479), Expect = 1e-46
Identities = 105/322 (32%), Positives = 183/322 (56%), Gaps = 16/322 (4%)
Query: 56 DEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVF 115
D +D +P ID S L G D R + I ++ AC+ GFF + NH +++++++ ++
Sbjct: 38 DSNSDVVPTIDVSRL-KGVDDDRREVIRELSLACQRLGFFQIVNHGINQNILDDALEVAK 96
Query: 116 AFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS----PDK 171
FF L ++K+ + +V I+Y TS DK +WR F+K HP P+
Sbjct: 97 GFFELPAKEKKKFMSNDVYAP-IRYSTSLKDGLDKIQYWRIFLKHYAHPLHRWIHLWPEN 155
Query: 172 PSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQ 231
P +RE ++ + KL E++ I+ESLGL NY+ M+ ++G+Q++A N YPPCP
Sbjct: 156 PPEYREKMGKFCEEVRKLSIEIMGAITESLGLGRNYMSSRMD-ENGMQVMAVNCYPPCPD 214
Query: 232 PDLAMGMPPHSDHGLLNLLIQNGVSGLQVLH------NGKWINVSSTSNCFLVLVSDHLE 285
P+ A+G+PPHSD+ + +L+QN ++GL++ + +G W+ V + V + DH+E
Sbjct: 215 PETALGLPPHSDYSCITILLQN-LAGLKIFNPMAHGGSGGWVVVPQVTGALKVHIGDHVE 273
Query: 286 IMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHID 345
++SNG YKSVVH+ ++ R+SLA++ + +D + EL+++E NP Y D
Sbjct: 274 VLSNGLYKSVVHKVTLNEENMRISLASLHSLGMDDKMSVPCELVNHE-NPVRYRESSFND 332
Query: 346 YMK-LQRNNQLYGKSVLNKVKI 366
++ L +N+ G ++ ++I
Sbjct: 333 FLDFLVKNDISQGDRFIDTLRI 354
>UniRef100_Q6V5C9 Fe2+ dioxygenase-like [Arabidopsis arenosa]
Length = 366
Score = 189 bits (479), Expect = 1e-46
Identities = 102/322 (31%), Positives = 183/322 (56%), Gaps = 16/322 (4%)
Query: 56 DEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVF 115
D ++ +P ID S L G D+R I ++ AC+ GFF + NH +++++++ ++
Sbjct: 48 DSNSEIVPTIDVSRL-KGGDDERRGVIRELSLACQHLGFFQIVNHGINQNILDDALEVAR 106
Query: 116 AFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS----PDK 171
FF L ++K+ + +V ++Y TS DK FWR F+K HP P+
Sbjct: 107 RFFELPAKEKKKFMSNDVYAP-VRYSTSLKDGLDKIQFWRIFLKHYAHPLHRWIHLWPEN 165
Query: 172 PSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQ 231
P G+RE ++ + +L E++ I+ESLGL +Y+ M+ ++G+Q++ N YPPCP
Sbjct: 166 PPGYREKMGKFCEEVKELSIEIMGAITESLGLGRDYLSSRMD-ENGMQVMTVNCYPPCPD 224
Query: 232 PDLAMGMPPHSDHGLLNLLIQNGVSGLQVLH------NGKWINVSSTSNCFLVLVSDHLE 285
P+ A+G+PPHSD+ + +L+QN ++GL++ +G+W+ V + V + DH+E
Sbjct: 225 PETALGLPPHSDYSCITILLQN-LAGLKIFDPMAHGGSGRWVAVPEVTGVLKVHIGDHVE 283
Query: 286 IMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHID 345
++SNG YKS+VH+ ++ TR+SLA++ + +D + EL+ +E NP Y D
Sbjct: 284 VLSNGLYKSIVHKVTLNEENTRISLASLHSLGMDDKMSVPCELVTDE-NPVRYKESSFKD 342
Query: 346 YMK-LQRNNQLYGKSVLNKVKI 366
++ L +N+ G ++ ++I
Sbjct: 343 FLDFLVKNDISQGDRFIDTLRI 364
>UniRef100_Q6V5J2 Fe2+ dioxygenase-like [Sisymbrium irio]
Length = 361
Score = 188 bits (478), Expect = 2e-46
Identities = 111/348 (31%), Positives = 190/348 (53%), Gaps = 26/348 (7%)
Query: 30 PDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKAC 89
PD +P SY P D D ++ +P ID S L G+ D+R I ++ AC
Sbjct: 27 PDCYVVPPSY----EPRDA------LDSNSEIVPTIDISRL-KGSDDERQGVIQELRSAC 75
Query: 90 EEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGD 149
+ +GFF + NH +++ ++++ + FF L ++K+ + +V ++Y TS D
Sbjct: 76 QLFGFFQIVNHGINQDILDEALVVANGFFQLPAKEKKKFMSNDVYAP-VRYTTSLKDGLD 134
Query: 150 KNLFWRDFIKIIVHPKFHS----PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEV 205
K FWR F+K HP P P G+RE ++ + L E++ I+ESLGL
Sbjct: 135 KIQFWRIFLKHYAHPLHRWIHLWPQNPPGYREKMGKFCEEVRSLSLEIMGAITESLGLGR 194
Query: 206 NYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLH--- 262
+Y+ M+ D G+Q++A N YPPCP P A+G+PPHSD+ + +L+QN ++GL++ +
Sbjct: 195 DYLSSRMDED-GMQVMAVNCYPPCPGPKTALGLPPHSDYSCITILLQN-LTGLEIFNLSA 252
Query: 263 ---NGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLD 319
+G+W+ V V + DH+E++SNG YKSVVH+ ++ TR+SLA++ + ++
Sbjct: 253 HGGSGRWVLVPEVKGILKVHIGDHVEVLSNGLYKSVVHKVTLNEEKTRISLASLHSLGME 312
Query: 320 TVVEPASELLDNESNPAAYVGMKHIDYMK-LQRNNQLYGKSVLNKVKI 366
+ +L++NE NP Y D++ L +N+ G ++ ++I
Sbjct: 313 DKMSVPYQLVNNE-NPLRYRESSFKDFLDFLVKNDISQGNRFIDTLRI 359
>UniRef100_Q9FLV0 Flavanone 3-hydroxylase-like protein [Arabidopsis thaliana]
Length = 341
Score = 183 bits (464), Expect = 7e-45
Identities = 108/324 (33%), Positives = 187/324 (57%), Gaps = 17/324 (5%)
Query: 35 IPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGF 94
I + + +TT P + ++D+ +++ + D+ L+ + D R+ I I +AC +GF
Sbjct: 6 ISTGFRHTTLPENYVRPISDRPRLSEVSQLEDFPLIDLSSTD-RSFLIQQIHQACARFGF 64
Query: 95 FILTNHSVSKSLMEKMVDQVFAFFNLKEEDK-QVYADKEVTDDSIKYGTSFNVSGDKNLF 153
F + NH V+K ++++MV FF++ E+K ++Y+D + + TSFNV ++
Sbjct: 65 FQVINHGVNKQIIDEMVSVAREFFSMSMEEKMKLYSDDPTK--TTRLSTSFNVKKEEVNN 122
Query: 154 WRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYI 208
WRD++++ +P H P P F+E ++YSR+ ++G ++ + ISESLGLE +Y+
Sbjct: 123 WRDYLRLHCYP-IHKYVNEWPSNPPSFKEIVSKYSREVREVGFKIEELISESLGLEKDYM 181
Query: 209 DKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNG-VSGLQVLHNGKWI 267
K + Q +A N YPPCP+P+L G+P H+D L +L+Q+ V GLQ+L +G+W
Sbjct: 182 KKVLGEQG--QHMAVNYYPPCPEPELTYGLPAHTDPNALTILLQDTTVCGLQILIDGQWF 239
Query: 268 NVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPAS- 326
V+ + F++ + D L+ +SNG YKSV HRA + R+S+A+ + P+ V+ PA
Sbjct: 240 AVNPHPDAFVINIGDQLQALSNGVYKSVWHRAVTNTENPRLSVASFLCPADCAVMSPAKP 299
Query: 327 --ELLDNESNPAAYVGMKHIDYMK 348
E D+E+ P Y + +Y K
Sbjct: 300 LWEAEDDETKP-VYKDFTYAEYYK 322
>UniRef100_Q6RIB1 Flavonol synthase [Ginkgo biloba]
Length = 340
Score = 180 bits (457), Expect = 5e-44
Identities = 115/340 (33%), Positives = 172/340 (49%), Gaps = 10/340 (2%)
Query: 21 TSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTK 80
T V+ ++ES +IP + N D + IPVID L + R K
Sbjct: 4 TRVQYVAESRP-QTIPLEFVRPVEERPINTTFNDDIGLGRQIPVIDMCSL--EAPELRVK 60
Query: 81 TIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKY 140
T +I +A +EWG F + NH++S L E + FF L +E+K+ YA S Y
Sbjct: 61 TFKEIARASKEWGIFQVINHAISPLLFESLETVGKQFFQLPQEEKEAYACTGEDGSSTGY 120
Query: 141 GTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRELLK 195
GT + D W DF ++ P P KPS + E + EYS+ + +LL
Sbjct: 121 GTKLACTTDGRQGWSDFFFHMLWPPSLRDFSKWPQKPSSYIEVTEEYSKGILGVLNKLLS 180
Query: 196 GISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGV 255
+S SL L+ + + + ++ L N YP CPQP++A G+ PH+D L +L N V
Sbjct: 181 ALSISLELQESALKDALGGENLEMELKINYYPTCPQPEVAFGVVPHTDMSALTILKPNDV 240
Query: 256 SGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIA 315
GLQV + KWI N ++ + D ++I+SNGK+KSV+HR+ V+ RMS +
Sbjct: 241 PGLQVWKDEKWITAHYVPNALIIHIGDQIQILSNGKFKSVLHRSLVNKEKVRMSWPVFCS 300
Query: 316 PSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQL 355
P LDTV+ P EL+D +SNP Y + +Y K ++ N+L
Sbjct: 301 PPLDTVIGPLKELID-DSNPPLYNARTYREY-KHRKINKL 338
>UniRef100_Q8LEJ4 Flavanone 3-hydroxylase-like protein [Arabidopsis thaliana]
Length = 341
Score = 180 bits (456), Expect = 6e-44
Identities = 107/324 (33%), Positives = 186/324 (57%), Gaps = 17/324 (5%)
Query: 35 IPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGF 94
I + + +TT P + ++D+ +++ + D+ L+ + D R+ I I +AC +GF
Sbjct: 6 ISTGFRHTTLPENYVRPISDRPRLSEVSQLEDFPLIDLSSTD-RSFLIQQIHQACARFGF 64
Query: 95 FILTNHSVSKSLMEKMVDQVFAFFNLKEEDK-QVYADKEVTDDSIKYGTSFNVSGDKNLF 153
F + NH V+K ++++MV FF++ E+K ++Y+D + + TSFNV ++
Sbjct: 65 FQVKNHGVNKQIIDEMVSVAREFFSMSMEEKMKLYSDDPTK--TTRLSTSFNVKKEEVNN 122
Query: 154 WRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYI 208
WRD++++ +P H P P F+E ++YSR+ ++G ++ + ISESLGLE +Y+
Sbjct: 123 WRDYLRLHCYP-IHKYVNEWPSNPPSFKEIVSKYSREVREVGFKIEELISESLGLEKDYM 181
Query: 209 DKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNG-VSGLQVLHNGKWI 267
K + Q +A N YPPCP+P+L G+P H+D L +L+Q+ V GLQ+L +G+W
Sbjct: 182 KKVLGEQG--QHMAVNYYPPCPEPELTYGLPAHTDPNALTILLQDTTVCGLQILIDGQWF 239
Query: 268 NVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPAS- 326
V+ + F++ + D L+ +SNG YKSV RA + R+S+A+ + P+ V+ PA
Sbjct: 240 AVNPHPDAFVINIGDQLQALSNGVYKSVWRRAVTNTENPRLSVASFLCPADCAVMSPAKP 299
Query: 327 --ELLDNESNPAAYVGMKHIDYMK 348
E D+E+ P Y + +Y K
Sbjct: 300 LWEAEDDETKP-VYKDFTYAEYYK 322
>UniRef100_Q5ZAT3 Putative ethylene-forming enzyme [Oryza sativa]
Length = 350
Score = 179 bits (455), Expect = 8e-44
Identities = 118/353 (33%), Positives = 179/353 (50%), Gaps = 19/353 (5%)
Query: 19 PFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQR 78
P SV+ + + + +P Y P D +E VA D IPV+D+ L G+ D+
Sbjct: 5 PVPSVQAMVAAIGIDHVPPRYL---RPTDADEPVAS-DGGEAEIPVVDFWRLQLGDGDEL 60
Query: 79 TKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSI 138
+ + AC++WGFF L NH+V + ++E M + FF L E K+ + +
Sbjct: 61 AR----LHIACQDWGFFQLVNHNVPEDVVEGMKASIKGFFELPAETKKQVVQEPGQLEG- 115
Query: 139 KYGTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGREL 193
YG F VS D+ L W D + + P PD+P+GFR Y L
Sbjct: 116 -YGQLFVVSEDQKLDWADILYVKTQPLQDRNLRFWPDQPAGFRMALDRYCAAVKITADGL 174
Query: 194 LKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ- 252
L ++ +LG+E I + G+Q + YPPC Q D +G+ PHSD L+ +L+Q
Sbjct: 175 LAAMATNLGVESEVIAERWV--GGVQSVRVQYYPPCGQADKVVGISPHSDADLVTILLQA 232
Query: 253 NGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLAT 312
N V GLQ+ G W+ V +V V D ++ +NG+YKSV HR V R+S+AT
Sbjct: 233 NEVDGLQIRRGGAWLPVRPLEGALIVNVGDIRQVFTNGRYKSVEHRVVVDGKKERLSMAT 292
Query: 313 VIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVK 365
+PS + +V P SE++++E + AAY M H + +KL +L GK+ LN +K
Sbjct: 293 FHSPSKNAIVGPLSEMVEHEDD-AAYTSMDHDELLKLFFAKKLEGKNFLNPIK 344
>UniRef100_Q8S208 Putative iron/ascorbate-dependent oxidoreductase [Oryza sativa]
Length = 344
Score = 179 bits (453), Expect = 1e-43
Identities = 120/353 (33%), Positives = 183/353 (50%), Gaps = 21/353 (5%)
Query: 19 PFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQR 78
P SV+++ + +P Y P DE VA E IPVID+ L G+ ++
Sbjct: 5 PVPSVQSMVAADGGAHVPPRYI---RPRDE--AVATDGETE--IPVIDFQRLQLGHDEEM 57
Query: 79 TKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSI 138
+ + +AC++WGFF L NHSV + +++ M FF L E K+ +A + D
Sbjct: 58 AR----LDRACQDWGFFQLINHSVPEDVVDGMKANARGFFELPAETKKQFAQERGQLDG- 112
Query: 139 KYGTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGREL 193
YG F VS D+ L W D + + P + P++ + FR +YS + L
Sbjct: 113 -YGQLFVVSEDQKLDWADILFLNTLPVQNRNFRFWPNQLAKFRSALDKYSAAVKSIVDFL 171
Query: 194 LKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQ- 252
L ++ +LG++ I D G+Q + N YPPC Q D +G PHSD LL L++Q
Sbjct: 172 LVTVANNLGVDPEVIANKCGTD-GIQAVRMNYYPPCVQADKVIGFSPHSDSDLLTLVLQV 230
Query: 253 NGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLAT 312
N V GLQ+ N W V F+V V D L+I +NG+YKS HRA V R+S+A
Sbjct: 231 NEVDGLQIKRNETWFPVRPLEGAFIVNVGDILQIFTNGRYKSAEHRAVVDMKKERLSIAA 290
Query: 313 VIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGKSVLNKVK 365
+PS+ V+ P E++ +E + A Y + H ++MKL +++L GKS L+++K
Sbjct: 291 FHSPSVHAVIGPLKEMVAHE-HEAVYRSIGHDEFMKLFFSSKLEGKSFLDRMK 342
>UniRef100_Q7FAL4 OSJNBa0064M23.14 protein [Oryza sativa]
Length = 340
Score = 178 bits (451), Expect = 2e-43
Identities = 107/294 (36%), Positives = 166/294 (56%), Gaps = 18/294 (6%)
Query: 62 IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLK 121
IP+ID + + + I +I +AC +GFF +TNH +++ L+EK++ FF L
Sbjct: 39 IPLIDLA------SPDKPRVIAEIAQACRTYGFFQVTNHGIAEELLEKVMAVALEFFRLP 92
Query: 122 EEDKQ-VYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS----PDKPSGFR 176
E+K+ +Y+D+ I+ TSFNV + WRD++++ HP P P+ F+
Sbjct: 93 PEEKEKLYSDEP--SKKIRLSTSFNVRKETVHNWRDYLRLHCHPLEEFVPEWPSNPAQFK 150
Query: 177 ETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAM 236
E + Y R+ +LG LL IS SLGLE +YI+K + Q +A N YP CP+PDL
Sbjct: 151 EIMSTYCREVRQLGLRLLGAISVSLGLEEDYIEKVLGEQE--QHMAVNYYPRCPEPDLTY 208
Query: 237 GMPPHSDHGLLNLLIQNG-VSGLQVLHNG-KWINVSSTSNCFLVLVSDHLEIMSNGKYKS 294
G+P H+D L +L+ + V+GLQVL +G +WI V+ N +V + D ++ +SN YKS
Sbjct: 209 GLPKHTDPNALTILLPDPHVAGLQVLRDGDQWIVVNPRPNALVVNLGDQIQALSNDAYKS 268
Query: 295 VVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMK 348
V HRA V+ RMS+A+ + P V+ PA +L+ + P Y + +Y K
Sbjct: 269 VWHRAVVNPVQERMSVASFMCPCNSAVISPARKLVADGDAP-VYRSFTYDEYYK 321
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 633,751,607
Number of Sequences: 2790947
Number of extensions: 27026795
Number of successful extensions: 63298
Number of sequences better than 10.0: 1255
Number of HSP's better than 10.0 without gapping: 959
Number of HSP's successfully gapped in prelim test: 296
Number of HSP's that attempted gapping in prelim test: 60171
Number of HSP's gapped (non-prelim): 1404
length of query: 366
length of database: 848,049,833
effective HSP length: 129
effective length of query: 237
effective length of database: 488,017,670
effective search space: 115660187790
effective search space used: 115660187790
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144517.6


