

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144515.12 - phase: 0
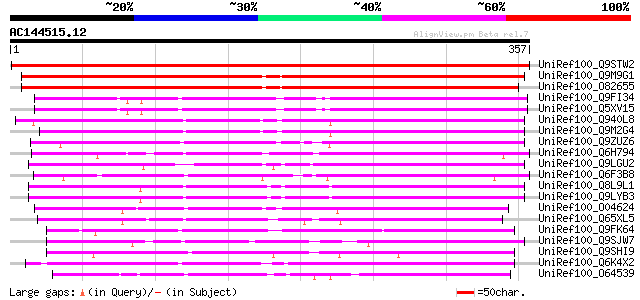
(357 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9STW2 Hypothetical protein T22A6.150 [Arabidopsis tha... 405 e-111
UniRef100_Q9M9G1 F14O23.23 protein [Arabidopsis thaliana] 270 3e-71
UniRef100_O82655 Hypothetical protein ORF2 [Arabidopsis thaliana] 267 4e-70
UniRef100_Q9FI34 Emb|CAB68190.1 [Arabidopsis thaliana] 215 2e-54
UniRef100_Q5XV15 Hypothetical protein [Arabidopsis thaliana] 213 7e-54
UniRef100_Q940L8 AT3g58520/F14P22_110 [Arabidopsis thaliana] 200 4e-50
UniRef100_Q9M2G4 Hypothetical protein F14P22.110 [Arabidopsis th... 196 1e-48
UniRef100_Q9ZUZ6 Hypothetical protein At2g39120 [Arabidopsis tha... 194 4e-48
UniRef100_Q6H794 Hypothetical protein P0458B05.12 [Oryza sativa] 186 7e-46
UniRef100_Q9LGU2 EST AU030398(E51008) corresponds to a region of... 158 2e-37
UniRef100_Q6F3B8 Expressed protein [Oryza sativa] 152 2e-35
UniRef100_Q8L9L1 Hypothetical protein [Arabidopsis thaliana] 146 1e-33
UniRef100_Q9LYB3 Hypothetical protein T20O10_190 [Arabidopsis th... 145 1e-33
UniRef100_O04624 A_IG002N01.30 protein [Arabidopsis thaliana] 138 2e-31
UniRef100_Q65XL5 Hypothetical protein OJ1735_C10.3 [Oryza sativa] 130 7e-29
UniRef100_Q9FK64 Tyrosine-specific protein phosphatase-like prot... 129 1e-28
UniRef100_Q9SJW7 Hypothetical protein At2g31290 [Arabidopsis tha... 126 1e-27
UniRef100_Q9SHI9 F12K11.21 [Arabidopsis thaliana] 121 3e-26
UniRef100_Q6K4X2 Tyrosine-specific protein phosphatase-like [Ory... 121 3e-26
UniRef100_O64539 YUP8H12R.26 protein [Arabidopsis thaliana] 114 4e-24
>UniRef100_Q9STW2 Hypothetical protein T22A6.150 [Arabidopsis thaliana]
Length = 395
Score = 405 bits (1040), Expect = e-111
Identities = 204/356 (57%), Positives = 255/356 (71%)
Query: 2 AQSTKLFHRFDHIRTFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLY 61
A +TKL RTFVNARVKWV D YLD AV +EKNLK VISLK+ IVSSPSKSL L
Sbjct: 3 AATTKLARALQQRRTFVNARVKWVSDHYLDEAVQREKNLKQVISLKDRIVSSPSKSLPLS 62
Query: 62 NASMLKASLNLPMITIKLVEKYHHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRT 121
+ S+LK +NL + +KY VF FQPSP P ++LT QAL LHKEE +H S
Sbjct: 63 SLSLLKPLVNLHITAAAFFQKYPSVFTTFQPSPSHPLHVRLTPQALTLHKEEETIHLSPP 122
Query: 122 NQEDAAQRLARLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPL 181
+ QRL + LML G LP+YV+++ ++D+GLP DY+T+L+ YP+YF+V ++D L
Sbjct: 123 QRNVTVQRLTKFLMLTGAGSLPLYVLDRFRFDLGLPRDYITSLIGDYPEYFEVTEIKDRL 182
Query: 182 SGKEVLALELVSWRKELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWV 241
+G++ LAL + S R L VSE+E+R + K+ I++ M PK + L KRVK WV
Sbjct: 183 TGEKTLALTISSRRNNLPVSEMERREATIDGSRVKKGLRIRYSMNFPKGYELHKRVKNWV 242
Query: 242 EGWQTLPYISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLG 301
E WQ LPYISPYENAFHL S SDQAEKW VA+LHELL LLVSKKTE++N+IC GE LG G
Sbjct: 243 EQWQNLPYISPYENAFHLGSYSDQAEKWAVAVLHELLFLLVSKKTETDNVICLGEYLGFG 302
Query: 302 SRFKKALVHHPGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLMTKA 357
RFKKALVHHPGIFY+S+KIRTQT VLREAY FL++ HP+MG R+ YI+LM+K+
Sbjct: 303 MRFKKALVHHPGIFYMSHKIRTQTVVLREAYHKVFLLEKHPLMGIRHQYIYLMSKS 358
>UniRef100_Q9M9G1 F14O23.23 protein [Arabidopsis thaliana]
Length = 470
Score = 270 bits (691), Expect = 3e-71
Identities = 143/347 (41%), Positives = 217/347 (62%), Gaps = 4/347 (1%)
Query: 9 HRFDHIRTFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKA 68
HR + V K+VRD LD AV +EKNL+ ++S+K+ I S P+KS+ + + K
Sbjct: 16 HRTFADTSAVEDTYKFVRDRGLDHAVEREKNLRPLLSIKDLIRSEPAKSVPISVITSQKD 75
Query: 69 SLNLPMITIKLVEKYHHVFMQFQPSP-GLPPRIKLTAQALALHKEEMEVHNSRTNQEDAA 127
SL +P+ I+ + + VF +F P G+ P I LT + L +E V+ S T ++ A
Sbjct: 76 SLRVPLRPIEFIRSFPSVFQEFLPGGIGIHPHISLTPEILNHDADEQLVYGSETYKQGLA 135
Query: 128 QRLARLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVL 187
RL +LLM+ + ++P+ +++ LKWD+GLP DYV T++ +PDYF V ++ L G
Sbjct: 136 DRLLKLLMINRINKIPLEILDLLKWDLGLPKDYVETMVPEFPDYFRV--IKSKLRGCSG- 192
Query: 188 ALELVSWRKELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTL 247
LELV W E +VS LEK+ + I FPM F ++K++K W++ WQ L
Sbjct: 193 ELELVCWSNEHAVSVLEKKARTLRKGEYTKGSAIAFPMKFSNGFVVDKKMKKWIDDWQKL 252
Query: 248 PYISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKA 307
PYISPYENA HL + SD+++KW A+LHE+++L VSKK E + ++ GE +GL SRFK+
Sbjct: 253 PYISPYENALHLSATSDESDKWAAAVLHEIMNLFVSKKVEKDAILHLGEFMGLRSRFKRV 312
Query: 308 LVHHPGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLM 354
L +HPGIFYLS+K+RT T VLR+ Y+ L++++ ++ +R Y+ LM
Sbjct: 313 LHNHPGIFYLSSKLRTHTVVLRDGYKRGMLIESNELVTSRNRYMKLM 359
>UniRef100_O82655 Hypothetical protein ORF2 [Arabidopsis thaliana]
Length = 470
Score = 267 bits (682), Expect = 4e-70
Identities = 141/343 (41%), Positives = 214/343 (62%), Gaps = 4/343 (1%)
Query: 9 HRFDHIRTFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKA 68
HR + V K+VRD LD AV +EKNL+ ++S+K+ I S P+KS+ + + K
Sbjct: 16 HRTFADTSAVEDTYKFVRDRGLDHAVEREKNLRPLLSIKDLIRSEPAKSVPISVITSQKD 75
Query: 69 SLNLPMITIKLVEKYHHVFMQFQPSP-GLPPRIKLTAQALALHKEEMEVHNSRTNQEDAA 127
SL +P+ I+ + + VF +F P G+ P I LT + L +E V+ S T ++ A
Sbjct: 76 SLRVPLRPIEFIRSFPSVFQEFLPGGIGIHPHISLTPEILNHDADEQLVYGSETYKQGLA 135
Query: 128 QRLARLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVL 187
RL +LLM+ + ++P+ +++ LKWD+GLP DYV T++ +PDYF V ++ L G
Sbjct: 136 DRLLKLLMINRINKIPLEILDLLKWDLGLPKDYVETMVPEFPDYFRV--IKSKLRGCSG- 192
Query: 188 ALELVSWRKELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTL 247
LELV W E +VS LEK+ + I FPM F ++K++K W++ WQ L
Sbjct: 193 ELELVCWSNEHAVSVLEKKARTLRKGEYTKGSAIAFPMKFSNGFVVDKKMKKWIDDWQKL 252
Query: 248 PYISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKA 307
PYISPYENA HL + SD+++KW A+LHE+++L VSKK E + ++ GE +GL SRFK+
Sbjct: 253 PYISPYENALHLSATSDESDKWAAAVLHEIMNLFVSKKVEKDAILHLGEFMGLRSRFKRV 312
Query: 308 LVHHPGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWY 350
L +HPGIFYLS+K+RT T VLR+ Y+ L++++ ++ +R Y
Sbjct: 313 LHNHPGIFYLSSKLRTHTVVLRDGYKRGMLIESNELVTSRNRY 355
>UniRef100_Q9FI34 Emb|CAB68190.1 [Arabidopsis thaliana]
Length = 422
Score = 215 bits (547), Expect = 2e-54
Identities = 125/345 (36%), Positives = 199/345 (57%), Gaps = 20/345 (5%)
Query: 18 VNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMITI 77
VN ++KWV+D LD V++EK+L+ V +L + I +SP L ++ + L LP +
Sbjct: 33 VNVKLKWVKDRELDAVVVREKHLRAVCNLVSVISASPDLRLPIFKLLPHRGQLGLPQ-EL 91
Query: 78 KL---VEKYHHVFMQ--FQPSPGLP-PRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLA 131
KL + +Y ++F++ + S G P LT + + L+ EE++V SR N+ D RL
Sbjct: 92 KLSAFIRRYPNIFVEHCYWDSAGTSVPCFGLTRETIDLYYEEVDV--SRVNERDVLVRLC 149
Query: 132 RLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALEL 191
+LLML L ++ I+ L+WD+GLP+DY +L+ +PD F + + L G L+L
Sbjct: 150 KLLMLTCERTLSLHSIDHLRWDLGLPYDYRDSLITKHPDLFSLVKLSSDLDG-----LKL 204
Query: 192 VSWRKELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTLPYIS 251
+ W + L+VS+++ R N D+R + FP+ + F L+++ W++ WQ LPY S
Sbjct: 205 IHWDEHLAVSQMQLREDVGN---DER---MAFPVKFTRGFGLKRKSIEWLQEWQRLPYTS 258
Query: 252 PYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHH 311
PY +A HLD +D +EK V + HELL L + KKTE +N+ + L +F K H
Sbjct: 259 PYVDASHLDPRTDLSEKRNVGVFHELLHLTIGKKTERKNVSNLRKPFALPQKFTKVFERH 318
Query: 312 PGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLMTK 356
PGIFY+S K TQT +LREAY L++ HP++ R + ++M +
Sbjct: 319 PGIFYISMKCDTQTVILREAYDRRHLIEKHPLVEVREKFANMMNE 363
>UniRef100_Q5XV15 Hypothetical protein [Arabidopsis thaliana]
Length = 422
Score = 213 bits (542), Expect = 7e-54
Identities = 124/345 (35%), Positives = 198/345 (56%), Gaps = 20/345 (5%)
Query: 18 VNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMITI 77
VN ++KWV+D LD V++EK+L+ V +L + I +SP L ++ + L LP +
Sbjct: 33 VNVKLKWVKDRELDAVVVREKHLRAVCNLVSVISASPDLRLPIFKLLPHRGQLGLPQ-EL 91
Query: 78 KL---VEKYHHVFMQ--FQPSPGLP-PRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLA 131
KL + +Y ++F++ + S G P LT + + L+ EE++V SR N+ D RL
Sbjct: 92 KLSAFIRRYPNIFVEHCYWDSAGTSVPCFGLTRETIDLYYEEVDV--SRVNERDVLVRLC 149
Query: 132 RLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALEL 191
+LLML L ++ I+ L+WD+G P+DY +L+ +PD F + + L G L+L
Sbjct: 150 KLLMLTCERTLSLHSIDHLRWDLGXPYDYRDSLITKHPDLFSLVKLSSDLDG-----LKL 204
Query: 192 VSWRKELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTLPYIS 251
+ W + L+VS+++ R N D+R + FP+ + F L+++ W++ WQ LPY S
Sbjct: 205 IHWDEHLAVSQMQLREDVGN---DER---MAFPVKFTRGFGLKRKSIEWLQEWQRLPYTS 258
Query: 252 PYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHH 311
PY +A HLD +D +EK V + HELL L + KKTE +N+ + L +F K H
Sbjct: 259 PYVDASHLDPRTDLSEKRNVGVFHELLHLTIGKKTERKNVSNLRKPFALPQKFTKVFERH 318
Query: 312 PGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLMTK 356
PGIFY+S K TQT +LREAY L++ HP++ R + ++M +
Sbjct: 319 PGIFYISMKCDTQTVILREAYDRRHLIEKHPLVEVREKFANMMNE 363
>UniRef100_Q940L8 AT3g58520/F14P22_110 [Arabidopsis thaliana]
Length = 418
Score = 200 bits (509), Expect = 4e-50
Identities = 124/362 (34%), Positives = 193/362 (53%), Gaps = 19/362 (5%)
Query: 5 TKLFHRFDHIR---TFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLY 61
+KLFH+ R R+KWV++ LD + E +LK LK+ I SP+ L+
Sbjct: 13 SKLFHQHQQWRGMGAMTKVRLKWVKNKNLDHVIDTETDLKAACILKDAIKRSPTGFLTAK 72
Query: 62 NASMLKASLNLPMITIKLVEKYHHVFMQFQPSPGLP-PRIKLTAQALALHKEEMEVHNSR 120
+ + + L L + ++ + +Y +F +F + P KLT AL L +E +H S
Sbjct: 73 SVADWQKLLGLTVPVLRFLRRYPTLFHEFPHARYASLPCFKLTDTALMLDSQEEIIHQS- 131
Query: 121 TNQEDAAQRLARLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDP 180
++ D +RL R+LM+ + + + LK+D+GLP +Y TL+ YPD+F C V+
Sbjct: 132 -HEADTVERLCRVLMMMRSKTVSLRSLHSLKFDLGLPDNYEKTLVMKYPDHF--CFVKAS 188
Query: 181 LSGKEVLALELVSWRKELSVSELEKRVMGFNYDADKRRH--------DIQFPMFLPKSFA 232
L+LV WR E + S L+KR + ++ R+ + FPM P+ +
Sbjct: 189 NGNP---CLKLVKWRDEFAFSALQKRNEDNDVSGEESRYREFKRGQSTLTFPMSFPRGYG 245
Query: 233 LEKRVKTWVEGWQTLPYISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLI 292
+K+VK W++ +Q LPYISPY++ ++D SD EK VA+LHELLSL + KKT+ L
Sbjct: 246 AQKKVKAWMDEFQKLPYISPYDDPSNIDPESDLMEKRAVAVLHELLSLTIHKKTKRNYLR 305
Query: 293 CYGECLGLGSRFKKALVHHPGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIH 352
L + +F + +PGIFYLS K +T T +L+E YR LV HP+ R + H
Sbjct: 306 SMRAELNIPHKFTRLFTRYPGIFYLSLKCKTTTVILKEGYRRGKLVDPHPLTRLRDKFYH 365
Query: 353 LM 354
+M
Sbjct: 366 VM 367
>UniRef100_Q9M2G4 Hypothetical protein F14P22.110 [Arabidopsis thaliana]
Length = 394
Score = 196 bits (497), Expect = 1e-48
Identities = 119/343 (34%), Positives = 186/343 (53%), Gaps = 16/343 (4%)
Query: 21 RVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMITIKLV 80
R+KWV++ LD + E +LK LK+ I SP+ L+ + + + L L + ++ +
Sbjct: 8 RLKWVKNKNLDHVIDTETDLKAACILKDAIKRSPTGFLTAKSVADWQKLLGLTVPVLRFL 67
Query: 81 EKYHHVFMQFQPSPGLP-PRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARLLMLAGM 139
+Y +F +F + P KLT AL L +E +H S ++ D +RL R+LM+
Sbjct: 68 RRYPTLFHEFPHARYASLPCFKLTDTALMLDSQEEIIHQS--HEADTVERLCRVLMMMRS 125
Query: 140 ARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWRKELS 199
+ + + LK+D+GLP +Y TL+ YPD+F C V+ L+LV WR E +
Sbjct: 126 KTVSLRSLHSLKFDLGLPDNYEKTLVMKYPDHF--CFVKASNGNP---CLKLVKWRDEFA 180
Query: 200 VSELEKRVMGFNYDADKRRH--------DIQFPMFLPKSFALEKRVKTWVEGWQTLPYIS 251
S L+KR + ++ R+ + FPM P+ + +K+VK W++ +Q LPYIS
Sbjct: 181 FSALQKRNEDNDVSGEESRYREFKRGQSTLTFPMSFPRGYGAQKKVKAWMDEFQKLPYIS 240
Query: 252 PYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHH 311
PY++ ++D SD EK VA+LHELLSL + KKT+ L L + +F + +
Sbjct: 241 PYDDPSNIDPESDLMEKRAVAVLHELLSLTIHKKTKRNYLRSMRAELNIPHKFTRLFTRY 300
Query: 312 PGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLM 354
PGIFYLS K +T T +L+E YR LV HP+ R + H+M
Sbjct: 301 PGIFYLSLKCKTTTVILKEGYRRGKLVDPHPLTRLRDKFYHVM 343
>UniRef100_Q9ZUZ6 Hypothetical protein At2g39120 [Arabidopsis thaliana]
Length = 387
Score = 194 bits (492), Expect = 4e-48
Identities = 123/347 (35%), Positives = 186/347 (53%), Gaps = 20/347 (5%)
Query: 15 RTFVNARVKWVRDDYLDTA--VLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNL 72
RT+V+ +KW RD Y D +L+ LK V+SLKN IV P++ + + S ++
Sbjct: 22 RTYVDVYMKWKRDPYFDNIEHILRSSQLKSVVSLKNCIVQEPNRCIPISAISKKTRQFDV 81
Query: 73 PMITIKLVEKYHHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLAR 132
+ K+ +F +F P +LT +A L ++E V+ +T+ +D RL +
Sbjct: 82 STKIAHFLRKFPSIFEEFVGPEYNLPWFRLTPEATELDRQERVVY--QTSADDLRDRLKK 139
Query: 133 LLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELV 192
L++++ LP+ +++ +KW +GLP DY+ F +ED + G LA++
Sbjct: 140 LILMSKDNVLPLSIVQGMKWYLGLPDDYLQFPDMNLDSSFRFVDMEDGVKG---LAVDYN 196
Query: 193 SWRKELSVSELEKRVMGFNYDADKRR-----HDIQFPMFLPKSFALEKRVKTWVEGWQTL 247
K LSV L+K M KRR +I+FP+F K L +++ W+ +Q L
Sbjct: 197 GGDKVLSV--LQKNAM------KKRRGEVSLEEIEFPLFPSKGCRLRVKIEDWLMEFQKL 248
Query: 248 PYISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKA 307
PY+SPY++ LD +SD AEK V LHELL L V E + L+C + GL + KA
Sbjct: 249 PYVSPYDDYSCLDPSSDIAEKRVVGFLHELLCLFVEHSAERKKLLCLKKHFGLPQKVHKA 308
Query: 308 LVHHPGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLM 354
HP IFYLS K +T TA+LRE YR++ V+ HPV+G R YI LM
Sbjct: 309 FERHPQIFYLSMKNKTCTAILREPYRDKASVETHPVLGVRKKYIQLM 355
>UniRef100_Q6H794 Hypothetical protein P0458B05.12 [Oryza sativa]
Length = 386
Score = 186 bits (473), Expect = 7e-46
Identities = 124/349 (35%), Positives = 182/349 (51%), Gaps = 22/349 (6%)
Query: 16 TFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLS----LYNASMLKASLN 71
T VN ++KWV+D LD AV +E++L+ L + + + P +S L ++S+ +A
Sbjct: 22 TLVNVKLKWVKDRALDGAVSRERDLRAAHHLLDVVSARPGHRVSRPELLADSSVRRAFGG 81
Query: 72 LPMITIKLVEKYHHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLA 131
+ + L +YH +F + + LT AL L + E++ ++ D RL
Sbjct: 82 VDGVDAFLA-RYHTLFALRRGGG-----VSLTDAALDLRRREVDCLVE--SEPDLVSRLR 133
Query: 132 RLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALEL 191
RLLML LP++ ++ L+WD+GLP DY ++L YPD+F + D G E + L L
Sbjct: 134 RLLMLTLPRSLPLHTVDLLRWDLGLPRDYRASILRRYPDHFAL----DQPEGDERVWLRL 189
Query: 192 VSWRKELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTLPYIS 251
+ W L+VSELEK G + FP+ K F L + W++ WQ LPY S
Sbjct: 190 LWWDDGLAVSELEKSTAG---GGGGDTTCLPFPVSFTKGFGLRSKCINWLKEWQALPYTS 246
Query: 252 PYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHH 311
PY + LD +D +EK V + HELL L V+K+TE N+ + LG+ +F K H
Sbjct: 247 PYADPSGLDRRTDVSEKRNVGVFHELLHLTVAKRTERRNVSNMRKLLGMPQKFTKVFERH 306
Query: 312 PGIFYLSNKIRTQTAVLREAYRNEFLV---KNHPVMGTRYWYIHLMTKA 357
PGIFYLS + TQT VLREAY L+ HP+ R Y +M A
Sbjct: 307 PGIFYLSRVLGTQTVVLREAYGGGSLLLAKHAHPLATIREEYSAVMRAA 355
>UniRef100_Q9LGU2 EST AU030398(E51008) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 498
Score = 158 bits (400), Expect = 2e-37
Identities = 114/352 (32%), Positives = 174/352 (49%), Gaps = 28/352 (7%)
Query: 14 IRTFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLP 73
+R + +V W RD LD A+L+++ + L ++ SP + L L S + + LP
Sbjct: 104 VRRISSLKVPWRRDAALDAAILRDRRYRLASRLVREVLLSPGRRLLLRYLSKRRQRIRLP 163
Query: 74 MITIKLVEKYHHVFMQF-------QPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDA 126
++ + +Y + PSP L ++ ++ ALH +
Sbjct: 164 VLVPTFLRRYPTLLSVSPPPNPVASPSPHLLSFLEFASRHHALHSPLL------------ 211
Query: 127 AQRLARLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDP---LSG 183
A RLA+LLM++ LP+ I K D GLP D++T+L+ YP F + V DP SG
Sbjct: 212 ASRLAKLLMISSTRALPVPKIAAAKRDFGLPDDFLTSLVPRYPHLFRL--VGDPGPDASG 269
Query: 184 KEVLALELVSWRKELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEG 243
L ELVSW +L+ S +E R D R F + LP+ F L+K ++ WV
Sbjct: 270 NAFL--ELVSWDDQLAKSVIELRA-DKEADVVGIRPRPNFTVKLPRGFYLKKEMREWVRD 326
Query: 244 WQTLPYISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSR 303
W LPY+SPY + L S +AEK + +LHE+LSL V ++ + + + L +
Sbjct: 327 WLELPYVSPYADTSGLHPASPEAEKRLIGVLHEVLSLSVERRMAVPIIGKFCDEFRLSNA 386
Query: 304 FKKALVHHPGIFYLSNKIRTQTAVLREAY-RNEFLVKNHPVMGTRYWYIHLM 354
F A HPGIFY+S K +TAVLREAY N LV P++ + ++ +M
Sbjct: 387 FANAFTRHPGIFYVSLKGGIKTAVLREAYDENGELVDKDPMIELKERFVAIM 438
>UniRef100_Q6F3B8 Expressed protein [Oryza sativa]
Length = 405
Score = 152 bits (383), Expect = 2e-35
Identities = 107/357 (29%), Positives = 182/357 (50%), Gaps = 29/357 (8%)
Query: 17 FVNARVKWVRDDYLDTAVL--KEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLP- 73
+V+ +++W +D D + + ++L+ ++SL + SP+ ++ S L+ L +P
Sbjct: 26 YVDVKMRWKKDASFDAVPVLSQARDLRPLVSLAGLLSPSPTPVSAV---SKLRIPLEVPD 82
Query: 74 MITIKLVEKYHHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARL 133
I + ++ F++ P +L+ A L +EE EV +R + D RL RL
Sbjct: 83 RRVISFLRRFPAAFVESVGPEHNHPWFRLSGSAAGLLQEEREVFAAR--RADITSRLGRL 140
Query: 134 LMLAGMARLPIYVIEKLKWDMGLPHDYVTTL-LAYYPDYF-------DVCVVEDPLSGKE 185
L++A RLP+ + + W +GLP DY D F VC E+ G+E
Sbjct: 141 LLMAPRRRLPLRAAQGMLWHLGLPEDYFRCRDYDIAQDGFRILTIGDSVCREEEEDDGRE 200
Query: 186 VLALELVSWRKELSVSELEKRVMGFNYDADKR---RHDIQFPMFLPKSFALEKRVKTWVE 242
++ ++ + E+ K V+ DA +R + P+F K L+++++ W+E
Sbjct: 201 LVLIDNGEHQ------EMPKSVL--QMDAIRRFGSMETVPIPLFQSKGLRLKQKIEAWLE 252
Query: 243 GWQTLPYISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGS 302
G+Q LPY+SPYE+ + +SD +EK V +LHELLSL V+ E L+C + LGL
Sbjct: 253 GFQKLPYVSPYEDFSGIYRDSDVSEKRVVGVLHELLSLFVTCSAERRRLLCLRQHLGLPQ 312
Query: 303 RFKKALVHHPGIFYLSNKIRTQTAVLREAY--RNEFLVKNHPVMGTRYWYIHLMTKA 357
+F + HP +FYL K +T VL+EAY R + ++ HP++ R Y LM ++
Sbjct: 313 KFHRVFERHPHVFYLLLKEKTCFVVLKEAYLARGDTAIEEHPMLVVRRKYAGLMEES 369
>UniRef100_Q8L9L1 Hypothetical protein [Arabidopsis thaliana]
Length = 404
Score = 146 bits (368), Expect = 1e-33
Identities = 101/345 (29%), Positives = 164/345 (47%), Gaps = 11/345 (3%)
Query: 14 IRTFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLP 73
IR + +V W +D LD A+ ++K K + +++ P + + L + L L
Sbjct: 28 IRWISSLKVVWKKDTRLDEAIEQDKRYKLCARVVKEVLNEPGQVIPLRYLEKRRERLRLT 87
Query: 74 MITIKLVEKYHHVFM----QFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQR 129
VE +F + +P ++ T + A EE +++ N+ +
Sbjct: 88 FKAKSFVEMNPSLFEIYYDRIKPKSDPVQFLRPTPRLRAFLDEEQRIYSE--NEPLLVSK 145
Query: 130 LARLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLAL 189
L +LLM+A + + +K D G P+D++ L+ YP+YF + + P GK L
Sbjct: 146 LCKLLMMAKDKVISADKLVHVKRDFGFPNDFLMKLVQKYPNYFRLTGL--PEEGKSFL-- 201
Query: 190 ELVSWRKELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTLPY 249
ELVSW + + S++E R R + + LP F L K ++ W W Y
Sbjct: 202 ELVSWNPDFAKSKIELRAEDETLKTGVRIRP-NYNVKLPSGFFLRKEMREWTRDWLEQDY 260
Query: 250 ISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALV 309
ISPYE+ HLD S + EK TV ++HELLSL + K+ L + + + F
Sbjct: 261 ISPYEDVSHLDQASKEMEKRTVGVVHELLSLSLLKRVPVPTLGKFCDEFRFSNAFSSVFT 320
Query: 310 HHPGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLM 354
H GIFYLS K +TAVLREAY+++ LV P++ + ++ L+
Sbjct: 321 RHSGIFYLSLKGGIKTAVLREAYKDDELVDRDPLLAIKDKFLRLL 365
>UniRef100_Q9LYB3 Hypothetical protein T20O10_190 [Arabidopsis thaliana]
Length = 404
Score = 145 bits (367), Expect = 1e-33
Identities = 101/345 (29%), Positives = 164/345 (47%), Gaps = 11/345 (3%)
Query: 14 IRTFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLP 73
IR + +V W +D LD A+ ++K K + +++ P + + L + L L
Sbjct: 28 IRWISSLKVVWKKDTRLDEAIEQDKRYKLCARVVKEVLNEPGQVIPLRYLEKRRERLRLT 87
Query: 74 MITIKLVEKYHHVFM----QFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQR 129
VE +F + +P ++ T + A EE +++ N+ +
Sbjct: 88 FKAKSFVEMNPSLFEIYYDRIKPKSDPVQFLRPTPRLRAFLDEEQRIYSE--NEPLLVSK 145
Query: 130 LARLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLAL 189
L +LLM+A + + +K D G P+D++ L+ YP+YF + + P GK L
Sbjct: 146 LCQLLMMAKDKVISADKLVHVKRDFGFPNDFLMKLVQKYPNYFRLTGL--PEEGKSFL-- 201
Query: 190 ELVSWRKELSVSELEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTLPY 249
ELVSW + + S++E R R + + LP F L K ++ W W Y
Sbjct: 202 ELVSWNPDFAKSKIELRAEDETLKTGVRIRP-NYNVKLPSGFFLRKEMREWTRDWLEQDY 260
Query: 250 ISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALV 309
ISPYE+ HLD S + EK TV ++HELLSL + K+ L + + + F
Sbjct: 261 ISPYEDVSHLDQASKEMEKRTVGVVHELLSLSLLKRVPVPTLGKFCDEFRFSNAFSSVFT 320
Query: 310 HHPGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLM 354
H GIFYLS K +TAVLREAY+++ LV P++ + ++ L+
Sbjct: 321 RHSGIFYLSLKGGIKTAVLREAYKDDELVDRDPLLAIKDKFLRLL 365
>UniRef100_O04624 A_IG002N01.30 protein [Arabidopsis thaliana]
Length = 878
Score = 138 bits (348), Expect = 2e-31
Identities = 103/334 (30%), Positives = 166/334 (48%), Gaps = 17/334 (5%)
Query: 18 VNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMIT- 76
V A VK ++ D+ V ++K LK V++++ +VS P + +SL + L L
Sbjct: 126 VRAAVKRRKELTFDSVVQRDKKLKLVLNIRKILVSQPDRMMSLRGLGKYRRDLGLKKRRR 185
Query: 77 -IKLVEKYHHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARLLM 135
I L+ KY VF + R K+T++A L+ +EM + N ++ +L +L+M
Sbjct: 186 FIALLRKYPGVF-EIVEEGAYSLRFKMTSEAERLYLDEMRIRNEL--EDVLVVKLRKLVM 242
Query: 136 LAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWR 195
++ R+ + I LK D+GLP ++ T+ YP YF V V P ALEL W
Sbjct: 243 MSIDKRILLEKISHLKTDLGLPLEFRDTICQRYPQYFRV--VPTPRGP----ALELTHWD 296
Query: 196 KELSVSELE-KRVMGFNYDADKRRHDIQFP-----MFLPKSFALEKRVKTWVEGWQTLPY 249
EL+VS E ++++R I P + LP+ L K + ++ + Y
Sbjct: 297 PELAVSAAELSEDDNRTRESEERNLIIDRPPKFNRVKLPRGLNLSKSETRKISQFRDMQY 356
Query: 250 ISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALV 309
ISPY++ HL S + + EK ++HELLSL K+T ++L + E + + L+
Sbjct: 357 ISPYKDFSHLRSGTLEKEKHACGVIHELLSLTTEKRTLVDHLTHFREEFRFSQQLRGMLI 416
Query: 310 HHPGIFYLSNKIRTQTAVLREAYRNEFLVKNHPV 343
HP +FY+S K + LREAYRN L+ P+
Sbjct: 417 RHPDLFYVSLKGERDSVFLREAYRNSELIDKDPL 450
>UniRef100_Q65XL5 Hypothetical protein OJ1735_C10.3 [Oryza sativa]
Length = 509
Score = 130 bits (326), Expect = 7e-29
Identities = 102/332 (30%), Positives = 164/332 (48%), Gaps = 25/332 (7%)
Query: 20 ARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMIT--I 77
A VK ++ D + ++K LK V+ L+N +VS+P + +SL + + L L I
Sbjct: 59 AAVKRRKEIPFDNVIQRDKKLKLVLKLRNILVSNPDRVMSLRDLGRFRRDLGLTRKRRLI 118
Query: 78 KLVEKYHHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARLLMLA 137
L++++ VF + R +LT A L+ +E+ + N ++ A +L +LLM++
Sbjct: 119 ALLKRFPGVFEVVEEGV-YSLRFRLTPAAERLYLDELHLKNE--SEGLAVTKLRKLLMMS 175
Query: 138 GMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWRKE 197
R+ I I LK D+GLP ++ T+ YP YF V ++ LEL W E
Sbjct: 176 QDKRILIEKIAHLKNDLGLPPEFRDTICLRYPQYFRVVQMD------RGPGLELTHWDPE 229
Query: 198 LSVS-----ELEKRVMGFNYDADKRRHDIQFPMF-----LPKSFALEKRVKTWVEGWQTL 247
L+VS E E R + +R I P+ LP+ L + V ++ +
Sbjct: 230 LAVSAAEVAEEENRAR----EEQERNLIIDRPLKFNRVKLPQGLKLSRGEARRVAQFKEM 285
Query: 248 PYISPYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKA 307
PYISPY + HL S S + EK ++HE+LSL + K+T ++L + E +
Sbjct: 286 PYISPYSDFSHLRSGSAEKEKHACGVVHEILSLTLEKRTLVDHLTHFREEFRFSQSLRGM 345
Query: 308 LVHHPGIFYLSNKIRTQTAVLREAYRNEFLVK 339
L+ HP +FY+S K + LREAY+N LV+
Sbjct: 346 LIRHPDMFYVSLKGDRDSVFLREAYKNSQLVE 377
>UniRef100_Q9FK64 Tyrosine-specific protein phosphatase-like protein [Arabidopsis
thaliana]
Length = 423
Score = 129 bits (324), Expect = 1e-28
Identities = 98/327 (29%), Positives = 164/327 (49%), Gaps = 18/327 (5%)
Query: 26 RDDYLDTAVLKEKNLKHVISLKNHIVSSPSKS--LSLYNASMLKASLNLPMITIKLVEKY 83
RD LD + + + L ++ + ++SS + +SL S K + L + + KY
Sbjct: 54 RDHQLDKIIPQIRKLNIILEISK-LMSSKKRGPFVSLQLMSRWKNLVGLNVNVGAFIGKY 112
Query: 84 HHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARLLMLAGMARLP 143
H F F K+T + L EE V R + DA +R+ +LL+L+ L
Sbjct: 113 PHAFEIFTHPFSKNLCCKITEKFKVLIDEEENV--VRECEVDAVKRVKKLLLLSKHGVLR 170
Query: 144 IYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWRKE-LSVSE 202
++ + ++ ++GLP D+ ++LA Y F + +E LELV E L V++
Sbjct: 171 VHALRLIRKELGLPEDFRDSILAKYSSEFRLVDLE---------TLELVDRDDESLCVAK 221
Query: 203 LEK-RVMGFNYD-ADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTLPYISPYENAFHLD 260
+E+ R + + K + FP+ LP F +EK + ++ WQ +PY+ PY+ +
Sbjct: 222 VEEWREVEYREKWLSKFETNYAFPIHLPTGFKIEKGFREELKNWQRVPYVKPYDRK-EIS 280
Query: 261 SNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHHPGIFYLSNK 320
++ EK VA++HELLSL V K E E L + + LG+ ++ ++ HPGIFY+S K
Sbjct: 281 RGLERFEKRVVAVIHELLSLTVEKMVEVERLAHFRKDLGIEVNVREVILKHPGIFYVSTK 340
Query: 321 IRTQTAVLREAYRNEFLVKNHPVMGTR 347
+QT LREAY L++ +P+ R
Sbjct: 341 GSSQTLFLREAYSKGCLIEPNPIYNVR 367
>UniRef100_Q9SJW7 Hypothetical protein At2g31290 [Arabidopsis thaliana]
Length = 415
Score = 126 bits (316), Expect = 1e-27
Identities = 101/338 (29%), Positives = 159/338 (46%), Gaps = 35/338 (10%)
Query: 26 RDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMITIKLVEKY-- 83
+D L++A+ + K LKN I+ P++ S+ +L+L + ++KY
Sbjct: 30 KDPDLESALSRNKRWIVNSRLKNIILRCPNQVASIKFLQKKFKTLDLQGKALNWLKKYPC 89
Query: 84 -HHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARLLMLAGMARL 142
HV+++ +LT + L +EE V + T + A RLA+LLML+ RL
Sbjct: 90 CFHVYLENDEY-----YCRLTKPMMTLVEEEELVKD--TQEPVLADRLAKLLMLSVNQRL 142
Query: 143 PIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWRKELSVSE 202
+ + + K G P DYV ++ P Y DV + + K + +EL+ W+ EL+VS
Sbjct: 143 NVVKLNEFKRSFGFPDDYVIRIV---PKYSDVFRLVNYSGRKSSMEIELLLWKPELAVSA 199
Query: 203 LEKRVMGFNYDADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQ------TLPYISPYENA 256
+E A K + F LP TW + W+ PYISPYE+
Sbjct: 200 VEAA-------AKKCGSEPSFSCSLPS---------TWTKPWERFMEFNAFPYISPYEDH 243
Query: 257 FHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHHPGIFY 316
L S ++EK +V ++HELLSL + KK L + GL + L+ HPGIFY
Sbjct: 244 GDLVEGSKESEKRSVGLVHELLSLTLWKKLSIVKLSHFKREFGLPEKLNGMLLKHPGIFY 303
Query: 317 LSNKIRTQTAVLREAYRNEFLVKNHPVMGTRYWYIHLM 354
+ NK + T +LRE Y L+ P++ + + LM
Sbjct: 304 VGNKYQVHTVILREGYNGSELIHKDPLVVVKDKFGELM 341
>UniRef100_Q9SHI9 F12K11.21 [Arabidopsis thaliana]
Length = 390
Score = 121 bits (304), Expect = 3e-26
Identities = 95/336 (28%), Positives = 152/336 (44%), Gaps = 22/336 (6%)
Query: 26 RDDYLDTAVLKEKNLKHVISLKNHIVSSPSK---SLSLYNASMLKASLNLPMITIKLVEK 82
RD + + K KNL VI++++ +++P+ SLS+ S L L+L + K
Sbjct: 36 RDPVFEKLMDKYKNLLKVIAIQDLTLANPTADPPSLSIEFLSRLSQKLHLNRGAASFLRK 95
Query: 83 YHHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARLLMLAGMARL 142
Y H+F P +LT A+ + ++E + + RL RLL ++ +
Sbjct: 96 YPHIFHVLYDPVKAEPFCRLTDVAMEISRQEALAITATLSL--VVDRLVRLLSMSISKSI 153
Query: 143 PIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDP------LSGKEVLALELVSWRK 196
P+ + K+ ++GLP D+ ++++ P F + + L +E LE + +
Sbjct: 154 PLRAVFKVWRELGLPDDFEDSVISKNPHLFKLSDGHESNTHILELVQEEEKRLEFEAAVE 213
Query: 197 ELSVSELEKRVMGFNYDADKRRHDIQFPM--FLPKSFALEKRVKTWVEGWQTLPYISPYE 254
+ V E K D R +IQF P L K K V+ WQ LPY+ PYE
Sbjct: 214 KWRVVECSKE------DCSVDRTEIQFSFKHSYPPGMRLSKTFKAKVKEWQRLPYVGPYE 267
Query: 255 NAFHLDSNSDQA---EKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHH 311
+ + EK VAI HE L+L V K E E + + +C G+ + + H
Sbjct: 268 DMVGKKKSRSGVMGIEKRAVAIAHEFLNLTVEKMVEVEKISHFRKCFGIDLNIRDLFLDH 327
Query: 312 PGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTR 347
PG+FY+S K + T LREAY L+ +PV R
Sbjct: 328 PGMFYVSTKGKRHTVFLREAYERGRLIDPNPVYDAR 363
>UniRef100_Q6K4X2 Tyrosine-specific protein phosphatase-like [Oryza sativa]
Length = 402
Score = 121 bits (303), Expect = 3e-26
Identities = 100/339 (29%), Positives = 160/339 (46%), Gaps = 17/339 (5%)
Query: 12 DHIRTFVNARVKWVRDDYLDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLN 71
D RT + R RD LD +++ KNL+ + L I +K SL S + +
Sbjct: 43 DSARTRLEGRT---RDHKLDKLMIQLKNLRLALDLHELISQQRNKYASLQLLSRWRHEVG 99
Query: 72 LPMITIKLVEKYHHVFMQFQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLA 131
L + ++KY H+F + K+T + L +E R N+ A+RL
Sbjct: 100 LNIEIGAFLKKYPHIFDIYVHPIKRNECCKVTPKMSELIAQEDAA--IRENEPAIAKRLK 157
Query: 132 RLLMLAGMARLPIYVIEKLKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALEL 191
+LL L+ L ++ + ++ ++GLP DY ++L + F LS + L L
Sbjct: 158 KLLTLSKDGTLNMHALWLVRRELGLPDDYRCSILPNHQSDFS-------LSSPDTLTL-- 208
Query: 192 VSWRKELSVSELEK-RVMGFNYD-ADKRRHDIQFPMFLPKSFALEKRVKTWVEGWQTLPY 249
++ + L+V+++E+ R + + FP+ P F +EK + + WQ LPY
Sbjct: 209 ITRDENLAVADVEEWRAKEYTEKWLAESETKYAFPINFPTGFKIEKGFRGKLGNWQRLPY 268
Query: 250 ISPYE-NAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKAL 308
YE N H N ++ EK V ILHELLSL + K E L + + + ++ +
Sbjct: 269 TKAYEKNELHPIRNIERLEKRIVGILHELLSLTIEKMIPLERLSHFRKPFEMEVNLRELI 328
Query: 309 VHHPGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVMGTR 347
+ HPGIFY+S K TQT +LRE+Y LV +PV R
Sbjct: 329 LKHPGIFYISTKGSTQTVLLRESYSKGCLVHPNPVYNVR 367
>UniRef100_O64539 YUP8H12R.26 protein [Arabidopsis thaliana]
Length = 413
Score = 114 bits (285), Expect = 4e-24
Identities = 98/333 (29%), Positives = 156/333 (46%), Gaps = 33/333 (9%)
Query: 30 LDTAVLKEKNLKHVISLKNHIVSSPSKSLSLYNASMLKASLNLPMITIKLVEKYHHVFMQ 89
LD A+ K ++ LK+ I + SL L + ++ + + +EKY +F
Sbjct: 68 LDKAIDLNKKPSLILQLKSIIQAQKHGSLLLRDLEKHVGFVHKWNL-MAAIEKYPTIFY- 125
Query: 90 FQPSPGLPPRIKLTAQALALHKEEMEVHNSRTNQEDAAQRLARLLMLAGMARLPIYVIEK 149
PP + LT +A + EE E S + L +LLM++ R+P+ +E
Sbjct: 126 VGGGHKEPPFVMLTEKAKKIAAEESEATESM--EPILVNNLRKLLMMSVDCRVPLEKVEF 183
Query: 150 LKWDMGLPHDYVTTLLAYYPDYFDVCVVEDPLSGKEVLALELVSWRKELSVSELEKRVM- 208
++ MGLP D+ +TL+ Y ++F + V+ +GK L LE +W L+++ E R+
Sbjct: 184 IQSAMGLPQDFKSTLIPKYTEFFSLKVI----NGKVNLVLE--NWDSSLAITAREDRLSR 237
Query: 209 -GFNYDADKRRH---------------DIQFPM-FLPKSFALEKRVKTWVEGWQTLPYIS 251
GF+ R+ I FP F P + LE+ E WQ + + S
Sbjct: 238 EGFSESVGDRKRVRITKDGNFLGRNAFKISFPPGFRPNASYLEE-----FEKWQKMEFPS 292
Query: 252 PYENAFHLDSNSDQAEKWTVAILHELLSLLVSKKTESENLICYGECLGLGSRFKKALVHH 311
PY NA D+ +A K VA+LHELLSL + K+ L + L SR L+ H
Sbjct: 293 PYLNARRFDAADPKARKRVVAVLHELLSLTMEKRVTCAQLDAFHSEYLLPSRLILCLIKH 352
Query: 312 PGIFYLSNKIRTQTAVLREAYRNEFLVKNHPVM 344
GIFY++NK T L++AY L++ P++
Sbjct: 353 QGIFYITNKGARGTVFLKDAYAGSNLIEKCPLL 385
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 584,541,219
Number of Sequences: 2790947
Number of extensions: 23344316
Number of successful extensions: 58052
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 57935
Number of HSP's gapped (non-prelim): 46
length of query: 357
length of database: 848,049,833
effective HSP length: 128
effective length of query: 229
effective length of database: 490,808,617
effective search space: 112395173293
effective search space used: 112395173293
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144515.12


