

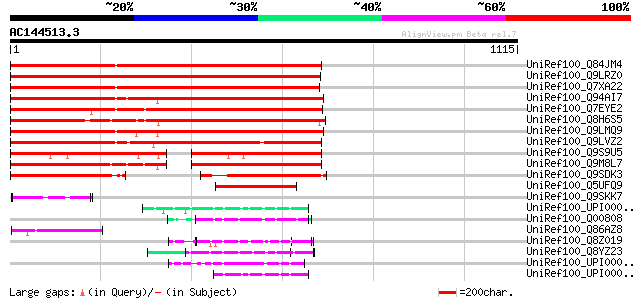
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.3 - phase: 0 /pseudo
(1115 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84JM4 Hypothetical protein At5g27030 [Arabidopsis tha... 1099 0.0
UniRef100_Q9LRZ0 Arabidopsis thaliana genomic DNA, chromosome 3,... 1048 0.0
UniRef100_Q7XA22 Putative beta transducin-like protein [Solanum ... 949 0.0
UniRef100_Q94AI7 Hypothetical protein At1g15750 [Arabidopsis tha... 925 0.0
UniRef100_Q7EYE2 WD-40 repeat protein-like [Oryza sativa] 924 0.0
UniRef100_Q8H6S5 CTV.2 [Poncirus trifoliata] 919 0.0
UniRef100_Q9LMQ9 F7H2.9 protein [Arabidopsis thaliana] 903 0.0
UniRef100_Q9LVZ2 Similarity to unknown protein [Arabidopsis thal... 880 0.0
UniRef100_Q9S9U5 F15P11.3 protein [Arabidopsis thaliana] 485 e-135
UniRef100_Q9M8L7 Hypothetical protein T21F11.18 [Arabidopsis tha... 421 e-116
UniRef100_Q9SDK3 EST AU078118(E3904) corresponds to a region of ... 399 e-109
UniRef100_Q5UFQ9 WD-40 repeat protein [Malus domestica] 271 6e-71
UniRef100_Q9SKK7 Hypothetical protein At2g25420 [Arabidopsis tha... 99 7e-19
UniRef100_UPI0000235C95 UPI0000235C95 UniRef100 entry 78 1e-12
UniRef100_Q00808 Vegetatible incompatibility protein HET-E-1 [Po... 78 2e-12
UniRef100_Q86AZ8 Similar to Anabaena sp. (Strain PCC 7120). Hypo... 73 4e-11
UniRef100_Q8Z019 WD-40 repeat protein [Anabaena sp.] 72 1e-10
UniRef100_Q8YZ23 WD-40 repeat protein [Anabaena sp.] 69 6e-10
UniRef100_UPI00003C1528 UPI00003C1528 UniRef100 entry 68 1e-09
UniRef100_UPI00002363EC UPI00002363EC UniRef100 entry 67 3e-09
>UniRef100_Q84JM4 Hypothetical protein At5g27030 [Arabidopsis thaliana]
Length = 1108
Score = 1099 bits (2842), Expect = 0.0
Identities = 540/688 (78%), Positives = 604/688 (87%), Gaps = 5/688 (0%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFLEEEKFKESVH+LEKESGFFFN KYF+EKV AGEW++VE YLSGF
Sbjct: 1 MSSLSRELVFLILQFLEEEKFKESVHRLEKESGFFFNTKYFDEKVLAGEWDDVETYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALDRQ+K KAVEILV DL+VFSTFNEELYKEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQEKAKAVEILVQDLRVFSTFNEELYKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTKTARGIML ELKKLIEANPLFRDKL+FPTL+SSRLRTLINQSLN
Sbjct: 121 LQNFRENEQLSKYGDTKTARGIMLGELKKLIEANPLFRDKLMFPTLRSSRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLGVGAHGPF 240
WQHQLCKNPRPNPDIKTLF DH+CT NGPLAP+ VN PV + KPAAY SLG H PF
Sbjct: 181 WQHQLCKNPRPNPDIKTLFTDHTCTLPNGPLAPSAVNQPVTTLTKPAAYPSLG--PHVPF 238
Query: 241 PPAAATANANALAGWMANASVSSSVQAAVVTASTIPVPHNQVSILKRPITPSTTPGMVEY 300
PP A ANA ALA WMA AS +S+VQAAVVT + +P P NQ+SILKRP TP TPG+V+Y
Sbjct: 239 PPGPAAANAGALASWMAAASGASAVQAAVVTPALMPQPQNQMSILKRPRTPPATPGIVDY 298
Query: 301 QSADHEQLMKRLRPAPSVEEVSYPSARQ-ASWSLDDLPRTVAMSLHQGSSVTSMDFHPSH 359
Q+ DHE LMKRLRPAPSVEEV+YP+ RQ A WSL+DLP A++LHQGS+VTSM+F+P
Sbjct: 299 QNPDHE-LMKRLRPAPSVEEVTYPAPRQQAPWSLEDLPTKAALALHQGSTVTSMEFYPMQ 357
Query: 360 QTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP-SVSRVTWSL 418
TLLLVGS GEI+LWEL RERLVS+PFKIWD+S CS FQA + K+TP SV+RV WS
Sbjct: 358 NTLLLVGSATGEITLWELAARERLVSRPFKIWDMSNCSHQFQALIAKETPISVTRVAWSP 417
Query: 419 DGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKL 478
DG+F+GVAFTKHLI +YA++G N+L Q EIDAH+G VNDLAFA+PN+QLCV+TCGDDKL
Sbjct: 418 DGNFIGVAFTKHLIQLYAFSGPNDLRQHTEIDAHVGAVNDLAFANPNRQLCVITCGDDKL 477
Query: 479 IKVWDLTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDA 538
IKVWD++GR+ F FEGH+APVYSICPH+KENIQFIFSTA+DGKIKAWLYDN+GSRVDYDA
Sbjct: 478 IKVWDVSGRKHFTFEGHDAPVYSICPHYKENIQFIFSTAIDGKIKAWLYDNLGSRVDYDA 537
Query: 539 PGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTT 598
PG WCT MLYSADGTRLFSCGTSKDGDSFLVEWNESEG+IKRTY F+KK AGVVQFDT+
Sbjct: 538 PGKWCTRMLYSADGTRLFSCGTSKDGDSFLVEWNESEGSIKRTYKEFQKKLAGVVQFDTS 597
Query: 599 QNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKIL 658
+N FLA GED QIKFWDM+N+N LTST+AEGGL LPHLRFNK+GNLLAVTTADNGFKIL
Sbjct: 598 KNHFLAVGEDGQIKFWDMNNINVLTSTDAEGGLPALPHLRFNKDGNLLAVTTADNGFKIL 657
Query: 659 ANAGGLRSLRTVETPAFEALRSPIESAA 686
AN G RSLR +ETPA E +R+P++ A
Sbjct: 658 ANPAGFRSLRAMETPASETMRTPVDFKA 685
>UniRef100_Q9LRZ0 Arabidopsis thaliana genomic DNA, chromosome 3, TAC clone:K20I9
[Arabidopsis thaliana]
Length = 1161
Score = 1048 bits (2709), Expect = 0.0
Identities = 518/685 (75%), Positives = 590/685 (85%), Gaps = 4/685 (0%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKESVHKLE+ESGFFFN+KYFEEK AGEW+EVEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKESVHKLEQESGFFFNIKYFEEKALAGEWDEVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALDR D+ KAVEIL DLKVF+TFNEELYKEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRNDRAKAVEILAKDLKVFATFNEELYKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTK+AR IM ELKKLIEANPLFR+KL FP+ K+SRLRTLINQSLN
Sbjct: 121 LENFRENEQLSKYGDTKSARSIMYTELKKLIEANPLFREKLAFPSFKASRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLGVGAHGPF 240
WQHQLCKNPRPNPDIKTLF+DHSC+PSNG A TPVNLPVAAVA+P+ + LGV GPF
Sbjct: 181 WQHQLCKNPRPNPDIKTLFLDHSCSPSNGARALTPVNLPVAAVARPSNFVPLGVHG-GPF 239
Query: 241 PPAAATA-NANALAGWMANASVSSSVQAAVVTASTIPVPHNQVSILKRPITPSTTPGMVE 299
A A NANALAGWMAN + SSSV + VV AS P+ +QV+ LK P PS + G+++
Sbjct: 240 QSNPAPAPNANALAGWMANPNPSSSVPSGVVAASPFPMQPSQVNELKHPRAPSNSLGLMD 299
Query: 300 YQSADHEQLMKRLRPAPSVEEVSYPSARQASWSLDDLPRTVAMSLHQGSSVTSMDFHPSH 359
YQSADHEQLMKRLR A + EV+YP+ SLDDLPR V ++ QGS V SMDFHPSH
Sbjct: 300 YQSADHEQLMKRLRSAQTSNEVTYPAHSHPPASLDDLPRNVVSTIRQGSVVISMDFHPSH 359
Query: 360 QTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP-SVSRVTWSL 418
TLL VG ++GE++LWE+G RE++V++PFKIW+++ACS+ FQ ++VK+ SV+RV WS
Sbjct: 360 HTLLAVGCSSGEVTLWEVGSREKVVTEPFKIWNMAACSVIFQGSIVKEPSISVTRVAWSP 419
Query: 419 DGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKL 478
DG+ +GV+FTKHLIH+YAY GS +L Q +EIDAH+G VNDLAFAHPNKQ+CVVTCGDDKL
Sbjct: 420 DGNLLGVSFTKHLIHVYAYQGS-DLRQHLEIDAHVGCVNDLAFAHPNKQMCVVTCGDDKL 478
Query: 479 IKVWDLTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDA 538
IKVWDL+G++LF FEGHEAPVYSICPH KENIQFIFSTA+DGKIKAWLYDN+GSRVDYDA
Sbjct: 479 IKVWDLSGKKLFTFEGHEAPVYSICPHQKENIQFIFSTALDGKIKAWLYDNVGSRVDYDA 538
Query: 539 PGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTT 598
PG WCTTMLYSADG+RLFSCGTSK+GDSFLVEWNESEGA+KRTY GFRKKSAGVVQFDTT
Sbjct: 539 PGQWCTTMLYSADGSRLFSCGTSKEGDSFLVEWNESEGALKRTYLGFRKKSAGVVQFDTT 598
Query: 599 QNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKIL 658
+NRFLA GED+QIKFW+MDN N LT EAEGGL LP LRFNK+GNLLAVTTADNGFKIL
Sbjct: 599 RNRFLAVGEDNQIKFWNMDNTNLLTVVEAEGGLPNLPRLRFNKDGNLLAVTTADNGFKIL 658
Query: 659 ANAGGLRSLRTVETPAFEALRSPIE 683
AN GLR+LR E +FEA ++ I+
Sbjct: 659 ANTDGLRTLRAFEARSFEASKASID 683
>UniRef100_Q7XA22 Putative beta transducin-like protein [Solanum bulbocastanum]
Length = 1078
Score = 949 bits (2454), Expect = 0.0
Identities = 473/685 (69%), Positives = 559/685 (81%), Gaps = 10/685 (1%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKE+VHKLE+ESGFFFNMKYF+E+VQAGEW+EVE+YL GF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKETVHKLEQESGFFFNMKYFDEQVQAGEWDEVERYLGGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKV+DNRYSMKIFFEIRKQKYLEALD+ D+ KAVEILV DLKVF++FNE+L+KEITQLLT
Sbjct: 61 TKVEDNRYSMKIFFEIRKQKYLEALDKHDRVKAVEILVKDLKVFASFNEDLFKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFR+NEQLSKYGDTK+AR IML+ELKKLIEANPLFRDKL FP+ K+SRLRTLINQSLN
Sbjct: 121 LDNFRQNEQLSKYGDTKSARNIMLVELKKLIEANPLFRDKLAFPSFKASRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAA-VAKPAAYTSLGVGAHGP 239
WQHQLCKNPRPNPDIKTLF DH+C+ SNG P P N P+A V KP A+ L GAH P
Sbjct: 181 WQHQLCKNPRPNPDIKTLFTDHTCSSSNGTRPPPPGNTPLAGPVPKPGAFPPL--GAHSP 238
Query: 240 FPPAAATANANALAGWMANASVSSSVQAAVVTASTIPVPHNQVSILKRPITPSTTPGMVE 299
F P + + +A+AGWM++A+ S S A + LK P PGM +
Sbjct: 239 FQP-VVSPSPSAIAGWMSSANPSMSHTAVAPGPPGLVQAPGAAGFLKHPRANPGGPGM-D 296
Query: 300 YQSADHEQLMKRLRPAPSVEEVSYPSARQA--SWSLDDLPRTVAMSLHQGSSVTSMDFHP 357
+Q A+ E LMKR+R S +EVS+ + +S DDLP+TV +L QGS+V SMDFHP
Sbjct: 297 FQMAESEHLMKRMRAGQS-DEVSFSGSTHPPNMYSPDDLPKTVVRNLSQGSNVMSMDFHP 355
Query: 358 SHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKD-TPSVSRVTW 416
QT+LLVG+N G+IS+WE+G RERL K FK+WDISACS+PFQ+A+VKD T SV+R W
Sbjct: 356 QQQTVLLVGTNVGDISIWEVGSRERLAHKSFKVWDISACSMPFQSALVKDATVSVNRCVW 415
Query: 417 SLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDD 476
DGS +GVAF+KH++ IY Y+ + EL Q +EIDAH GGVND+AF+HPNKQLCVVTCGDD
Sbjct: 416 GPDGSILGVAFSKHIVQIYTYSPAGELRQHLEIDAHAGGVNDIAFSHPNKQLCVVTCGDD 475
Query: 477 KLIKVWD-LTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVD 535
K IKVWD ++GRR FEGHEAPVYS+CPH+KE+IQFIFSTA+DGKIKAWLYD MGSRVD
Sbjct: 476 KTIKVWDAVSGRRQHMFEGHEAPVYSVCPHYKESIQFIFSTAIDGKIKAWLYDGMGSRVD 535
Query: 536 YDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQF 595
YDAPGHWCTTM YSADGTRLFSCGTSK+G+S LVEWNESEGAIKRT++GFRK+S GVVQF
Sbjct: 536 YDAPGHWCTTMAYSADGTRLFSCGTSKEGESHLVEWNESEGAIKRTFSGFRKRSLGVVQF 595
Query: 596 DTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGF 655
DTT+NRFLAAG++ QIKFW+MDN N LT+T+ +GGL P LRFNKEG+LLAVTT+DNG
Sbjct: 596 DTTRNRFLAAGDEFQIKFWEMDNTNMLTATDGDGGLPASPRLRFNKEGSLLAVTTSDNGI 655
Query: 656 KILANAGGLRSLRTVETPAFEALRS 680
K+LAN G R LR +E+ AFE R+
Sbjct: 656 KVLANTDGQRMLRMLESRAFEGSRA 680
>UniRef100_Q94AI7 Hypothetical protein At1g15750 [Arabidopsis thaliana]
Length = 1131
Score = 925 bits (2391), Expect = 0.0
Identities = 471/706 (66%), Positives = 552/706 (77%), Gaps = 24/706 (3%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKE+VHKLE+ESGFFFNMKYFE++V G W+EVEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKETVHKLEQESGFFFNMKYFEDEVHNGNWDEVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALD+ D+PKAV+ILV DLKVFSTFNEEL+KEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDKHDRPKAVDILVKDLKVFSTFNEELFKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTK+AR IML+ELKKLIEANPLFRDKL FPTL++SRLRTLINQSLN
Sbjct: 121 LENFRENEQLSKYGDTKSARAIMLVELKKLIEANPLFRDKLQFPTLRNSRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLP-VAAVAKPAAYTSLGVGAHGP 239
WQHQLCKNPRPNPDIKTLF+DHSC P NG AP+PVN P + + K + L GAHGP
Sbjct: 181 WQHQLCKNPRPNPDIKTLFVDHSCGPPNGARAPSPVNNPLLGGIPKAGGFPPL--GAHGP 238
Query: 240 FPPAAATANANALAGWMANASVSSSVQAAVVTASTIPVPHNQV-SILKRPITPSTTPGMV 298
F P A+ LAGWM S SSV V+A I + + + LK P TP T +
Sbjct: 239 FQPTASPV-PTPLAGWM---SSPSSVPHPAVSAGAIALGGPSIPAALKHPRTPPTNASL- 293
Query: 299 EYQSADHEQLMKRLRPAPSVEEVSY-------------PSARQASWSLDDLPRTVAMSLH 345
+Y SAD E + KR RP +EV+ A + DDLP+TVA +L
Sbjct: 294 DYPSADSEHVSKRTRPMGISDEVNLGVNMLPMSFSGQAHGHSPAFKAPDDLPKTVARTLS 353
Query: 346 QGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVV 405
QGSS SMDFHP QTLLLVG+N G+I LWE+G RERLV K FK+WD+S CS+P QAA+V
Sbjct: 354 QGSSPMSMDFHPIKQTLLLVGTNVGDIGLWEVGSRERLVQKTFKVWDLSKCSMPLQAALV 413
Query: 406 KD-TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHP 464
K+ SV+RV WS DGS GVA+++H++ +Y+Y+G ++ Q +EIDAH+GGVND++F+ P
Sbjct: 414 KEPVVSVNRVIWSPDGSLFGVAYSRHIVQLYSYHGGEDMRQHLEIDAHVGGVNDISFSTP 473
Query: 465 NKQLCVVTCGDDKLIKVWD-LTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIK 523
NKQLCV+TCGDDK IKVWD TG + FEGHEAPVYS+CPH+KENIQFIFSTA+DGKIK
Sbjct: 474 NKQLCVITCGDDKTIKVWDAATGVKRHTFEGHEAPVYSVCPHYKENIQFIFSTALDGKIK 533
Query: 524 AWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYN 583
AWLYDNMGSRVDYDAPG WCTTM YSADGTRLFSCGTSKDG+SF+VEWNESEGA+KRTY
Sbjct: 534 AWLYDNMGSRVDYDAPGRWCTTMAYSADGTRLFSCGTSKDGESFIVEWNESEGAVKRTYQ 593
Query: 584 GFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEG 643
GF K+S GVVQFDTT+NR+LAAG+D IKFWDMD V LT+ + +GGLQ P +RFNKEG
Sbjct: 594 GFHKRSLGVVQFDTTKNRYLAAGDDFSIKFWDMDAVQLLTAIDGDGGLQASPRIRFNKEG 653
Query: 644 NLLAVTTADNGFKILANAGGLRSLRTVETPAFEALRSPIESAANKA 689
+LLAV+ +N KI+AN+ GLR L T E + E+ + I S A A
Sbjct: 654 SLLAVSGNENVIKIMANSDGLRLLHTFENISSESSKPAINSIAAAA 699
>UniRef100_Q7EYE2 WD-40 repeat protein-like [Oryza sativa]
Length = 1150
Score = 924 bits (2388), Expect = 0.0
Identities = 467/711 (65%), Positives = 555/711 (77%), Gaps = 28/711 (3%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKE+VHKLE+ES F+FNMK+FE+ VQ GEW+EVEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKETVHKLEQESAFYFNMKHFEDLVQGGEWDEVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKV+DNRYSMKIFFEIRKQKYLEALDR D+ KAVEILV DLKVF++FNEEL+KEITQLLT
Sbjct: 61 TKVEDNRYSMKIFFEIRKQKYLEALDRHDRAKAVEILVKDLKVFASFNEELFKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQS-- 178
L NFR+NEQLSKYGDTK+AR IML+ELKKLIEANPLFRDKL FP K SRLRTLINQ
Sbjct: 121 LENFRQNEQLSKYGDTKSARNIMLMELKKLIEANPLFRDKLNFPPFKVSRLRTLINQRDV 180
Query: 179 ---------------LNWQHQLCKNPRPNPDIKTLFIDHSCT-PSNGPLAPTPVNLP-VA 221
LNWQHQLCKNPRPNPDIKTLF DHSC P+NG AP P N P V
Sbjct: 181 ICMNNNVNIQIGNAPLNWQHQLCKNPRPNPDIKTLFTDHSCAAPTNGARAPPPANGPLVG 240
Query: 222 AVAKPAAYTSLGVGAHGPFPPAAATANANALAGWMANASVSSSVQAAVVTASTIPVPHNQ 281
+ K AA+ +G AH PF P + + NA+AGWM NA+ S A + P N
Sbjct: 241 PIPKSAAFPPMG--AHAPFQPVVSPS-PNAIAGWMTNANPSLPHAAVAQGPPGLVQPPNT 297
Query: 282 VSILKRPITPSTTPGMVEYQSADHEQLMKRLRPAPSVEEVSYPSARQAS--WSLDDLPRT 339
+ LK P TP++ P ++YQSAD E LMKR+R +EVS+ A + ++ DDLP+
Sbjct: 298 AAFLKHPRTPTSAPA-IDYQSADSEHLMKRMRVGQP-DEVSFSGASHPANIYTQDDLPKQ 355
Query: 340 VAMSLHQGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLP 399
V +L+QGS+V S+DFHP QT+LLVG+N G+I +WE+G RER+ K FK+WDIS+C+LP
Sbjct: 356 VVRNLNQGSNVMSLDFHPVQQTILLVGTNVGDIGIWEVGSRERIAHKTFKVWDISSCTLP 415
Query: 400 FQAAVVKDTP-SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVND 458
QAA++KD SV+R WS DGS +GVAF+KH++ YA+ + EL Q+ EIDAHIGGVND
Sbjct: 416 LQAALMKDAAISVNRCLWSPDGSILGVAFSKHIVQTYAFVLNGELRQQAEIDAHIGGVND 475
Query: 459 LAFAHPNKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTA 517
+AF+HPNK L ++TCGDDKLIKVWD TG++ + FEGHEAPVYS+CPH+KE+IQFIFSTA
Sbjct: 476 IAFSHPNKTLSIITCGDDKLIKVWDAQTGQKQYTFEGHEAPVYSVCPHYKESIQFIFSTA 535
Query: 518 VDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGA 577
+DGKIKAWLYD +GSRVDYDAPGHWCTTM YSADGTRLFSCGTSKDGDS LVEWNE+EGA
Sbjct: 536 IDGKIKAWLYDCLGSRVDYDAPGHWCTTMAYSADGTRLFSCGTSKDGDSHLVEWNETEGA 595
Query: 578 IKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHL 637
IKRTYNGFRK+S GVVQFDTT+NRFLAAG++ +KFWDMDN N LT+T+ +GGL P L
Sbjct: 596 IKRTYNGFRKRSLGVVQFDTTRNRFLAAGDEFVVKFWDMDNTNILTTTDCDGGLPASPRL 655
Query: 638 RFNKEGNLLAVTTADNGFKILANAGGLRSLRTVETPAFEALRSPIESAANK 688
RFN+EG+LLAVT +NG KILAN G R LR +E+ A+E R P + K
Sbjct: 656 RFNREGSLLAVTANENGIKILANTDGQRLLRMLESRAYEGSRGPPQQINTK 706
>UniRef100_Q8H6S5 CTV.2 [Poncirus trifoliata]
Length = 1127
Score = 919 bits (2375), Expect = 0.0
Identities = 469/718 (65%), Positives = 551/718 (76%), Gaps = 38/718 (5%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKE+VHKLE+ESGFFFNMKYFE++V G W++VEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKETVHKLEQESGFFFNMKYFEDEVHNGNWDDVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALD+ D+ KAVEILV DLKVFSTFNEEL+KEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDKHDRAKAVEILVKDLKVFSTFNEELFKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTK+AR IML+ELKKLIEANPLFRDKL FP LK N SLN
Sbjct: 121 LENFRENEQLSKYGDTKSARSIMLVELKKLIEANPLFRDKLQFPNLK--------NSSLN 172
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLGVGAHGPF 240
WQHQLCKNPRPNPDIKTLF+DH+C NG AP+P N P+ + P A +GAHGPF
Sbjct: 173 WQHQLCKNPRPNPDIKTLFVDHTCGQPNGARAPSPANNPLLG-SLPKAGVFPPLGAHGPF 231
Query: 241 PPAAATANANALAGWMANASVSS--SVQAAVVTASTIPVPHNQVSILKRPITPSTTPGMV 298
P A LAGWM+N + +V + + +P + LK P TP T P V
Sbjct: 232 QPTPAPV-PTPLAGWMSNPPTVTHPAVSGGAIGLGSPSIP---AAALKHPRTPPTNPS-V 286
Query: 299 EYQSADHEQLMKRLRPAPSVEEVSYP-----------SARQASWSLDDLPRTVAMSLHQG 347
+Y S D + L KR RP +E++ P S QA + +DLP+TV +L+QG
Sbjct: 287 DYPSGDSDHLSKRTRPIGISDEINLPVNVLPVSFTGHSHSQAFSAPEDLPKTVTRTLNQG 346
Query: 348 SSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKD 407
SS SMDFHP QTLLLVG+N G+I LWE+G RERLV + FK+WD+ ACS+P QAA+VKD
Sbjct: 347 SSPMSMDFHPVQQTLLLVGTNVGDIGLWEVGSRERLVLRNFKVWDLGACSMPLQAALVKD 406
Query: 408 TP-SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNK 466
SV+RV WS DGS GVA+++H++ IY+Y+G +E+ Q +EIDAH+GGVND+AF+HPNK
Sbjct: 407 PGVSVNRVIWSPDGSLFGVAYSRHIVQIYSYHGGDEVRQHLEIDAHVGGVNDIAFSHPNK 466
Query: 467 QLCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAW 525
QLCV+TCGDDK IKVWD T G + + FEGHEAPVYS+CPHHKENIQFIFSTA+DGKIKAW
Sbjct: 467 QLCVITCGDDKTIKVWDATNGAKQYIFEGHEAPVYSVCPHHKENIQFIFSTALDGKIKAW 526
Query: 526 LYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGF 585
LYDN+GSRVDY+APG WCTTM YSADGTRLFSCGTSKDG+SF+VEWNESEGA+KRTY GF
Sbjct: 527 LYDNLGSRVDYEAPGRWCTTMAYSADGTRLFSCGTSKDGESFIVEWNESEGAVKRTYQGF 586
Query: 586 RKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNL 645
RK+S GVVQFDTT+NRFLAAG+D IKFWDMD+V LTS +A+GGL P +RFNK+G L
Sbjct: 587 RKRSLGVVQFDTTKNRFLAAGDDFSIKFWDMDSVQLLTSIDADGGLPASPRIRFNKDGCL 646
Query: 646 LAVTTADNGFKILANAGGLRSLRTVETPAFEALR---------SPIESAANKAVKWKG 694
LAV+T DNG KILA + G+R LRT E A++A R SPI +AA A G
Sbjct: 647 LAVSTNDNGIKILATSDGIRLLRTFENLAYDASRTSENSKPTISPISAAAAAAATSAG 704
>UniRef100_Q9LMQ9 F7H2.9 protein [Arabidopsis thaliana]
Length = 1153
Score = 903 bits (2334), Expect = 0.0
Identities = 467/725 (64%), Positives = 550/725 (75%), Gaps = 40/725 (5%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKE+VHKLE+ESGFFFNMKYFE++V G W+EVEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKETVHKLEQESGFFFNMKYFEDEVHNGNWDEVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALD+ D+PKAV+ILV DLKVFSTFNEEL+KEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDKHDRPKAVDILVKDLKVFSTFNEELFKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTK+AR IML+ELKKLIEANPLFRDKL FPTL++SRLRTLINQSLN
Sbjct: 121 LENFRENEQLSKYGDTKSARAIMLVELKKLIEANPLFRDKLQFPTLRNSRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLP-VAAVAKPAAYTSLGVGAHGP 239
WQHQLCKNPRPNPDIKTLF+DHSC P NG AP+PVN P + + K + L GAHGP
Sbjct: 181 WQHQLCKNPRPNPDIKTLFVDHSCGPPNGARAPSPVNNPLLGGIPKAGGFPPL--GAHGP 238
Query: 240 FPPAAATANANALAGWMANAS----VSSSVQAAVVTASTIP---------------VPHN 280
F P A+ LAGWM++ S + S A + +IP P
Sbjct: 239 FQPTASPV-PTPLAGWMSSPSSVPHPAVSAGAIALGGPSIPDTLCVIYAICLQWNSFPIF 297
Query: 281 QVSILKRPITPSTTPGMVEYQSADHEQLMKRLRPAPSVEEVSY-------------PSAR 327
++ LK P TP T + +Y SAD E + KR RP +EV+
Sbjct: 298 GIAALKHPRTPPTNASL-DYPSADSEHVSKRTRPMGISDEVNLGVNMLPMSFSGQAHGHS 356
Query: 328 QASWSLDDLPRTVAMSLHQGSSVTSMDFHPSHQTLLL-VGSNNGEISLWELGMRERLVSK 386
A + DDLP+TVA +L QGSS SMDFHP QTLLL V +I LWE+G RERLV K
Sbjct: 357 PAFKAPDDLPKTVARTLSQGSSPMSMDFHPIKQTLLLGVFLALCDIGLWEVGSRERLVQK 416
Query: 387 PFKIWDISACSLPFQAAVVKD-TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQ 445
FK+WD+S CS+P QAA+VK+ SV+RV WS DGS GVA+++H++ +Y+Y+G ++ Q
Sbjct: 417 TFKVWDLSKCSMPLQAALVKEPVVSVNRVIWSPDGSLFGVAYSRHIVQLYSYHGGEDMRQ 476
Query: 446 RVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWD-LTGRRLFNFEGHEAPVYSICP 504
+EIDAH+GGVND++F+ PNKQLCV+TCGDDK IKVWD TG + FEGHEAPVYS+CP
Sbjct: 477 HLEIDAHVGGVNDISFSTPNKQLCVITCGDDKTIKVWDAATGVKRHTFEGHEAPVYSVCP 536
Query: 505 HHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDG 564
H+KENIQFIFSTA+DGKIKAWLYDNMGSRVDYDAPG WCTTM YSADGTRLFSCGTSKDG
Sbjct: 537 HYKENIQFIFSTALDGKIKAWLYDNMGSRVDYDAPGRWCTTMAYSADGTRLFSCGTSKDG 596
Query: 565 DSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTS 624
+SF+VEWNESEGA+KRTY GF K+S GVVQFDTT+NR+LAAG+D IKFWDMD V LT+
Sbjct: 597 ESFIVEWNESEGAVKRTYQGFHKRSLGVVQFDTTKNRYLAAGDDFSIKFWDMDAVQLLTA 656
Query: 625 TEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKILANAGGLRSLRTVETPAFEALRSPIES 684
+ +GGLQ P +RFNKEG+LLAV+ +N KI+AN+ GLR L T E + E+ + I S
Sbjct: 657 IDGDGGLQASPRIRFNKEGSLLAVSGNENVIKIMANSDGLRLLHTFENISSESSKPAINS 716
Query: 685 AANKA 689
A A
Sbjct: 717 IAAAA 721
>UniRef100_Q9LVZ2 Similarity to unknown protein [Arabidopsis thaliana]
Length = 1128
Score = 880 bits (2275), Expect = 0.0
Identities = 452/705 (64%), Positives = 548/705 (77%), Gaps = 33/705 (4%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFK++VH+LEKESGFFFNM+YFE+ V AGEW++VEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKDTVHRLEKESGFFFNMRYFEDSVTAGEWDDVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALD++D KAV+ILV +LKVFSTFNEEL+KEIT LLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDKKDHAKAVDILVKELKVFSTFNEELFKEITMLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
LTNFRENEQLSKYGDTK+ARGIML ELKKLIEANPLFRDKL FP+LK+SRLRTLINQSLN
Sbjct: 121 LTNFRENEQLSKYGDTKSARGIMLGELKKLIEANPLFRDKLQFPSLKNSRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTP-VNLPVAAVAKPAAYTSLGVGAHGP 239
WQHQLCKNPRPNPDIKTLF+DH+C NG P+P N + +V K + L GAHGP
Sbjct: 181 WQHQLCKNPRPNPDIKTLFVDHTCGHPNGAHTPSPTTNHLMGSVPKVGGFPPL--GAHGP 238
Query: 240 FPPAAATANANALAGWMANASVSSSVQAAVVTASTIPV--PHNQVSILK--RPITPSTTP 295
F P A +LAGWM N SVQ V+A I + P++ VS+LK RP +P T
Sbjct: 239 FQPTPAPL-TTSLAGWMPN----PSVQHPTVSAGPIGLGAPNSAVSMLKRERPRSPPTNS 293
Query: 296 GMVEYQSADHEQLMKRLRP----------APSVEEVSYP--SARQASWSLDDLPRTVAMS 343
++YQ+AD E ++KR RP +V V+YP S A++S DDLP+ V+
Sbjct: 294 LSMDYQTADSESVLKRPRPFGISDGVNNLPVNVLPVTYPGQSHAHATYSTDDLPKNVSRI 353
Query: 344 LHQGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAA 403
L QGS++ SMDFHP QT+LLVG+N G+I++WE+G RE+LVS+ FK+WD++ C++ QA+
Sbjct: 354 LSQGSAIKSMDFHPVQQTMLLVGTNLGDIAIWEVGSREKLVSRSFKVWDLATCTVNLQAS 413
Query: 404 VVKD-TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFA 462
+ + T +V+RV WS DG +GVA++KH++HIY+Y+G +L +EIDAH G VNDLAF+
Sbjct: 414 LASEYTAAVNRVVWSPDGGLLGVAYSKHIVHIYSYHGGEDLRNHLEIDAHAGNVNDLAFS 473
Query: 463 HPNKQLCVVTCGDDKLIKVWD-LTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGK 521
PN+QLCVVTCG+DK IKVWD +TG +L FEGHEAPVYS+CPH KENIQFIFSTAVDGK
Sbjct: 474 QPNQQLCVVTCGEDKTIKVWDAVTGNKLHTFEGHEAPVYSVCPHQKENIQFIFSTAVDGK 533
Query: 522 IKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRT 581
IKAWLYDNMGSRVDYDAPG CT+M Y AD GTSK+G+SF+VEWNESEGA+KRT
Sbjct: 534 IKAWLYDNMGSRVDYDAPGRSCTSMAYCAD-------GTSKEGESFIVEWNESEGAVKRT 586
Query: 582 YNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNK 641
Y G K+S GVVQFDT +N+FL AG++ Q+KFWDMD+V+ L+ST AEGGL P LR NK
Sbjct: 587 YLGLGKRSVGVVQFDTMKNKFLVAGDEFQVKFWDMDSVDLLSSTAAEGGLPSSPCLRINK 646
Query: 642 EGNLLAVTTADNGFKILANAGGLRSLRTVETPAFEALRSPIESAA 686
EG LLAV+T DNG KILANA G R L ++ ++ R+P S A
Sbjct: 647 EGTLLAVSTTDNGIKILANAEGSRILHSMANRGLDSSRAPPGSVA 691
Score = 35.4 bits (80), Expect = 9.8
Identities = 30/125 (24%), Positives = 53/125 (42%), Gaps = 6/125 (4%)
Query: 493 EGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADG 552
EG++ V C +N ++ S A GKI + + + AP T++ +
Sbjct: 834 EGNKEDVVP-CFALSKNDSYVMS-ASGGKISLFNMMTFKTMTTFMAPPPAATSLAFHPQD 891
Query: 553 TRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIK 612
+ + G DS + +N +K G +K+ G+ F N +++G DSQ+
Sbjct: 892 NNIIAIGMD---DSSIQIYNVRVDEVKSKLKGHQKRVTGLA-FSNVLNVLVSSGADSQLC 947
Query: 613 FWDMD 617
W MD
Sbjct: 948 VWSMD 952
>UniRef100_Q9S9U5 F15P11.3 protein [Arabidopsis thaliana]
Length = 892
Score = 485 bits (1248), Expect = e-135
Identities = 272/405 (67%), Positives = 294/405 (72%), Gaps = 61/405 (15%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFLEEEKFKESVH+LEKESGFFFN KYF+EKV AGEW++VE YLSGF
Sbjct: 1 MSSLSRELVFLILQFLEEEKFKESVHRLEKESGFFFNTKYFDEKVLAGEWDDVETYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDR-------------QDKPKAVEILVGDLKVFSTF 107
TKVDDNRYSMKIFFEIRKQKYLEALDR Q+K KAVEILV DL+VFSTF
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRFVVLEFDVLCCYRQEKAKAVEILVQDLRVFSTF 120
Query: 108 NEELYKEITQLLTLTNFR---------------------------ENEQLSKYGDTKTAR 140
NEELYKEITQLLTL NFR ENEQLSKYGDTKTAR
Sbjct: 121 NEELYKEITQLLTLQNFRNMRLGLGSVQVEFVMLCRLFLFGCERGENEQLSKYGDTKTAR 180
Query: 141 GIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLNWQHQLCKNPRPNPDIKTLFI 200
GIML ELKKLIEANPLFRDKL+FPTL+SSRLRTLINQSLNWQHQLCKNPRPNPDIKTLF
Sbjct: 181 GIMLGELKKLIEANPLFRDKLMFPTLRSSRLRTLINQSLNWQHQLCKNPRPNPDIKTLFT 240
Query: 201 DHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLG---VGAHGPFPPAAATANANALAGWMA 257
DH+CT NGPLAP+ VN PV + KPAAY SLG V PFPP A ANA ALA WMA
Sbjct: 241 DHTCTLPNGPLAPSAVNQPVTTLTKPAAYPSLGPHVVRNLDPFPPGPAAANAGALASWMA 300
Query: 258 NASVSSSVQAAVVTASTIPVPHNQ------------VSILKRPITPSTTPGMVEYQSADH 305
AS +S+VQAAVVT + +P P NQ VSILKRP TP TPG+V+YQ+ DH
Sbjct: 301 AASGASAVQAAVVTPALMPQPQNQISIKNLLFFFQKVSILKRPRTPPATPGIVDYQNPDH 360
Query: 306 EQLMKRLRPAPSVEEVSYP-----SARQASWSLDDLPRTVAMSLH 345
E LMKRLRPAPSVEE S + +WS D VA + H
Sbjct: 361 E-LMKRLRPAPSVEEALIAKETPISVTRVAWSPDGNFIGVAFTKH 404
Score = 452 bits (1162), Expect = e-125
Identities = 221/329 (67%), Positives = 255/329 (77%), Gaps = 43/329 (13%)
Query: 401 QAAVVKDTP-SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDL 459
+A + K+TP SV+RV WS DG+F+GVAFTKHLI +YA++G N+L Q EIDAH+G VNDL
Sbjct: 374 EALIAKETPISVTRVAWSPDGNFIGVAFTKHLIQLYAFSGPNDLRQHTEIDAHVGAVNDL 433
Query: 460 AFAHPNKQLCVVTCGDDKLIK---------VWDLTGRRLFNFEGHEAPVYSICPHHKENI 510
AFA+PN+QLCV+TCGDDKLIK VWD++GR+ F FEGH+APVYSICPH+KENI
Sbjct: 434 AFANPNRQLCVITCGDDKLIKLANLINVEQVWDVSGRKHFTFEGHDAPVYSICPHYKENI 493
Query: 511 Q---------------------------------FIFSTAVDGKIKAWLYDNMGSRVDYD 537
Q FIFSTA+DGKIKAWLYDN+GSRVDYD
Sbjct: 494 QVRQGLNQTIFYINYYTVLPIRLKNNKVDDTFHIFIFSTAIDGKIKAWLYDNLGSRVDYD 553
Query: 538 APGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDT 597
APG WCT MLYSADGTRLFSCGTSKDGDSFLVEWNESEG+IKRTY F+KK AGVVQFDT
Sbjct: 554 APGKWCTRMLYSADGTRLFSCGTSKDGDSFLVEWNESEGSIKRTYKEFQKKLAGVVQFDT 613
Query: 598 TQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKI 657
++N FLA GED QIKFWDM+N+N LTST+AEGGL LPHLRFNK+GNLLAVTTADNGFKI
Sbjct: 614 SKNHFLAVGEDGQIKFWDMNNINVLTSTDAEGGLPALPHLRFNKDGNLLAVTTADNGFKI 673
Query: 658 LANAGGLRSLRTVETPAFEALRSPIESAA 686
LAN G RSLR +ETPA E +R+P++ A
Sbjct: 674 LANPAGFRSLRAMETPASETMRTPVDFKA 702
>UniRef100_Q9M8L7 Hypothetical protein T21F11.18 [Arabidopsis thaliana]
Length = 1073
Score = 421 bits (1083), Expect = e-116
Identities = 230/351 (65%), Positives = 261/351 (73%), Gaps = 12/351 (3%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKE+VHKLE+ESGFFFNMKYFE++V G W+EVEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKETVHKLEQESGFFFNMKYFEDEVHNGNWDEVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALDR D+PKAV+ILV DLKVFSTFNEEL+KEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRHDRPKAVDILVKDLKVFSTFNEELFKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTK+AR IML+ELKKLIEANPLFRDKL FPTL++SRLRTLINQSLN
Sbjct: 121 LENFRENEQLSKYGDTKSARAIMLVELKKLIEANPLFRDKLQFPTLRTSRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLGVGAHGPF 240
WQHQLCKNPRPNPDIKTLF+DHSC N AP+PVN P+ + P A +GAHGPF
Sbjct: 181 WQHQLCKNPRPNPDIKTLFVDHSCRLPNDARAPSPVNNPLLG-SLPKAEGFPPLGAHGPF 239
Query: 241 PPAAATANANALAGWMANASVSSSVQAAVVTASTIPVPHNQV-SILKRPITPSTTPGMVE 299
P + LAGWM S SSV V+ I + + + LK P TP + V+
Sbjct: 240 QPTPSPV-PTPLAGWM---SSPSSVPHPAVSGGPIALGAPSIQAALKHPRTPPSN-SAVD 294
Query: 300 YQSADHEQLMKRLRPAPSVEEVS-----YPSARQASWSLDDLPRTVAMSLH 345
Y S D + + KR RP +E + S + WS D VA S H
Sbjct: 295 YPSGDSDHVSKRTRPMGISDEAALVKEPVVSVNRVIWSPDGSLFGVAYSRH 345
Score = 418 bits (1074), Expect = e-115
Identities = 195/286 (68%), Positives = 238/286 (83%), Gaps = 2/286 (0%)
Query: 401 QAAVVKD-TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDL 459
+AA+VK+ SV+RV WS DGS GVA+++H++ +Y+Y+G ++ Q +EIDAH+GGVND+
Sbjct: 315 EAALVKEPVVSVNRVIWSPDGSLFGVAYSRHIVQLYSYHGGEDMRQHLEIDAHVGGVNDI 374
Query: 460 AFAHPNKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAV 518
AF+ PNKQLCV TCGDDK IKVWD TG + + FEGHEAPVYSICPH+KENIQFIFSTA+
Sbjct: 375 AFSTPNKQLCVTTCGDDKTIKVWDAATGVKRYTFEGHEAPVYSICPHYKENIQFIFSTAL 434
Query: 519 DGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAI 578
DGKIKAWLYDNMGSRVDY+APG WCTTM YSADGTRLFSCGTSKDG+SF+VEWNESEGA+
Sbjct: 435 DGKIKAWLYDNMGSRVDYEAPGRWCTTMAYSADGTRLFSCGTSKDGESFIVEWNESEGAV 494
Query: 579 KRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLR 638
KRTY GF K+S GVVQFDTT+NR+LAAG+D IKFWDMD + LT+ +A+GGLQ P +R
Sbjct: 495 KRTYQGFHKRSLGVVQFDTTKNRYLAAGDDFSIKFWDMDTIQLLTAIDADGGLQASPRIR 554
Query: 639 FNKEGNLLAVTTADNGFKILANAGGLRSLRTVETPAFEALRSPIES 684
FNKEG+LLAV+ DN K++AN+ GLR L TVE + E+ + I S
Sbjct: 555 FNKEGSLLAVSANDNMIKVMANSDGLRLLHTVENLSSESSKPAINS 600
>UniRef100_Q9SDK3 EST AU078118(E3904) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 930
Score = 399 bits (1025), Expect = e-109
Identities = 204/255 (80%), Positives = 222/255 (87%), Gaps = 15/255 (5%)
Query: 1 MTSLSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGF 60
M+SLSRELVFLILQFL+EEKFKE+VHKLE+ESGFFFNMKYFEEKV AGEW+EVEKYLSGF
Sbjct: 1 MSSLSRELVFLILQFLDEEKFKETVHKLEQESGFFFNMKYFEEKVHAGEWDEVEKYLSGF 60
Query: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLT 120
TKVDDNRYSMKIFFEIRKQKYLEALDR D+ KAV+ILV DLKVFSTFNEELYKEITQLLT
Sbjct: 61 TKVDDNRYSMKIFFEIRKQKYLEALDRHDRAKAVDILVKDLKVFSTFNEELYKEITQLLT 120
Query: 121 LTNFRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLN 180
L NFRENEQLSKYGDTK+AR IML+ELKKLIEANPLFR+KLVFPTLK+SRLRTLINQSLN
Sbjct: 121 LENFRENEQLSKYGDTKSARSIMLIELKKLIEANPLFREKLVFPTLKASRLRTLINQSLN 180
Query: 181 WQHQLCKNPRPNPDIKTLFIDHSCTPSNGPLAPTPVNLPVAAVAKPAAYTSLGVGAHGPF 240
WQHQLCKNPRPNPDIKTLF DH+CTP NG A +PV++P+AAV K A G +
Sbjct: 181 WQHQLCKNPRPNPDIKTLFTDHTCTPPNGARA-SPVSVPLAAVPK----------AGGTY 229
Query: 241 PPAAATANANALAGW 255
PP A+ GW
Sbjct: 230 PP----LTAHTRVGW 240
Score = 365 bits (936), Expect = 5e-99
Identities = 187/281 (66%), Positives = 213/281 (75%), Gaps = 39/281 (13%)
Query: 421 SFV--GVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKL 478
SFV GVAF KHLIH++AY NE Q +
Sbjct: 268 SFVVSGVAFAKHLIHLHAYQQPNETRQ--------------------------------V 295
Query: 479 IKVWDLTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDA 538
++VWD+ G++LF+FEGHEAPVYSICPHHKE+IQFIFST++DGKIKAWLYD+MGSRVDYDA
Sbjct: 296 LEVWDMHGQKLFSFEGHEAPVYSICPHHKESIQFIFSTSLDGKIKAWLYDHMGSRVDYDA 355
Query: 539 PGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSA--GVVQFD 596
PG WCTTMLYSADGTRLFSCGTSKDGDS+LVEWNESEG+IKRTY+GFRKKSA GVVQFD
Sbjct: 356 PGKWCTTMLYSADGTRLFSCGTSKDGDSYLVEWNESEGSIKRTYSGFRKKSAGVGVVQFD 415
Query: 597 TTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFK 656
T QN LAAGED+QIKFWD+DN L+STEA+GGL GLP LRFNKEGNLLAVTT DNGFK
Sbjct: 416 TAQNHILAAGEDNQIKFWDVDNTTMLSSTEADGGLPGLPRLRFNKEGNLLAVTTVDNGFK 475
Query: 657 ILANAGGLRSLRTVETPAFEALRSPIESAANKAVKWKGAPL 697
ILANA GLR+LR FEA RS E++ ++K GAP+
Sbjct: 476 ILANADGLRTLRAFGNRPFEAFRSQYEAS---SMKVSGAPV 513
>UniRef100_Q5UFQ9 WD-40 repeat protein [Malus domestica]
Length = 182
Score = 271 bits (694), Expect = 6e-71
Identities = 123/179 (68%), Positives = 150/179 (83%), Gaps = 1/179 (0%)
Query: 454 GGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICPHHKENIQF 512
G VNDLAF +P K+LC +TCGDDK IKVW+ T G +L+ FEGH+A V+S+CPH+KEN+ F
Sbjct: 2 GSVNDLAFCNPAKKLCAITCGDDKAIKVWNATTGSKLYTFEGHDAAVHSVCPHNKENVHF 61
Query: 513 IFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWN 572
FST+V+GKIKAWLYDNMG RVDYDAPGH TTM YSADG RLFSCGTSKDG+S +VEWN
Sbjct: 62 FFSTSVNGKIKAWLYDNMGYRVDYDAPGHSITTMAYSADGKRLFSCGTSKDGESHVVEWN 121
Query: 573 ESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGL 631
E++G IKR Y GF+K S+GVVQFDT++N++LA G+D IK WDMD +N LT+ +AEGGL
Sbjct: 122 ENDGVIKRNYVGFQKPSSGVVQFDTSENKYLAVGDDYAIKVWDMDKINILTTIDAEGGL 180
>UniRef100_Q9SKK7 Hypothetical protein At2g25420 [Arabidopsis thaliana]
Length = 730
Score = 99.0 bits (245), Expect = 7e-19
Identities = 57/171 (33%), Positives = 90/171 (52%), Gaps = 18/171 (10%)
Query: 6 RELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGFTKVDD 65
+ LVFLILQF +EE ++ES+H LE++SG FF+ Y + G W++ + YLS FT +
Sbjct: 9 KNLVFLILQFFDEEGYEESLHLLEQDSGVFFDFSYLSNAILNGNWKDADDYLSAFTSPEA 68
Query: 66 NRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLTLTNFR 125
N +S K+FF L + +AV+I DL+ ++ + ++ +++ T
Sbjct: 69 NTFSRKMFF---------GLFKSGGSEAVKIFSKDLRRIPVLKDDSFDDLVEVIAET--- 116
Query: 126 ENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLIN 176
D R + ++L KL E+NP R KL FP+L S L LI+
Sbjct: 117 ------CCVDKAPGRAKLCVDLHKLAESNPSLRGKLDFPSLNKSALLALIS 161
Score = 92.0 bits (227), Expect = 9e-17
Identities = 55/178 (30%), Positives = 92/178 (50%), Gaps = 38/178 (21%)
Query: 4 LSRELVFLILQFLEEEKFKESVHKLEKESGFFFNMKYFEEKVQAGEWEEVEKYLSGFTKV 63
L +L+ LILQFL E K+K ++HKLE+E+ FFN+ Y E ++ GE+ + E+YL FT
Sbjct: 174 LKEDLICLILQFLYEAKYKNTLHKLEQETKVFFNLNYLAEVMKLGEYGKAEEYLGAFTNW 233
Query: 64 DDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFSTFNEELYKEITQLLTLTN 123
+DN+YS +F E++K L++ + ++ T +
Sbjct: 234 EDNKYSKAMFLELQKLICLQSTE-----------------------------WEVATPSG 264
Query: 124 FRENEQLSKYGDTKTARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLRTLINQSLNW 181
+N L K + +L K NP+ +D+L FP+++ SRL TL+ Q+++W
Sbjct: 265 SLDNMSLK----IKLHASVAMLAKK-----NPVLKDELKFPSMEKSRLLTLVKQTMDW 313
>UniRef100_UPI0000235C95 UPI0000235C95 UniRef100 entry
Length = 1364
Score = 78.2 bits (191), Expect = 1e-12
Identities = 105/408 (25%), Positives = 164/408 (39%), Gaps = 67/408 (16%)
Query: 292 STTPGMVEYQSADHEQLMKRLRPAPSVEEVSYPSARQ-ASWSLDDL-----PRT----VA 341
S P + + SA+ + L L P SV V P +Q S S DD P T
Sbjct: 730 SQLPKVEQTWSAEQQTLENHLGPVESV--VFSPDGKQLVSGSYDDTVKIWDPATGELLQT 787
Query: 342 MSLHQGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVS---------------- 385
+ H G+ V S+ F P + LL GS + I LW+ E L +
Sbjct: 788 LDGHSGT-VESLAFSPDGK-LLASGSYDNTIDLWDSATGELLQTFEGHPHSIWSVAFAPD 845
Query: 386 ----------KPFKIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIY 435
KIWD++ L Q + + SV V +S DG + + I ++
Sbjct: 846 GKELASASDDSTIKIWDLATGEL--QQTLDSHSQSVRSVAFSPDGKLLASSSLDSTIKVW 903
Query: 436 AYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDK-LIKVWD-LTGRRLFNFE 493
+ EL Q +E G V +AF+ K+L G +K +K+W+ TG L E
Sbjct: 904 N-PATGELQQSLE--GRSGWVKSVAFSPDGKKLA---SGSEKNTVKLWNPATGELLQTLE 957
Query: 494 GHEAPVYSIC--PHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGH--WCTTMLYS 549
GH V S+ P K+ + S++ D IK W ++ + GH W + +S
Sbjct: 958 GHSQSVRSVAFSPDGKQ----LASSSSDTTIKLW--NSTTGELQQTFKGHDLWIRAVAFS 1011
Query: 550 ADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDS 609
DG L S D+ + W+ + ++++ +S V F + ++ DS
Sbjct: 1012 PDGKHL----VSGSDDNTIKLWDLATSELQQSLED-HSRSVHAVAFSPDDKQLASSSLDS 1066
Query: 610 QIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKI 657
IK WD + EG QG+ + F+ +G LLA + D K+
Sbjct: 1067 TIKLWD--SATGELQRTLEGHSQGVRSVTFSPDGKLLASNSYDGTIKL 1112
>UniRef100_Q00808 Vegetatible incompatibility protein HET-E-1 [Podospora anserina]
Length = 1356
Score = 77.8 bits (190), Expect = 2e-12
Identities = 87/323 (26%), Positives = 131/323 (39%), Gaps = 44/323 (13%)
Query: 347 GSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISA--CSLPFQAAV 404
GSSV S+ F P Q V S +G+ K KIWD ++ C+ +
Sbjct: 967 GSSVLSVAFSPDGQR---VASGSGD--------------KTIKIWDTASGTCTQTLEG-- 1007
Query: 405 VKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHP 464
SV V +S DG V I I+ S Q +E H G V + F+
Sbjct: 1008 --HGGSVWSVAFSPDGQRVASGSDDKTIKIWD-TASGTCTQTLE--GHGGWVQSVVFSPD 1062
Query: 465 NKQLCVVTCGDDKLIKVWD-LTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIK 523
++ V + DD IK+WD ++G EGH V+S+ + Q + S ++DG IK
Sbjct: 1063 GQR--VASGSDDHTIKIWDAVSGTCTQTLEGHGDSVWSVA--FSPDGQRVASGSIDGTIK 1118
Query: 524 AWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYN 583
W + + G W ++ +S DG R+ S D + W+ + G +T
Sbjct: 1119 IWDAASGTCTQTLEGHGGWVHSVAFSPDGQRVASGSI----DGTIKIWDAASGTCTQTLE 1174
Query: 584 GFRKKSAGVVQ---FDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFN 640
G G VQ F R + D IK WD + + E GG + + F+
Sbjct: 1175 GH----GGWVQSVAFSPDGQRVASGSSDKTIKIWDTASGTCTQTLEGHGG--WVQSVAFS 1228
Query: 641 KEGNLLAVTTADNGFKILANAGG 663
+G +A ++DN KI A G
Sbjct: 1229 PDGQRVASGSSDNTIKIWDTASG 1251
Score = 62.4 bits (150), Expect = 7e-08
Identities = 67/249 (26%), Positives = 102/249 (40%), Gaps = 15/249 (6%)
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
SV V +S DG V I I+ S Q +E H G V +AF+ ++
Sbjct: 843 SVLSVAFSADGQRVASGSDDKTIKIWD-TASGTGTQTLE--GHGGSVWSVAFSPDRER-- 897
Query: 470 VVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYD 528
V + DDK IK+WD +G EGH V S+ + Q + S + D IK W
Sbjct: 898 VASGSDDKTIKIWDAASGTCTQTLEGHGGRVQSVA--FSPDGQRVASGSDDHTIKIWDAA 955
Query: 529 NMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKK 588
+ + G ++ +S DG R+ S GD + W+ + G +T G
Sbjct: 956 SGTCTQTLEGHGSSVLSVAFSPDGQRV----ASGSGDKTIKIWDTASGTCTQTLEG-HGG 1010
Query: 589 SAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAV 648
S V F R + +D IK WD + + E GG + + F+ +G +A
Sbjct: 1011 SVWSVAFSPDGQRVASGSDDKTIKIWDTASGTCTQTLEGHGG--WVQSVVFSPDGQRVAS 1068
Query: 649 TTADNGFKI 657
+ D+ KI
Sbjct: 1069 GSDDHTIKI 1077
Score = 39.3 bits (90), Expect = 0.68
Identities = 31/119 (26%), Positives = 51/119 (42%), Gaps = 7/119 (5%)
Query: 545 TMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLA 604
++ +SADG R+ S D + W+ + G +T G S V F + R +
Sbjct: 846 SVAFSADGQRV----ASGSDDKTIKIWDTASGTGTQTLEG-HGGSVWSVAFSPDRERVAS 900
Query: 605 AGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKILANAGG 663
+D IK WD + + E GG + + F+ +G +A + D+ KI A G
Sbjct: 901 GSDDKTIKIWDAASGTCTQTLEGHGG--RVQSVAFSPDGQRVASGSDDHTIKIWDAASG 957
>UniRef100_Q86AZ8 Similar to Anabaena sp. (Strain PCC 7120). Hypothetical WD-repeat
protein alr2800 [Dictyostelium discoideum]
Length = 730
Score = 73.2 bits (178), Expect = 4e-11
Identities = 55/213 (25%), Positives = 105/213 (48%), Gaps = 17/213 (7%)
Query: 4 LSRELVFLILQFLEEEKFKESVHKLEKESGFFFN--------MKYFEEKVQAGEWEEVEK 55
+ +++ +I+Q+L++E + S+ ++ E+ FN MK ++ + G+W EVEK
Sbjct: 153 IKEDIIRMIIQYLQDEGYTASLLTVQDEANVKFNEHVYKITQMKKMKKAILEGDWNEVEK 212
Query: 56 YLSGFTKVDDNRYSMKIFFEIRKQKYLEALDRQDKPKAVEILVGDLKVFS--TFNEELYK 113
T + + + + KQ YLE L++Q+ KA L LK N + +K
Sbjct: 213 ----LTAKNTFKNHQSFLYAVYKQAYLELLEKQEYQKAFSYLTKRLKPLEGRQNNADEFK 268
Query: 114 EITQLLTLTNFRENEQLSKYGDTK-TARGIMLLELKKLIEANPLFRDKLVFPTLKSSRLR 172
++ LLT + +E + +K T+R ++ + + ++E +F + RL
Sbjct: 269 DLCYLLTCRSVQELNSFKSWDGSKGTSREKLVEQFQSMLELENTKSTGALF-RVPPHRLI 327
Query: 173 TLINQSLNWQHQLCK-NPRPNPDIKTLFIDHSC 204
+L QS+ +Q + + +P+ P IKTL D+SC
Sbjct: 328 SLFKQSVAYQIEFGRYHPKVMPKIKTLLDDYSC 360
>UniRef100_Q8Z019 WD-40 repeat protein [Anabaena sp.]
Length = 1711
Score = 71.6 bits (174), Expect = 1e-10
Identities = 66/252 (26%), Positives = 103/252 (40%), Gaps = 55/252 (21%)
Query: 409 PSVSRVTWSLDGSFVGVAFTKHLIHIYAYNG--------------SNELAQRVEIDA--- 451
P V+ ++++ D V +A H IH+Y G S + +I A
Sbjct: 1435 PDVTSISFTPDNKIVALASPDHTIHLYNRQGGLLRSLPGHNHWITSLSFSPNKQILASGS 1494
Query: 452 --------------------HIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFN 491
H G V D+ F+ K +V+ DK IK+W L GR +
Sbjct: 1495 ADKTIKLWSVNGRLLKTLLGHNGWVTDIKFSADGKN--IVSASADKTIKIWSLDGRLIRT 1552
Query: 492 FEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGH--WCTTMLYS 549
+GH A V+S+ + + Q + ST+ D IK W N+ + Y GH + +S
Sbjct: 1553 LQGHSASVWSV--NLSPDGQTLASTSQDETIKLW---NLNGELIYTLRGHSDVVYNLSFS 1607
Query: 550 ADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGV--VQFDTTQNRFLAAGE 607
DG + S S DG L WN G + +T+ G R GV V F + G
Sbjct: 1608 PDGKTIAS--ASDDGTIKL--WNVPNGTLLKTFQGHR---GGVRSVSFSPDGKILASGGH 1660
Query: 608 DSQIKFWDMDNV 619
D+ +K W+++ +
Sbjct: 1661 DTTVKVWNLEGI 1672
Score = 57.8 bits (138), Expect = 2e-06
Identities = 60/256 (23%), Positives = 110/256 (42%), Gaps = 22/256 (8%)
Query: 411 VSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCV 470
V+ V++S DG + + IH++ +G + H GVN ++F+ P+ ++ +
Sbjct: 1109 VTSVSYSPDGEVIASGSVDNTIHLWRRDGK----LLTTLTGHNDGVNSVSFS-PDGEI-L 1162
Query: 471 VTCGDDKLIKVWDLTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNM 530
+ D IK+W G+ + +GH+ V S+ N + I S + D I W +
Sbjct: 1163 ASASADSTIKLWQRNGQLITTLKGHDQGVKSV--SFSPNGEIIASGSSDHTINLW---SR 1217
Query: 531 GSRVDYDAPGH--WCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKK 588
++ GH ++ +S +G + S S DG L W+ +G T K+
Sbjct: 1218 AGKLLLSLNGHSQGVNSIKFSPEGDTIAS--ASDDGTIRL--WS-LDGRPLITIPSHTKQ 1272
Query: 589 SAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAV 648
V F ++AG D+ +K W N T EG + + + F+ +G L+A
Sbjct: 1273 VLAVT-FSPDGQTIVSAGADNTVKLWSR---NGTLLTTLEGHNEAVWQVIFSPDGRLIAT 1328
Query: 649 TTADNGFKILANAGGL 664
+AD + + G +
Sbjct: 1329 ASADKTITLWSRDGNI 1344
Score = 56.2 bits (134), Expect = 5e-06
Identities = 70/321 (21%), Positives = 132/321 (40%), Gaps = 43/321 (13%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
V S+ F P+ + ++ GS++ I+LW + L ++ +
Sbjct: 1191 VKSVSFSPNGE-IIASGSSDHTINLWSRAGKLLL-------------------SLNGHSQ 1230
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
V+ + +S +G + A I +++ +G + I +H V + F+ P+ Q
Sbjct: 1231 GVNSIKFSPEGDTIASASDDGTIRLWSLDGR----PLITIPSHTKQVLAVTFS-PDGQT- 1284
Query: 470 VVTCGDDKLIKVWDLTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDN 529
+V+ G D +K+W G L EGH V+ + + + I + + D I W D
Sbjct: 1285 IVSAGADNTVKLWSRNGTLLTTLEGHNEAVWQVI--FSPDGRLIATASADKTITLWSRDG 1342
Query: 530 --MGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRK 587
+G+ + H ++ +S DG L S D+ + W + + +T+ G K
Sbjct: 1343 NILGT---FAGHNHEVNSLSFSPDGNIL----ASGSDDNTVRLWTVNR-TLPKTFYG-HK 1393
Query: 588 KSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLA 647
S V+F + + DS +K W +D L T L + + F + ++A
Sbjct: 1394 GSVSYVRFSNDGKKITSLSTDSTMKTWSLD--GKLLQT-LSSPLPDVTSISFTPDNKIVA 1450
Query: 648 VTTADNGFKILANAGG-LRSL 667
+ + D+ + GG LRSL
Sbjct: 1451 LASPDHTIHLYNRQGGLLRSL 1471
>UniRef100_Q8YZ23 WD-40 repeat protein [Anabaena sp.]
Length = 934
Score = 69.3 bits (168), Expect = 6e-10
Identities = 84/374 (22%), Positives = 149/374 (39%), Gaps = 49/374 (13%)
Query: 303 ADHEQLMKRLRPAPSVEEVSYPSARQAS--WSLDDLPRTVAMSLHQGSSVTSMDFHPSHQ 360
A+H+ +++ + +P + ++ S + WSLD + V + +G S+ F P
Sbjct: 322 AEHDGMLESVSFSPDSKFIATASRDKTVKIWSLDGKKQLVVLREEKGEGFNSVAFSPDG- 380
Query: 361 TLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTPSVSRVTWSLDG 420
TL+ GS + +W + K +V V +S D
Sbjct: 381 TLMATGSWDNTAKIWSREGKRLHTLDGHK-------------------EAVLEVAFSPDS 421
Query: 421 SFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIK 480
+ A + + +++ G +L +E H VN + F+ P+ QL + T G D +K
Sbjct: 422 QLLATASWDNTVKLWSREG--KLLHTLE--GHKDKVNSITFS-PDGQL-IATVGWDNTMK 475
Query: 481 VWDLTGRRLFNFEGHEAPVYSI--CPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDA 538
+W+L G+ L F GH+ ++S+ P K+ I + + D +K W D +
Sbjct: 476 LWNLDGKELRTFRGHQDMIWSVSFSPDGKQ----IATASGDRTVKLWSLDGKELQT---L 528
Query: 539 PGH--WCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFD 596
GH ++ +S DG + + GD + WN ++ Y + V F
Sbjct: 529 RGHQNGVNSVTFSPDGKLI----ATASGDRTVKLWNSKGQELETLYG--HTDAVNSVAFS 582
Query: 597 TTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFK 656
AG D K W +++ N S G + L F+ G +A + D K
Sbjct: 583 PDGTSIATAGNDKTAKIWKLNSPN---SIIVRGHEDEVFDLVFSPNGKYIATASWDKTAK 639
Query: 657 ILANAGG-LRSLRT 669
+ + G L+ LRT
Sbjct: 640 LWSIVGDKLQELRT 653
Score = 64.3 bits (155), Expect = 2e-08
Identities = 67/284 (23%), Positives = 119/284 (41%), Gaps = 19/284 (6%)
Query: 386 KPFKIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQ 445
K KIW +++ P V V + +S +G ++ A +++ G ++L +
Sbjct: 595 KTAKIWKLNS---PNSIIVRGHEDEVFDLVFSPNGKYIATASWDKTAKLWSIVG-DKLQE 650
Query: 446 RVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFNFEGHEAPVYSICPH 505
+ H G VN L+F+ K + T DK K+W+L G GH+ V+S+ +
Sbjct: 651 LRTFNGHQGRVNKLSFSPDGKY--IATTSWDKTAKLWNLDGTLQKTLTGHKDTVWSV--N 706
Query: 506 HKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGD 565
+ Q I + + D +K W D + + ++S DG + + G K
Sbjct: 707 FSPDGQLIATASEDKTVKLWNRDGELLKT-LPRQSSVVNSAVFSPDGKLIATAGWDK--- 762
Query: 566 SFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTST 625
V+ +G +++T G V F +A D+ +K W++D T
Sbjct: 763 --TVKIWSIDGRLQKTLTG-HTSGINSVTFSPDGKLIASASWDNTVKIWNLDGKELRT-- 817
Query: 626 EAEGGLQGLPHLRFNKEGNLLAVTTADNGFKIL-ANAGGLRSLR 668
G + ++ F+ +G L+A + DN KI N LR+LR
Sbjct: 818 -LRGHKNVVHNVTFSPDGKLIATASGDNTVKIWNINGQELRTLR 860
Score = 49.7 bits (117), Expect = 5e-04
Identities = 55/233 (23%), Positives = 94/233 (39%), Gaps = 23/233 (9%)
Query: 386 KPFKIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQ 445
K K+W++ Q + +V V +S DG + A + ++ N EL +
Sbjct: 680 KTAKLWNLDGT---LQKTLTGHKDTVWSVNFSPDGQLIATASEDKTVKLW--NRDGELLK 734
Query: 446 RVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFNFEGHEAPVYSICPH 505
+ + + VN F+ K + T G DK +K+W + GR GH + + S+
Sbjct: 735 TLPRQSSV--VNSAVFSPDGK--LIATAGWDKTVKIWSIDGRLQKTLTGHTSGINSVT-- 788
Query: 506 HKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGH--WCTTMLYSADGTRLFSCGTSKD 563
+ + I S + D +K W D R GH + +S DG + +
Sbjct: 789 FSPDGKLIASASWDNTVKIWNLDGKELRT---LRGHKNVVHNVTFSPDGKLI----ATAS 841
Query: 564 GDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDM 616
GD+ + WN G RT G+ K + ++F + + LA G I W +
Sbjct: 842 GDNTVKIWN-INGQELRTLRGY-KDAVWSLRF-SLDGKTLATGSRYDIVVWHL 891
>UniRef100_UPI00003C1528 UPI00003C1528 UniRef100 entry
Length = 1116
Score = 68.2 bits (165), Expect = 1e-09
Identities = 72/302 (23%), Positives = 130/302 (42%), Gaps = 34/302 (11%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
V S+DFHP+ + LL G +G +++W + K F++ ++ + F
Sbjct: 18 VKSLDFHPT-EPWLLAGLYSGSVNIWNY--ETGAIVKTFEVTNVPVRCVKF--------- 65
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
++R W + GS + + YN +L + +AH + LA HP+
Sbjct: 66 -IARKNWFVAGS------DDFQLRAFNYNTHEKL---ISFEAHPDYIRCLA-VHPSGSY- 113
Query: 470 VVTCGDDKLIKVWDL--TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLY 527
V+T DD IK+WD R + FEGH + ++C + K++ F S+++D +K W
Sbjct: 114 VITGSDDMSIKMWDWDKNWRLVQTFEGHTHYIMNLCFNPKDSNSFA-SSSLDRTVKVWTL 172
Query: 528 DNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVE-WNESEGAIKRTYNGFR 586
+ + +DA + Y G + + GD V+ W+ + +T G
Sbjct: 173 GSSVANFTFDAHDKGVNYVEYYHGGEKPYMLTV---GDDRTVKVWDYLSKSCVQTLTGHT 229
Query: 587 KKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLL 646
+ + F ++ ED +K W N L ST + GL+ + + + K GN +
Sbjct: 230 SNVSFAI-FHPCLPLIISGSEDGTVKLWH-SNTYRLEST-LDYGLERVWCVAYKKSGNDV 286
Query: 647 AV 648
A+
Sbjct: 287 AI 288
>UniRef100_UPI00002363EC UPI00002363EC UniRef100 entry
Length = 2088
Score = 67.0 bits (162), Expect = 3e-09
Identities = 54/212 (25%), Positives = 92/212 (42%), Gaps = 17/212 (8%)
Query: 449 IDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHK 507
++ H V ++++H ++ L + DD+ +K+WD TG EGH V S+ H
Sbjct: 271 LEGHEAAVLSVSYSHDSRLLA--SASDDRTVKIWDTETGSLQHTLEGHSDLVRSVIFSHD 328
Query: 508 ENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGH--WCTTMLYSADGTRLFSCGTSKDGD 565
+ ++A D +K W D + + GH W ++++S D L S D
Sbjct: 329 SR---LLASASDSTVKIW--DTGTGSLQHTLEGHRDWVRSVIFSHDSQLL----ASASDD 379
Query: 566 SFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTST 625
S + W+ G+++ T G R V+ F +A +DS +K WD L T
Sbjct: 380 STVKIWDTGTGSLQHTLEGHRDWVRSVI-FSHDSQLLASASDDSTVKIWD-TGTGSLQHT 437
Query: 626 EAEGGLQGLPHLRFNKEGNLLAVTTADNGFKI 657
EG + + F+ + LLA + D +I
Sbjct: 438 -LEGHRDWVRSVIFSHDSRLLASASDDRTVRI 468
Score = 41.2 bits (95), Expect = 0.18
Identities = 43/195 (22%), Positives = 75/195 (38%), Gaps = 36/195 (18%)
Query: 358 SHQTLLLVGSNNGEISLWELG--------------MRERLVS------------KPFKIW 391
SH + LL +++ + +W+ G +R + S KIW
Sbjct: 326 SHDSRLLASASDSTVKIWDTGTGSLQHTLEGHRDWVRSVIFSHDSQLLASASDDSTVKIW 385
Query: 392 DISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDA 451
D SL +D V V +S D + A + I+ G+ L +E
Sbjct: 386 DTGTGSLQHTLEGHRDW--VRSVIFSHDSQLLASASDDSTVKIWD-TGTGSLQHTLE--G 440
Query: 452 HIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENI 510
H V + F+H ++ L + DD+ +++WD G EGH + V S+ H +
Sbjct: 441 HRDWVRSVIFSHDSRLL--ASASDDRTVRIWDTEKGSHKHTLEGHSSLVTSVSFSHDSRL 498
Query: 511 QFIFSTAVDGKIKAW 525
+ S + D ++ W
Sbjct: 499 --LASASNDQTVRIW 511
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.333 0.143 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,795,713,553
Number of Sequences: 2790947
Number of extensions: 74567905
Number of successful extensions: 310672
Number of sequences better than 10.0: 2060
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 2057
Number of HSP's that attempted gapping in prelim test: 303686
Number of HSP's gapped (non-prelim): 7302
length of query: 1115
length of database: 848,049,833
effective HSP length: 138
effective length of query: 977
effective length of database: 462,899,147
effective search space: 452252466619
effective search space used: 452252466619
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 80 (35.4 bits)
Medicago: description of AC144513.3


