

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144515.9 + phase: 0
(207 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
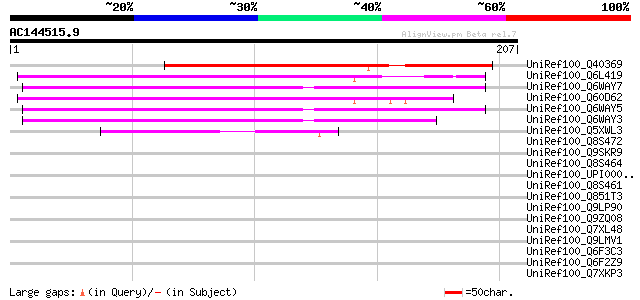
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q40369 RPE15 protein [Medicago sativa] 188 7e-47
UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum] 100 2e-20
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 100 4e-20
UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solan... 99 9e-20
UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum] 98 1e-19
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 97 3e-19
UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum] 63 5e-09
UniRef100_Q8S472 Putative gypsy-type retrotransposon RIRE2 [Zea ... 43 0.004
UniRef100_Q9SKR9 Putative Athila retroelement ORF1 protein [Arab... 43 0.004
UniRef100_Q8S464 Putative gag-pol-orf1 [Zea mays] 43 0.006
UniRef100_UPI0000437FD5 UPI0000437FD5 UniRef100 entry 42 0.013
UniRef100_Q8S461 Putative prpol [Zea mays] 41 0.017
UniRef100_Q851T3 Putative gag-pol [Oryza sativa] 41 0.017
UniRef100_Q9LP90 T32E20.30 [Arabidopsis thaliana] 41 0.022
UniRef100_Q9ZQ08 Putative Athila retroelement ORF1 protein [Arab... 40 0.050
UniRef100_Q7XL48 OSJNBa0056L23.9 protein [Oryza sativa] 39 0.065
UniRef100_Q9LMV1 F5M15.26 [Arabidopsis thaliana] 39 0.065
UniRef100_Q6F3C3 Putative gag-pol [Oryza sativa] 39 0.085
UniRef100_Q6F2Z9 Putative polyprotein [Oryza sativa] 39 0.085
UniRef100_Q7XKP3 OSJNBa0032N05.6 protein [Oryza sativa] 39 0.11
>UniRef100_Q40369 RPE15 protein [Medicago sativa]
Length = 219
Score = 188 bits (478), Expect = 7e-47
Identities = 92/135 (68%), Positives = 109/135 (80%), Gaps = 7/135 (5%)
Query: 64 AAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFE 123
AAAR++P LDEEE+TQTFLKTLKK++ ERMI++APNNFS+MVTMGTRLEEAVR+GIIV E
Sbjct: 13 AAARVSPRLDEEELTQTFLKTLKKEHKERMIVSAPNNFSDMVTMGTRLEEAVREGIIVLE 72
Query: 124 KAESSVNASKRYGNGHHKKKET-EVGMVSAGAGQSMATVAPINAAQMPPSYPYVPYSQHP 182
K E S +A K+YGNGHHK+KE+ +VGMVS P+NA Q+P Y Y+ SQHP
Sbjct: 73 KGECSASAPKKYGNGHHKRKESKKVGMVSGNHD------TPVNATQLPLPYQYMQISQHP 126
Query: 183 FFPPFYHQYPLPPGQ 197
FFPPFY QYPLPPGQ
Sbjct: 127 FFPPFYQQYPLPPGQ 141
>UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum]
Length = 421
Score = 100 bits (249), Expect = 2e-20
Identities = 59/194 (30%), Positives = 99/194 (50%), Gaps = 21/194 (10%)
Query: 4 AAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRG 63
A++W+ + HT+D+LA F + +N + P+R L ++ +K E+FREYA RWR
Sbjct: 244 ASKWFIDQDIANWHTWDDLARCFVQQFQYNIDIVPDRSSLANMRKKTTENFREYAVRWRE 303
Query: 64 AAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFE 123
AAR+ P + E EM FL+ + DY ++ A F+E++ +G +E ++ G IV +
Sbjct: 304 QAARVKPPMKELEMIDVFLQAQEPDYFHYLLSAVGKTFTEVIKVGEMVENGIKSGKIVSQ 363
Query: 124 KAESSVNASKRYGNGH---HKKKETEVGMVSAGAGQSMATVAPINAAQMPPSYPYVPYSQ 180
A + + G+G+ K++E +VSA P +Y ++Q
Sbjct: 364 AALKATTQALPNGSGNIGGKKRREDVATVVSA-----------------PRTYAQDNHTQ 406
Query: 181 HPFFPPFYHQYPLP 194
H +FPP QY +P
Sbjct: 407 H-YFPPQIPQYLVP 419
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 99.8 bits (247), Expect = 4e-20
Identities = 60/189 (31%), Positives = 94/189 (48%), Gaps = 4/189 (2%)
Query: 6 EWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRGAA 65
+WY L +D + + +L AF Y N + P+R L+SL QK ESF+EYAQRWR A
Sbjct: 218 DWYMELKRDSIRCWKDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWRELA 277
Query: 66 ARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFEKA 125
AR+ P + E E+T F+ TL+ +++RM +FS++V G R E ++ G I
Sbjct: 278 ARVQPPMLERELTDMFIGTLQGVFMDRMGSCPFVSFSDVVICGERTESLIKTGKI----Q 333
Query: 126 ESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAAQMPPSYPYVPYSQHPFFP 185
++ ++SK+ G +++E E V Q+ + + A +P P Q P
Sbjct: 334 DAGSSSSKKPFAGAPRRREGETNAVQYRRDQNRSQRCQVAAVTIPAPQPRQQQQQRVQQP 393
Query: 186 PFYHQYPLP 194
Q P
Sbjct: 394 QQQQQQQRP 402
>UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solanum demissum]
Length = 1784
Score = 98.6 bits (244), Expect = 9e-20
Identities = 60/197 (30%), Positives = 98/197 (49%), Gaps = 19/197 (9%)
Query: 4 AAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRG 63
A+EW+ + HT+D LA F + +N + P+R L ++ +K E+FREYA RWR
Sbjct: 736 ASEWFIDQDIANWHTWDNLARCFVQQFQYNIDIVPDRSSLANMRKKTTENFREYAVRWRE 795
Query: 64 AAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFE 123
AAR+ ++ E EM FL+ + DY ++ A F E++ +G +E ++ G IV +
Sbjct: 796 QAARVKLSMKESEMIDVFLQAQEPDYFHYLLSAVGKTFVEVIKVGEMVENGIKSGKIVSQ 855
Query: 124 KAESSVNASKRYGNGH--HKKKETEVGMVSAGA-------GQSMAT----------VAPI 164
A + + + G+G+ KK+ +V V GA G+S A+ ++PI
Sbjct: 856 AALKATTQALQNGSGNIGGKKRREDVATVGNGAKDEFTPIGESYASLFQKLRTLNVLSPI 915
Query: 165 NAAQMPPSYPYVPYSQH 181
P + YSQH
Sbjct: 916 ERKMSNPPPRNLDYSQH 932
>UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 98.2 bits (243), Expect = 1e-19
Identities = 60/189 (31%), Positives = 92/189 (47%), Gaps = 4/189 (2%)
Query: 6 EWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRGAA 65
+WY L +D + + +L AF Y N + P+R L+SL QK ESF+EYAQRWR A
Sbjct: 218 DWYMELKRDSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWRELA 277
Query: 66 ARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFEKA 125
AR+ P + E E+T F+ TL+ +++RM +FS++V G R E ++ G I
Sbjct: 278 ARVQPPMLERELTDMFIGTLQGVFMDRMGSCPFGSFSDVVICGERTESLIKTGKI----Q 333
Query: 126 ESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAAQMPPSYPYVPYSQHPFFP 185
+ ++SK+ G +++E E +V Q+ A +P P Q P
Sbjct: 334 DVGSSSSKKPFAGAPRRREGETNVVQHRRDQNRIEYRQAAAVTIPAPQPRQQQQQRVQQP 393
Query: 186 PFYHQYPLP 194
Q P
Sbjct: 394 QQQQQQQRP 402
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 97.1 bits (240), Expect = 3e-19
Identities = 56/169 (33%), Positives = 86/169 (50%), Gaps = 4/169 (2%)
Query: 6 EWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRGAA 65
+WY L D + + +L AF Y N + P+R L+SL QK ESF+EYAQRWR A
Sbjct: 217 DWYMELKSDSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSNESFKEYAQRWRELA 276
Query: 66 ARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFEKA 125
AR+ P + E E+T F+ TL+ +++RM +FS++V G R E ++ G I
Sbjct: 277 ARVQPPMLERELTDMFIGTLQGVFMDRMGSCPFGSFSDVVICGERTESLIKTGKI----Q 332
Query: 126 ESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAAQMPPSYP 174
+ ++SK+ G +++E E V Q+ A +P P
Sbjct: 333 DVGSSSSKKPFAGAPRRREGETNAVQHRRDQNRIEYRQAAAVTIPAPQP 381
>UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum]
Length = 1716
Score = 62.8 bits (151), Expect = 5e-09
Identities = 35/99 (35%), Positives = 55/99 (55%), Gaps = 16/99 (16%)
Query: 38 PNREFLRSLSQKKEESFREYAQRWRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAA 97
P+R L + QK ES+RE+A RWR AAR+ P + E+E+ + F++ K
Sbjct: 299 PDRYSLEKMKQKSTESYREFAYRWRKEAARVRPPMSEKEIVEVFVRVAK----------- 347
Query: 98 PNNFSEMVTMGTRLEEAVRDGIIVFEKA--ESSVNASKR 134
F+E+V +G +E+ +R G IV+ A ESS+ K+
Sbjct: 348 ---FAEIVKIGETIEDGLRTGKIVYVAASPESSILLKKK 383
>UniRef100_Q8S472 Putative gypsy-type retrotransposon RIRE2 [Zea mays]
Length = 1082
Score = 43.1 bits (100), Expect = 0.004
Identities = 39/179 (21%), Positives = 71/179 (38%), Gaps = 12/179 (6%)
Query: 2 EDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREF-LRSLSQKKEESFREYAQR 60
+ A W L + +D+L AF ++ T ++P + LRS Q+ ES R+Y +R
Sbjct: 578 DTARAWLEHLPPGQISNWDDLVQAFAGNFQ-GTYVRPGNSWDLRSCRQQPGESLRDYIRR 636
Query: 61 WRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGII 120
+ + D + + T +D V ++ P SE++ + T+
Sbjct: 637 FSKQRTELPNITDSDVIGAFLAGTTCRDLVSKLGRKTPTRASELMDIATKFASGQEAVEA 696
Query: 121 VFEK--------AESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAAQMPP 171
+F K +E + AS G KK+++ +A A + A + PP
Sbjct: 697 IFRKDKQPQGRPSEEAPEASTPRGAKKKGKKKSQAKRDAADA--DLVAAAEYKNPRKPP 753
>UniRef100_Q9SKR9 Putative Athila retroelement ORF1 protein [Arabidopsis thaliana]
Length = 929
Score = 43.1 bits (100), Expect = 0.004
Identities = 56/240 (23%), Positives = 90/240 (37%), Gaps = 60/240 (25%)
Query: 7 WYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRGAAA 66
W +L K+ + T+D+ AF + + N+R R + + K+ ESF E +R++G
Sbjct: 134 WEKTLPKNSITTWDDCKMAFLAKFFSNSRTARLRNEISGFTLKQNESFCEAWERFKGYQ- 192
Query: 67 RITPALDEEEMTQTFLKTLKKDYVE--RMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFEK 124
T + L TL + + RM++ +N G L++ + +G + E
Sbjct: 193 --TKCPHHGFSQASLLNTLYRGVLPKIRMLLDTASN-------GNFLKKDIEEGWELVEN 243
Query: 125 -AESSVNASKRYGNG----------HHKK----------------------KETEVGMVS 151
A+S N ++ YG HH++ E E V
Sbjct: 244 LAQSGGNYNEDYGRSIYTSSNTDDKHHREMKALNDKLDKIIQMQQKHVHFISEDEPFQVQ 303
Query: 152 AGAGQSMATVA---------PINAAQMPPSYPYVPYSQHPFFPP------FYHQYPLPPG 196
G A + P AA P+ P+VPY+Q F P Y Q PPG
Sbjct: 304 EGENDQCAEIRYVHNQGPSLPSAAAAAEPTKPFVPYNQSLGFVPKQQFQGGYQQQQPPPG 363
>UniRef100_Q8S464 Putative gag-pol-orf1 [Zea mays]
Length = 1066
Score = 42.7 bits (99), Expect = 0.006
Identities = 44/201 (21%), Positives = 80/201 (38%), Gaps = 13/201 (6%)
Query: 2 EDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREF-LRSLSQKKEESFREYAQR 60
+ A W L + +D+L AF ++ T ++P + LRS Q+ ES R+Y +R
Sbjct: 430 DTARAWLEHLPPGQISNWDDLVQAFAGNFQ-GTYVRPGNSWDLRSCRQQPGESLRDYIRR 488
Query: 61 WRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGII 120
+ + D + + T +D V ++ P SE++ + T+
Sbjct: 489 FSKQRTELPNITDSDVIGAFLAGTTFRDLVSKLGRKTPTRASELMDIATKFASGQEAVEA 548
Query: 121 VFEK--------AESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAAQMPPS 172
+F K +E + AS G KK+++ +A A + A + PP
Sbjct: 549 IFRKDKQPQGRPSEGAPEASAPRGAKKKGKKKSQSKRDAADA--DLVAAAEYKNPRKPPG 606
Query: 173 YPYVPYSQHPFFPPFYHQYPL 193
+ + + P YHQ P+
Sbjct: 607 GANL-FDKMLKEPCPYHQGPV 626
>UniRef100_UPI0000437FD5 UPI0000437FD5 UniRef100 entry
Length = 1364
Score = 41.6 bits (96), Expect = 0.013
Identities = 23/103 (22%), Positives = 47/103 (45%), Gaps = 1/103 (0%)
Query: 43 LRSLSQKKEESFREYAQRWRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFS 102
+R Q +ES R +A ++R R+ P++ E E+ Q L+ + ++
Sbjct: 684 IRERKQGVDESIRTFAYQYRALCLRLKPSMTEREILQAALRNCNPR-IASILRGTVTTVD 742
Query: 103 EMVTMGTRLEEAVRDGIIVFEKAESSVNASKRYGNGHHKKKET 145
E+V +GT +E+ + + + + NA GN K +++
Sbjct: 743 ELVRVGTLIEKDINEERSFWRQMHQEANAKSTEGNKFFKGRQS 785
>UniRef100_Q8S461 Putative prpol [Zea mays]
Length = 1850
Score = 41.2 bits (95), Expect = 0.017
Identities = 28/124 (22%), Positives = 52/124 (41%), Gaps = 2/124 (1%)
Query: 2 EDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREF-LRSLSQKKEESFREYAQR 60
+ A W L + +D+L AF ++ T ++P + LRS Q+ ES R+Y +R
Sbjct: 578 DTARAWLEHLPPGQISNWDDLVQAFAGNFQ-GTYVRPGNSWDLRSCRQQPGESLRDYIRR 636
Query: 61 WRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGII 120
+ + D + + T +D V ++ P SE++ + T+
Sbjct: 637 FSKQRTELPNITDSDVIGAFLAGTTCRDLVSKLGRKTPTRASELMDIATKFASGQEAVEA 696
Query: 121 VFEK 124
+F K
Sbjct: 697 IFRK 700
>UniRef100_Q851T3 Putative gag-pol [Oryza sativa]
Length = 971
Score = 41.2 bits (95), Expect = 0.017
Identities = 36/170 (21%), Positives = 68/170 (39%), Gaps = 19/170 (11%)
Query: 4 AAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRG 63
A WY +L + ++++++L F ++ ++ L + Q+ ES REY +R+
Sbjct: 418 ARSWYFNLPANSIYSWEQLRDVFVLNFRGTYEEPKTQQHLLGIRQRPGESIREYMRRFSQ 477
Query: 64 AAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFE 123
A ++ + + L+ + ++ P +V + DG F
Sbjct: 478 ARCQVEDITEASVINAASAGLLEGELTRKIANKEPQTLEHLVR--------IIDG---FA 526
Query: 124 KAESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAAQMPPSY 173
K+E SKR + +TE S A Q+ A V P Q P ++
Sbjct: 527 KSEED---SKR-----RQAIQTEYDKASIAAAQAQAQVKPPRQGQAPMTW 568
>UniRef100_Q9LP90 T32E20.30 [Arabidopsis thaliana]
Length = 1397
Score = 40.8 bits (94), Expect = 0.022
Identities = 32/135 (23%), Positives = 66/135 (48%), Gaps = 5/135 (3%)
Query: 3 DAAEWYT-SLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRW 61
+A WY ++S+ D ++ +L + +G N +L+ + L + K+ S EY QR+
Sbjct: 172 EALSWYNWAISRGDFVSWLKLKSGLMLRFG-NLKLRGPSQSLFCI--KQTGSVAEYVQRF 228
Query: 62 RGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIV 121
++++ LD++++ FL L + E + + P N EMV + +E +V ++
Sbjct: 229 EDLSSQVG-GLDDQKLEGIFLNGLTGEMQELVHMHKPQNLPEMVAVARSMETSVMRRVVQ 287
Query: 122 FEKAESSVNASKRYG 136
E ++ +R G
Sbjct: 288 KELQLVKLDEKRRKG 302
>UniRef100_Q9ZQ08 Putative Athila retroelement ORF1 protein [Arabidopsis thaliana]
Length = 750
Score = 39.7 bits (91), Expect = 0.050
Identities = 34/140 (24%), Positives = 64/140 (45%), Gaps = 5/140 (3%)
Query: 7 WYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRGAAA 66
W +L ++ + T+D+ AF + + N+R R + +QK+ ESF E +R++G
Sbjct: 102 WEKTLPQNSITTWDDCKKAFFAKFFSNSRTARLRNEISGFTQKQNESFCEAWERFKGYQT 161
Query: 67 RIT-PALDEEEMTQTFLKTLKKDYVERMIIAAPNNF-SEMVTMGTRLEE--AVRDGIIVF 122
+ P + + T + + + A+ NF ++ V G L E A DG
Sbjct: 162 KCPHPGFKQASLLSTLYRGVLPKLRMLLDTASNGNFLNKDVEKGWELVENLAQSDG-NYN 220
Query: 123 EKAESSVNASKRYGNGHHKK 142
E + S++ S + HH++
Sbjct: 221 ENYDKSISTSSESDDKHHRE 240
>UniRef100_Q7XL48 OSJNBa0056L23.9 protein [Oryza sativa]
Length = 836
Score = 39.3 bits (90), Expect = 0.065
Identities = 49/206 (23%), Positives = 86/206 (40%), Gaps = 24/206 (11%)
Query: 2 EDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREF-LRSLSQKKEESFREYAQR 60
+ A W L + + ++ EL F +++ T +P +F L ++ QK ES R+Y +R
Sbjct: 403 DSARSWLHGLPRGTIGSWAELRDHFIANFQ-GTFERPGTQFDLYNVIQKSGESLRDYIRR 461
Query: 61 WRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGII 120
+ +I+ D+ + F K ++ + + P T+ E+A
Sbjct: 462 FSEQCNKISDITDDV-IIAAFTKGIRHEELVGKFGRKPPK-----TVKLMFEKANE---- 511
Query: 121 VFEKAESSVNASKRYGNGHHKKKETEVGMVSAGAGQSMATVAPINAAQ--MPPSYPYVPY 178
+ KAE +V ASK+ G+ KK+T + G G + + +N M P+ P
Sbjct: 512 -YAKAEDAVIASKQSGSSWKPKKDTP----ATGGGGRSSKRSRVNTFDKIMNSQCPHHPK 566
Query: 179 SQHP-----FFPPFYHQYPLPPGQNS 199
S H + F QY +NS
Sbjct: 567 SNHAAKDCFVYKQFAEQYTKTTRKNS 592
>UniRef100_Q9LMV1 F5M15.26 [Arabidopsis thaliana]
Length = 1838
Score = 39.3 bits (90), Expect = 0.065
Identities = 25/125 (20%), Positives = 54/125 (43%)
Query: 4 AAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFREYAQRWRG 63
A W++ L + + +F +L ++F HY + + L S+SQ +ES R + R++
Sbjct: 267 AHNWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFKL 326
Query: 64 AAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFE 123
IT + + + + + + AP+ + + +R E + +I+
Sbjct: 327 VVTNITVPDEAAIVALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELEEEKLILAR 386
Query: 124 KAESS 128
K S+
Sbjct: 387 KHNST 391
>UniRef100_Q6F3C3 Putative gag-pol [Oryza sativa]
Length = 1512
Score = 38.9 bits (89), Expect = 0.085
Identities = 35/146 (23%), Positives = 64/146 (42%), Gaps = 15/146 (10%)
Query: 2 EDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREF-LRSLSQKKEESFREYAQR 60
+ A W L + + ++ EL F +++ T +P +F L ++ QK ES R+Y +R
Sbjct: 430 DSAQSWLNGLPRGTIGSWAELRDHFIANFQ-GTFERPGTQFDLYNVVQKSGESLRDYIRR 488
Query: 61 WRGAAARITPALDEEEMTQTFLKTLK-KDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGI 119
+ +I+ D+ + F K ++ +D V + P +M
Sbjct: 489 FSEQRNKISDITDDV-IIAAFTKGIRHEDLVGKFGCKPPRTVKQMFEKANE--------- 538
Query: 120 IVFEKAESSVNASKRYGNGHHKKKET 145
+ KAE ++ ASK+ G KK+T
Sbjct: 539 --YAKAEDAITASKQSGTTWKPKKDT 562
>UniRef100_Q6F2Z9 Putative polyprotein [Oryza sativa]
Length = 973
Score = 38.9 bits (89), Expect = 0.085
Identities = 35/146 (23%), Positives = 64/146 (42%), Gaps = 15/146 (10%)
Query: 2 EDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREF-LRSLSQKKEESFREYAQR 60
+ A W L +D + ++ EL F +++ T +P F L ++ QK ES R+Y +R
Sbjct: 485 DSAWSWLHGLPRDTIGSWAELRDHFIANFQ-GTFERPGTHFDLYNIVQKSGESLRDYIRR 543
Query: 61 WRGAAARITPALDEEEMTQTFLKTLK-KDYVERMIIAAPNNFSEMVTMGTRLEEAVRDGI 119
+ +I+ D+ + F K ++ +D V++ P +M
Sbjct: 544 FSEQRNKISDITDDV-IIAAFTKGIRHEDLVDKFGRKPPKTVKQMFEKANE--------- 593
Query: 120 IVFEKAESSVNASKRYGNGHHKKKET 145
+ KAE ++ SK+ G KK+T
Sbjct: 594 --YAKAEDAITTSKQSGTTWKPKKDT 617
>UniRef100_Q7XKP3 OSJNBa0032N05.6 protein [Oryza sativa]
Length = 371
Score = 38.5 bits (88), Expect = 0.11
Identities = 47/168 (27%), Positives = 73/168 (42%), Gaps = 25/168 (14%)
Query: 4 AAEWYTSLSKDDVH----TFDELAAAFK-SHYGFNTRLKPNREFLRSLSQKKEESFREYA 58
A+EW+ + + T+ E AFK +H REF R+L QK + + EY
Sbjct: 109 ASEWWDHIRMNRAEGQPITWAEFTEAFKKTHIPAGVVALKKREF-RALKQK-DHTVAEYL 166
Query: 59 QRWRGAAARITP--ALDEEEMTQTFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAV- 115
+ AR P +EE + FL+ LK + +I +F E+V RLE+
Sbjct: 167 HEFN-RLARYAPEDVRTDEERQEKFLEGLKDELSVMLISHDYEDFQELVDKAIRLEDKKN 225
Query: 116 -----RDGIIVFEKAESSVNASKRYGNGHHKKKETEVG-MVSAGAGQS 157
+ G IVF++A+ S + + K ++G S G GQS
Sbjct: 226 RMDNRKRGRIVFQEAQDS--------SQRQRIKPLQIGESFSTGQGQS 265
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 348,955,410
Number of Sequences: 2790947
Number of extensions: 13324923
Number of successful extensions: 52537
Number of sequences better than 10.0: 166
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 139
Number of HSP's that attempted gapping in prelim test: 52437
Number of HSP's gapped (non-prelim): 228
length of query: 207
length of database: 848,049,833
effective HSP length: 122
effective length of query: 85
effective length of database: 507,554,299
effective search space: 43142115415
effective search space used: 43142115415
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 72 (32.3 bits)
Medicago: description of AC144515.9


