

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.17 + phase: 0
(319 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
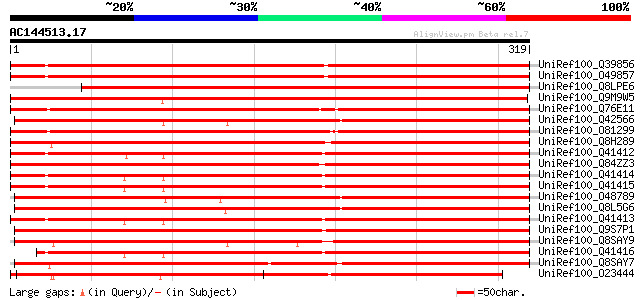
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q39856 Epoxide hydrolase [Glycine max] 561 e-159
UniRef100_O49857 Epoxide hydrolase [Glycine max] 560 e-158
UniRef100_Q8LPE6 Epoxide hydrolase [Cicer arietinum] 515 e-145
UniRef100_Q9M9W5 Putative epoxide hydrolase [Arabidopsis thaliana] 427 e-118
UniRef100_Q76E11 Soluble epoxide hydrolase [Citrus jambhiri] 413 e-114
UniRef100_Q42566 Putative epoxide hydrolase ATsEH [Arabidopsis t... 402 e-111
UniRef100_O81299 T14P8.15 protein [Arabidopsis thaliana] 399 e-110
UniRef100_Q8H289 Epoxide hydrolase [Ananas comosus] 394 e-108
UniRef100_Q41412 Epoxide hydrolase [Solanum tuberosum] 394 e-108
UniRef100_Q84ZZ3 Soluble epoxide hydrolase [Euphorbia lagascae] 392 e-108
UniRef100_Q41414 Epoxide hydrolase [Solanum tuberosum] 390 e-107
UniRef100_Q41415 Epoxide hydrolase [Solanum tuberosum] 389 e-107
UniRef100_O48789 Putative epoxide hydrolase [Arabidopsis thaliana] 388 e-106
UniRef100_Q8L5G6 Soluble epoxide hydrolase [Brassica napus] 380 e-104
UniRef100_Q41413 Epoxide hydrolase [Solanum tuberosum] 374 e-102
UniRef100_Q9S7P1 ESTs AU065732 [Oryza sativa] 353 3e-96
UniRef100_Q8SAY9 Putative hydrolase [Oryza sativa] 350 4e-95
UniRef100_Q41416 Epoxide hydrolase [Solanum tuberosum] 348 1e-94
UniRef100_Q8SAY7 Putative hydrolase [Oryza sativa] 347 2e-94
UniRef100_O23444 Putative epoxide hydrolase [Arabidopsis thaliana] 342 8e-93
>UniRef100_Q39856 Epoxide hydrolase [Glycine max]
Length = 341
Score = 561 bits (1446), Expect = e-159
Identities = 262/319 (82%), Positives = 290/319 (90%), Gaps = 3/319 (0%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+I+HRTVEVNGIKMH+AEKG EGPVVLFLHGFPELWYSWRHQI +L SLGYRAVAPDL
Sbjct: 26 MEQIKHRTVEVNGIKMHVAEKG-EGPVVLFLHGFPELWYSWRHQILSLSSLGYRAVAPDL 84
Query: 61 RGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIK 120
RGYGDT+ P SISSY CFH+VGD+V+LID LGV+QVFLV HD GAIIGWYLCMFRP+++K
Sbjct: 85 RGYGDTEAPPSISSYNCFHIVGDLVALIDSLGVQQVFLVAHDWGAIIGWYLCMFRPDKVK 144
Query: 121 AYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTT 180
AYVCLSVP L R+P IRTVDGMRA+YGDDYY+CRFQ+PGEMEAQMAEVGT YV+KNILTT
Sbjct: 145 AYVCLSVPLLRRDPNIRTVDGMRALYGDDYYVCRFQKPGEMEAQMAEVGTEYVLKNILTT 204
Query: 181 RKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
R GPPI PKG + FNP+ P+ LPSWLTE+DLAY+VSKFEKTGFTG LNYYRNFNLNW
Sbjct: 205 RNPGPPILPKGRF--QFNPEMPNTLPSWLTEEDLAYYVSKFEKTGFTGPLNYYRNFNLNW 262
Query: 241 ELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHFN 300
ELTAPWTG +IKVPVK+ITGELDMVY S NLKEYIHGGGFK+DVPNLE+VI+QKGVAHFN
Sbjct: 263 ELTAPWTGGQIKVPVKYITGELDMVYNSLNLKEYIHGGGFKQDVPNLEQVIVQKGVAHFN 322
Query: 301 NQEAEEEISNYIYEFIMKF 319
NQEA EEI NYIY+FI KF
Sbjct: 323 NQEAAEEIDNYIYDFINKF 341
>UniRef100_O49857 Epoxide hydrolase [Glycine max]
Length = 341
Score = 560 bits (1442), Expect = e-158
Identities = 261/319 (81%), Positives = 290/319 (90%), Gaps = 3/319 (0%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+I+HRTVEVNGIKMH+AEKG EGPVVLFLHGFPELWYSWRHQI +L SLGYRAVAPDL
Sbjct: 26 MEQIKHRTVEVNGIKMHVAEKG-EGPVVLFLHGFPELWYSWRHQILSLSSLGYRAVAPDL 84
Query: 61 RGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIK 120
RGYGDT+ P SISSY CFH+VGD+V+LID LGV+QVFLV HD GAIIGWYLCMFRP+++K
Sbjct: 85 RGYGDTEAPPSISSYNCFHIVGDLVALIDSLGVQQVFLVAHDWGAIIGWYLCMFRPDKVK 144
Query: 121 AYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTT 180
AYVCLSVP L R+P IRTVDGMRA+YGDDYY+CRFQ+PGEMEAQMAEVGT YV++NILTT
Sbjct: 145 AYVCLSVPLLRRDPNIRTVDGMRALYGDDYYVCRFQKPGEMEAQMAEVGTEYVLENILTT 204
Query: 181 RKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
R GPPI PKG + FNP+ P+ LPSWLTE+DLAY+VSKFEKTGFTG LNYYRNFNLNW
Sbjct: 205 RNPGPPILPKGRF--QFNPEMPNTLPSWLTEEDLAYYVSKFEKTGFTGPLNYYRNFNLNW 262
Query: 241 ELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHFN 300
ELTAPWTG +IKVPVK+ITGELDMVY S NLKEYIHGGGFK+DVPNLE+VI+QKGVAHFN
Sbjct: 263 ELTAPWTGGQIKVPVKYITGELDMVYNSLNLKEYIHGGGFKQDVPNLEQVIVQKGVAHFN 322
Query: 301 NQEAEEEISNYIYEFIMKF 319
NQEA EEI NYIY+FI KF
Sbjct: 323 NQEAAEEIDNYIYDFINKF 341
>UniRef100_Q8LPE6 Epoxide hydrolase [Cicer arietinum]
Length = 275
Score = 515 bits (1327), Expect = e-145
Identities = 242/275 (88%), Positives = 254/275 (92%)
Query: 45 IAALGSLGYRAVAPDLRGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMG 104
IAALGSLGYRAVAPDLRGYGDTD P S++SYT FH+VGD+V+LIDLLGVEQVFLV HD G
Sbjct: 1 IAALGSLGYRAVAPDLRGYGDTDAPGSVNSYTGFHIVGDLVALIDLLGVEQVFLVAHDWG 60
Query: 105 AIIGWYLCMFRPERIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQ 164
AIIGWY CMFRPERIKAYVCLSVP LHRNPKI+TVD MRA YGDDYYICRFQEPG+MEAQ
Sbjct: 61 AIIGWYFCMFRPERIKAYVCLSVPLLHRNPKIKTVDAMRAAYGDDYYICRFQEPGKMEAQ 120
Query: 165 MAEVGTTYVMKNILTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKT 224
MAEVGT YV+KNILTTRKTGPPI PKGEYGTGFNPDTPD LPSWLTE DLAYFVSKFEKT
Sbjct: 121 MAEVGTAYVLKNILTTRKTGPPILPKGEYGTGFNPDTPDTLPSWLTEADLAYFVSKFEKT 180
Query: 225 GFTGGLNYYRNFNLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDV 284
GFTGGLNYYRN NLNWEL APW G K+ VPVKFITGELDMVYTS N+KEYIHGGGFKEDV
Sbjct: 181 GFTGGLNYYRNLNLNWELMAPWRGAKVYVPVKFITGELDMVYTSLNMKEYIHGGGFKEDV 240
Query: 285 PNLEEVIIQKGVAHFNNQEAEEEISNYIYEFIMKF 319
PNLEEVI+QKGVAHFNNQEA EEISN+IYEFI KF
Sbjct: 241 PNLEEVIVQKGVAHFNNQEAAEEISNHIYEFIKKF 275
>UniRef100_Q9M9W5 Putative epoxide hydrolase [Arabidopsis thaliana]
Length = 331
Score = 427 bits (1099), Expect = e-118
Identities = 199/323 (61%), Positives = 250/323 (76%), Gaps = 5/323 (1%)
Query: 1 MERIEHRTVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPD 59
ME I+HR V VNGI MHIAEKG KEGPVVL LHGFP+LWY+WRHQI+ L SLGYRAVAPD
Sbjct: 1 MEGIDHRMVSVNGITMHIAEKGPKEGPVVLLLHGFPDLWYTWRHQISGLSSLGYRAVAPD 60
Query: 60 LRGYGDTDVPSSISSYTCFHVVGDIVSLIDLLG--VEQVFLVGHDMGAIIGWYLCMFRPE 117
LRGYGD+D P S S YTC +VVGD+V+L+D + E+VFLVGHD GAIIGW+LC+FRPE
Sbjct: 61 LRGYGDSDSPESFSEYTCLNVVGDLVALLDSVAGNQEKVFLVGHDWGAIIGWFLCLFRPE 120
Query: 118 RIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNI 177
+I +VCLSVP+ RNPK++ V G +AV+GDDYYICRFQEPG++E ++A ++N+
Sbjct: 121 KINGFVCLSVPYRSRNPKVKPVQGFKAVFGDDYYICRFQEPGKIEGEIASADPRIFLRNL 180
Query: 178 LTTRKTGPPIFPKGE-YGTGFNPDTPD-NLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRN 235
T R GPPI PK +G NP++ + LP W ++ DL ++VSKFEK GFTGGLNYYR
Sbjct: 181 FTGRTLGPPILPKDNPFGEKPNPNSENIELPEWFSKKDLDFYVSKFEKAGFTGGLNYYRA 240
Query: 236 FNLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKG 295
+LNWELTAPWTG KI+VPVKF+TG+ DMVYT+ +KEYIHGGGF DVP L+E+++ +
Sbjct: 241 MDLNWELTAPWTGAKIQVPVKFMTGDFDMVYTTPGMKEYIHGGGFAADVPTLQEIVVIED 300
Query: 296 VAHFNNQEAEEEISNYIYEFIMK 318
HF NQE +E++ +I +F K
Sbjct: 301 AGHFVNQEKPQEVTAHINDFFTK 323
>UniRef100_Q76E11 Soluble epoxide hydrolase [Citrus jambhiri]
Length = 316
Score = 413 bits (1062), Expect = e-114
Identities = 193/319 (60%), Positives = 241/319 (75%), Gaps = 3/319 (0%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+IEH TV NGI MH+A G GP VLF+HGFPELWYSWR+Q+ L S GYRA+APDL
Sbjct: 1 MEKIEHTTVATNGINMHVASIGT-GPAVLFIHGFPELWYSWRNQLLYLSSRGYRAIAPDL 59
Query: 61 RGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIK 120
RGYGDTD P S++SYT H+VGD++ L+D LG+ QVFLVGHD GA+I WY C+FRP+R+K
Sbjct: 60 RGYGDTDAPPSVTSYTALHLVGDLIGLLDKLGIHQVFLVGHDWGALIAWYFCLFRPDRVK 119
Query: 121 AYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTT 180
A V +SVPF RNP +R ++ RAVYGDDYYICRFQEPGE+E + A++ T +MK L
Sbjct: 120 ALVNMSVPFPPRNPAVRPLNNFRAVYGDDYYICRFQEPGEIEEEFAQIDTARLMKKFLCL 179
Query: 181 RKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
R P PK + G PD P LPSWL+E+D+ Y+ SKF + GFTG +NYYR ++LNW
Sbjct: 180 RIPKPLCIPK-DTGLSTLPD-PSALPSWLSEEDVNYYASKFNQKGFTGPVNYYRCWDLNW 237
Query: 241 ELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHFN 300
EL APWTGV+IKVPVK+I G+ D+VY + KEYIH GGFK+ VP L++V++ +GVAHF
Sbjct: 238 ELMAPWTGVQIKVPVKYIVGDQDLVYNNKGTKEYIHNGGFKKYVPYLQDVVVMEGVAHFI 297
Query: 301 NQEAEEEISNYIYEFIMKF 319
NQE EE+ +IYEFI KF
Sbjct: 298 NQEKAEEVGAHIYEFIKKF 316
>UniRef100_Q42566 Putative epoxide hydrolase ATsEH [Arabidopsis thaliana]
Length = 321
Score = 402 bits (1032), Expect = e-111
Identities = 198/322 (61%), Positives = 243/322 (74%), Gaps = 7/322 (2%)
Query: 4 IEHRTVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+EHR V NGI +H+A +G +GP+VL LHGFPELWYSWRHQI L + GYRAVAPDLRG
Sbjct: 1 MEHRKVRGNGIDIHVAIQGPSDGPIVLLLHGFPELWYSWRHQIPGLAARGYRAVAPDLRG 60
Query: 63 YGDTDVPSSISSYTCFHVVGDIVSLIDLLGV---EQVFLVGHDMGAIIGWYLCMFRPERI 119
YGD+D P+ ISSYTCF++VGD++++I L E+VF+VGHD GA+I WYLC+FRP+R+
Sbjct: 61 YGDSDAPAEISSYTCFNIVGDLIAVISALTASEDEKVFVVGHDWGALIAWYLCLFRPDRV 120
Query: 120 KAYVCLSVPFLHR--NPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNI 177
KA V LSVPF R +P ++ VD MRA YGDDYYICRFQE G++EA++AEVGT VMK +
Sbjct: 121 KALVNLSVPFSFRPTDPSVKPVDRMRAFYGDDYYICRFQEFGDVEAEIAEVGTERVMKRL 180
Query: 178 LTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFN 237
LT R GP I PK + G +T LPSWLTE+D+AYFVSKFE+ GF+G +NYYRNFN
Sbjct: 181 LTYRTPGPVIIPKDKSFWGSKGETIP-LPSWLTEEDVAYFVSKFEEKGFSGPVNYYRNFN 239
Query: 238 LNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVA 297
N EL PW G KI+VP KF+ GELD+VY +KEYIHG FKEDVP LEE ++ +GVA
Sbjct: 240 RNNELLGPWVGSKIQVPTKFVIGELDLVYYMPGVKEYIHGPQFKEDVPLLEEPVVMEGVA 299
Query: 298 HFNNQEAEEEISNYIYEFIMKF 319
HF NQE +EI I +FI KF
Sbjct: 300 HFINQEKPQEILQIILDFISKF 321
>UniRef100_O81299 T14P8.15 protein [Arabidopsis thaliana]
Length = 324
Score = 399 bits (1025), Expect = e-110
Identities = 184/319 (57%), Positives = 235/319 (72%), Gaps = 3/319 (0%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+IEH T+ NGI MH+A G GPV+LF+HGFP+LWYSWRHQ+ + +LGYRA+APDL
Sbjct: 1 MEKIEHTTISTNGINMHVASIGS-GPVILFVHGFPDLWYSWRHQLVSFAALGYRAIAPDL 59
Query: 61 RGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIK 120
RGYGD+D P S SYT H+VGD+V L+D LGV++VFLVGHD GAI+ W+LCM RP+R+
Sbjct: 60 RGYGDSDAPPSRESYTILHIVGDLVGLLDSLGVDRVFLVGHDWGAIVAWWLCMIRPDRVN 119
Query: 121 AYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTT 180
A V SV F RNP ++ VD RA++GDDYYICRFQEPGE+E A+V T ++ T+
Sbjct: 120 ALVNTSVVFNPRNPSVKPVDAFRALFGDDYYICRFQEPGEIEEDFAQVDTKKLITRFFTS 179
Query: 181 RKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
R PP PK G PD P +LP+WLTE D+ ++ KF + GFTGGLNYYR NL+W
Sbjct: 180 RNPRPPCIPKSVGFRGL-PD-PPSLPAWLTEQDVRFYGDKFSQKGFTGGLNYYRALNLSW 237
Query: 241 ELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHFN 300
ELTAPWTG++IKVPVKFI G+LD+ Y KEYIH GG K+ VP L+EV++ +GV HF
Sbjct: 238 ELTAPWTGLQIKVPVKFIVGDLDITYNIPGTKEYIHEGGLKKHVPFLQEVVVMEGVGHFL 297
Query: 301 NQEAEEEISNYIYEFIMKF 319
+QE +E++++IY F KF
Sbjct: 298 HQEKPDEVTDHIYGFFKKF 316
>UniRef100_Q8H289 Epoxide hydrolase [Ananas comosus]
Length = 318
Score = 394 bits (1012), Expect = e-108
Identities = 187/321 (58%), Positives = 234/321 (72%), Gaps = 5/321 (1%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKE--GPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAP 58
ME I HRTVE+NGI +H+AEKG + VL LHGFPELWYSWRHQI L + GYRA+AP
Sbjct: 1 MEGIVHRTVEINGIAVHVAEKGGDDAAAAVLLLHGFPELWYSWRHQIVGLAARGYRAIAP 60
Query: 59 DLRGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPER 118
DLRGYGDT P S++SYT FH+VGD+V+L+D L + QVF+VGHD GA I W LCM RP+R
Sbjct: 61 DLRGYGDTSAPPSVNSYTLFHLVGDVVALLDALELPQVFVVGHDWGAAIAWTLCMIRPDR 120
Query: 119 IKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNIL 178
+KA V SV + RNP + V ++ +YGD+ Y+CRFQEPG EA+ AEVGT V++ IL
Sbjct: 121 VKALVNTSVAHMPRNPSVSPVHQIKHLYGDNIYVCRFQEPGVAEAEFAEVGTKNVLRKIL 180
Query: 179 TTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNL 238
T R P ++G + LPSWL+E+DL Y+ SKFEKTGFTGG+NYYR NL
Sbjct: 181 TMRDPRPSSLTHKDWG---STGEEIALPSWLSEEDLDYYASKFEKTGFTGGMNYYRCMNL 237
Query: 239 NWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAH 298
NWELTAPW G KI+VP KFI G+LD+ Y N+++YIH GGFK +VP LEEV++ +GV H
Sbjct: 238 NWELTAPWAGAKIQVPTKFIVGDLDLTYHYPNIQDYIHKGGFKNEVPLLEEVVVLEGVGH 297
Query: 299 FNNQEAEEEISNYIYEFIMKF 319
F QE EE++++IY FI KF
Sbjct: 298 FIQQERAEEVTDHIYNFIKKF 318
>UniRef100_Q41412 Epoxide hydrolase [Solanum tuberosum]
Length = 321
Score = 394 bits (1012), Expect = e-108
Identities = 189/323 (58%), Positives = 241/323 (74%), Gaps = 6/323 (1%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+IEH+ V VNG+ MHIAE G +GP +LFLHGFPELWYSWRHQ+ L GYRAVAPDL
Sbjct: 1 MEKIEHKMVAVNGLNMHIAELG-QGPTILFLHGFPELWYSWRHQMVYLAECGYRAVAPDL 59
Query: 61 RGYGDTDVPS--SISSYTCFHVVGDIVSLIDLLGV--EQVFLVGHDMGAIIGWYLCMFRP 116
RGYGDT S S ++ H+VGD+V+L++ + E+VF+V HD GA+I W+LC+FRP
Sbjct: 60 RGYGDTTGASLNDPSKFSILHLVGDVVALLEAIAPNEEKVFVVAHDWGALIAWHLCLFRP 119
Query: 117 ERIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKN 176
+++KA V LSV FL RNPK+ TV+ ++A+YG+D+YI RFQ PGE+EA+ A +G ++K
Sbjct: 120 DKVKALVNLSVHFLPRNPKMNTVEWLKAIYGEDHYISRFQVPGEIEAEFAPIGAKSILKK 179
Query: 177 ILTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNF 236
ILT R P FPKG+ G PD P L SWL+E++L Y+ +KFE+TGFTGG+NYYR
Sbjct: 180 ILTYRDPAPFYFPKGK-GLEALPDAPVALSSWLSEEELDYYANKFEQTGFTGGVNYYRAL 238
Query: 237 NLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGV 296
++WELTAPWTG ++KVP KFI GE D+VY KEYIH GGFKEDVP LEEV++ +G
Sbjct: 239 PISWELTAPWTGAQVKVPTKFIVGEFDLVYHIPGAKEYIHNGGFKEDVPLLEEVVVLEGS 298
Query: 297 AHFNNQEAEEEISNYIYEFIMKF 319
AHF NQE EIS +IY+FI KF
Sbjct: 299 AHFVNQERPHEISKHIYDFIQKF 321
>UniRef100_Q84ZZ3 Soluble epoxide hydrolase [Euphorbia lagascae]
Length = 321
Score = 392 bits (1008), Expect = e-108
Identities = 184/320 (57%), Positives = 232/320 (72%), Gaps = 4/320 (1%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+IEH V NGI MHIA G +GPV+LFLHGFP+LWYSWRHQ+ L S+GYR +APDL
Sbjct: 5 MEKIEHSMVSTNGINMHIASIGTQGPVILFLHGFPDLWYSWRHQLLYLSSVGYRCIAPDL 64
Query: 61 RGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIK 120
RGYGDTD P +I+ YT FH++GD+V L+D LG++QVFLVGHD GAII WY C+ P RIK
Sbjct: 65 RGYGDTDAPPAINQYTVFHILGDLVGLLDSLGIDQVFLVGHDWGAIISWYFCLLMPFRIK 124
Query: 121 AYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTT 180
A V SV F R+P+ +TV+ R G+D+YICRFQE GE+E A+ GT ++ LT+
Sbjct: 125 ALVNASVVFTPRDPRCKTVEKYRKELGEDFYICRFQEVGEIEDDFAQAGTAKIITKFLTS 184
Query: 181 RKTGPPIFPKGEYGTGFNP-DTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLN 239
R PP PK TG+ P ++PSWL++DD+ Y+VSK+ K GF+GGLNYYR +LN
Sbjct: 185 RHIRPPCIPK---ETGYRSLREPSHIPSWLSQDDINYYVSKYNKKGFSGGLNYYRCLDLN 241
Query: 240 WELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHF 299
WELTAPWTGV+IKVPVKFI G+ D Y +KE+IH GG K+ VP L+E++I +G AHF
Sbjct: 242 WELTAPWTGVQIKVPVKFIVGDQDATYHLPGVKEFIHNGGLKKHVPFLQEIVILEGAAHF 301
Query: 300 NNQEAEEEISNYIYEFIMKF 319
QE EEIS +I +F KF
Sbjct: 302 LQQEKPEEISAHILDFFEKF 321
>UniRef100_Q41414 Epoxide hydrolase [Solanum tuberosum]
Length = 321
Score = 390 bits (1001), Expect = e-107
Identities = 188/323 (58%), Positives = 240/323 (74%), Gaps = 6/323 (1%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+IEH+ V VNG+ MHIAE G +GP +LFLHGFPELWYSWRHQ+ L GYRAVAP L
Sbjct: 1 MEKIEHKMVAVNGLNMHIAELG-QGPTILFLHGFPELWYSWRHQMVYLAERGYRAVAPVL 59
Query: 61 RGYGDTDVP--SSISSYTCFHVVGDIVSLIDLLGV--EQVFLVGHDMGAIIGWYLCMFRP 116
RGYGDT + S ++ +VGD+V+L++ + E+VF+V HD GA+I W+LC+FRP
Sbjct: 60 RGYGDTTGAPLNDPSKFSILQLVGDVVALLEAIAPNEEKVFVVAHDWGALIAWHLCLFRP 119
Query: 117 ERIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKN 176
+++KA V SV FL RNPK+ TV+G++AVYG+D+YI RFQ PGE+EA+ A +G V+K
Sbjct: 120 DKVKALVNSSVHFLPRNPKMNTVEGLKAVYGEDHYISRFQVPGEIEAEFAPIGAKSVLKK 179
Query: 177 ILTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNF 236
ILT R P FPKG+ G PD P L SWL+E++L Y+ +KFE+TGFTGG+NYYR
Sbjct: 180 ILTFRDPAPFYFPKGK-GLEALPDAPVALSSWLSEEELDYYANKFEQTGFTGGVNYYRAL 238
Query: 237 NLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGV 296
++NWELTAPWTG ++KVP KFI GE D+VY KEYIH GGFK+DVP LEEV++ +G
Sbjct: 239 SINWELTAPWTGAQVKVPTKFIVGEFDLVYHIPGAKEYIHNGGFKKDVPLLEEVVVLEGA 298
Query: 297 AHFNNQEAEEEISNYIYEFIMKF 319
AHF NQE EIS +IY+FI KF
Sbjct: 299 AHFVNQERPHEISKHIYDFIQKF 321
>UniRef100_Q41415 Epoxide hydrolase [Solanum tuberosum]
Length = 321
Score = 389 bits (998), Expect = e-107
Identities = 185/323 (57%), Positives = 239/323 (73%), Gaps = 6/323 (1%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+IEH+ V VNG+ MH+AE G EGP +LF+HGFPELWYSWRHQ+ L GYRAVAPDL
Sbjct: 1 MEKIEHKMVAVNGLNMHLAELG-EGPTILFIHGFPELWYSWRHQMVYLAERGYRAVAPDL 59
Query: 61 RGYGDTDVP--SSISSYTCFHVVGDIVSLIDLLGV--EQVFLVGHDMGAIIGWYLCMFRP 116
RGYGDT + S ++ H+VGD+V+L++ + E+VF+V HD GA+I W+LC+FRP
Sbjct: 60 RGYGDTTGAPLNDPSKFSILHLVGDVVALLEAIAPNEEKVFVVAHDWGALIAWHLCLFRP 119
Query: 117 ERIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKN 176
+++KA V LSV F RNPK+ V+G++A+YG+D+YI RFQ PGE+EA+ A +G V+K
Sbjct: 120 DKVKALVNLSVHFSKRNPKMNVVEGLKAIYGEDHYISRFQVPGEIEAEFAPIGAKSVLKK 179
Query: 177 ILTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNF 236
ILT R P FPKG+ G PD P L SWL+E++L Y+ +KFE+TGFTG +NYYR
Sbjct: 180 ILTYRDPAPFYFPKGK-GLEAIPDAPVALSSWLSEEELDYYANKFEQTGFTGAVNYYRAL 238
Query: 237 NLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGV 296
+NWELTAPWTG ++KVP KFI GE D+VY KEYIH GGFK+DVP LEEV++ +G
Sbjct: 239 PINWELTAPWTGAQVKVPTKFIVGEFDLVYHIPGAKEYIHNGGFKKDVPLLEEVVVLEGA 298
Query: 297 AHFNNQEAEEEISNYIYEFIMKF 319
AHF +QE EIS +IY+FI KF
Sbjct: 299 AHFVSQERPHEISKHIYDFIQKF 321
>UniRef100_O48789 Putative epoxide hydrolase [Arabidopsis thaliana]
Length = 320
Score = 388 bits (997), Expect = e-106
Identities = 191/321 (59%), Positives = 241/321 (74%), Gaps = 6/321 (1%)
Query: 4 IEHRTVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+EHR V NGI +H+A +G +G +VL LHGFPELWYSWRHQI+ L + GYRAVAPDLRG
Sbjct: 1 MEHRNVRGNGIDIHVAIQGPSDGTIVLLLHGFPELWYSWRHQISGLAARGYRAVAPDLRG 60
Query: 63 YGDTDVPSSISSYTCFHVVGDIVSLIDLLGVE--QVFLVGHDMGAIIGWYLCMFRPERIK 120
YGD+D P+ ISS+TCF++VGD+V++I L E +VF+VGHD GA+I WYLC+FRP+++K
Sbjct: 61 YGDSDAPAEISSFTCFNIVGDLVAVISTLIKEDKKVFVVGHDWGALIAWYLCLFRPDKVK 120
Query: 121 AYVCLSVP--FLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNIL 178
A V LSVP F +P ++ VD MRAVYG+DYY+CRFQE G++EA++AEVGT VMK +L
Sbjct: 121 ALVNLSVPLSFWPTDPSVKPVDRMRAVYGNDYYVCRFQEVGDIEAEIAEVGTERVMKRLL 180
Query: 179 TTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNL 238
T R GP I PK + G +T LPSWLTE+D+AYFVSKF++ GF G +NYYRNFN
Sbjct: 181 TYRTPGPLIIPKDKSFWGSKGETIP-LPSWLTEEDVAYFVSKFKEKGFCGPVNYYRNFNR 239
Query: 239 NWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAH 298
N EL PW G KI+VP KF+ GELD+VY +KEYIHG FKEDVP +EE ++ +GVAH
Sbjct: 240 NNELLGPWVGSKIQVPTKFVIGELDLVYYMPGVKEYIHGPQFKEDVPLIEEPVVMEGVAH 299
Query: 299 FNNQEAEEEISNYIYEFIMKF 319
F NQE +EI I +FI F
Sbjct: 300 FLNQEKPQEILQIILDFISTF 320
>UniRef100_Q8L5G6 Soluble epoxide hydrolase [Brassica napus]
Length = 318
Score = 380 bits (977), Expect = e-104
Identities = 183/319 (57%), Positives = 238/319 (74%), Gaps = 4/319 (1%)
Query: 4 IEHRTVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+EHR + NG+ +H+A +G +GPVVL +HGFP LWYSWRHQI L +LGYRAVAPDLRG
Sbjct: 1 MEHRKLRGNGVDIHVAIQGPSDGPVVLLIHGFPTLWYSWRHQIPGLAALGYRAVAPDLRG 60
Query: 63 YGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAY 122
YGD+D PS ISSYTCFH+VGD++++I L ++VF+VGHD GA+I WYLC+FRP+++KA
Sbjct: 61 YGDSDAPSEISSYTCFHLVGDMIAVISALTEDKVFVVGHDWGALIAWYLCLFRPDKVKAL 120
Query: 123 VCLSVPFLH--RNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTT 180
V LSVPF ++P ++ VD +R YGDD+Y+CRFQE GE+EA+++EVG V++ ILT
Sbjct: 121 VNLSVPFSFGPKDPTVKPVDVLRKFYGDDFYMCRFQEVGEIEAEISEVGVERVVRRILTY 180
Query: 181 RKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
R P I PK + G +T LPSWLTE+D+AY+VSKF++ G+TGG+NYYRNF+ N
Sbjct: 181 RTPRPLILPKDKSFWGPKDETIP-LPSWLTEEDVAYYVSKFQEKGYTGGVNYYRNFDRNN 239
Query: 241 ELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHFN 300
EL APW G KI+VP KF GE D+VY +EYIHG FKE+VP LEE ++ +G AHF
Sbjct: 240 ELFAPWVGCKIQVPTKFAIGEQDLVYHFPGAREYIHGPKFKEEVPLLEEPVVIEGAAHFV 299
Query: 301 NQEAEEEISNYIYEFIMKF 319
NQE +EI I +FI KF
Sbjct: 300 NQEKPQEILQLIVDFISKF 318
>UniRef100_Q41413 Epoxide hydrolase [Solanum tuberosum]
Length = 321
Score = 374 bits (959), Expect = e-102
Identities = 178/323 (55%), Positives = 235/323 (72%), Gaps = 6/323 (1%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
ME+IEH+ V VNG+ MHIAE G +GP +LF+HGFPELWYSWRHQ+ L GYRAVAPDL
Sbjct: 1 MEKIEHKMVAVNGLNMHIAELG-QGPTILFIHGFPELWYSWRHQMVYLAERGYRAVAPDL 59
Query: 61 RGYGDTDVP--SSISSYTCFHVVGDIVSLIDLLGV--EQVFLVGHDMGAIIGWYLCMFRP 116
RGYGDT + S ++ FH+VGD+V+L++ + ++VF+V HD GA+I W+LC+FRP
Sbjct: 60 RGYGDTTGAPINDPSKFSIFHLVGDVVALLEAIAPNEDKVFVVAHDWGALIAWHLCLFRP 119
Query: 117 ERIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKN 176
+++KA V LSV + RN + ++G++A+YG+DYYICRFQ PGE+EA+ A +G V+K
Sbjct: 120 DKVKALVNLSVHYHPRNSNMNPIEGLKALYGEDYYICRFQVPGEIEAEFAPIGAKSVLKK 179
Query: 177 ILTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNF 236
+LT R P FPKG+ G D P L +WL+E++L Y+ +KFE+TGFTG LNYYR
Sbjct: 180 MLTYRDPAPFYFPKGK-GLEAIADAPIVLSTWLSEEELDYYANKFEQTGFTGALNYYRAL 238
Query: 237 NLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGV 296
++N ELTAPWTG ++ VP KFI GE D+ Y KEYIH GGFK+ VP LEEV++ +G
Sbjct: 239 SINSELTAPWTGAQVNVPTKFIVGEFDLAYHMRGAKEYIHNGGFKKYVPLLEEVVVLEGA 298
Query: 297 AHFNNQEAEEEISNYIYEFIMKF 319
AHF NQE EIS +IY+FI KF
Sbjct: 299 AHFVNQERPHEISKHIYDFIQKF 321
>UniRef100_Q9S7P1 ESTs AU065732 [Oryza sativa]
Length = 322
Score = 353 bits (907), Expect = 3e-96
Identities = 172/319 (53%), Positives = 222/319 (68%), Gaps = 5/319 (1%)
Query: 4 IEHRTVEV-NGIKMHIAEKGKE-GPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLR 61
+ HRTVEV +G+++H+AE G E GP VL +HGFPELWYSWRHQ+ AL + G+RAVAPDLR
Sbjct: 6 VRHRTVEVASGVRLHVAEAGPEDGPAVLLVHGFPELWYSWRHQMRALAARGFRAVAPDLR 65
Query: 62 GYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKA 121
GYGD+D P SYT H+VGD+V+LI +G +VF+ HD GA + W LC+ RP+ + A
Sbjct: 66 GYGDSDAPPGRDSYTVLHLVGDLVALIADVGQPRVFVAAHDWGAAVAWQLCLLRPDLVTA 125
Query: 122 YVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTTR 181
+V LSV + RNP V +RAV GD +YIC FQ+PG EA+ ++K R
Sbjct: 126 FVALSVEYHPRNPTRSPVQTLRAVCGDGHYICFFQKPGVAEAEFGRGDIKCLLKKFYGMR 185
Query: 182 KTGPPIFPKGEYGTGFNP-DTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
K P I P G+ T F+ D+ P+WL+E+D++Y+ KFEKTGFTGGLNYYR +LNW
Sbjct: 186 KAAPLIIPPGK--TLFDSIDSDGTCPAWLSEEDISYYAEKFEKTGFTGGLNYYRCIDLNW 243
Query: 241 ELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHFN 300
ELTAPWTGV IKVP KFI G+ D+ Y +K+YIH GG K VPNLE+V+I +GVAHF
Sbjct: 244 ELTAPWTGVPIKVPTKFIVGDQDLTYNIPGVKDYIHKGGLKACVPNLEDVVIMEGVAHFI 303
Query: 301 NQEAEEEISNYIYEFIMKF 319
NQE +E+S++I F KF
Sbjct: 304 NQEKPDEVSDHICGFFSKF 322
>UniRef100_Q8SAY9 Putative hydrolase [Oryza sativa]
Length = 338
Score = 350 bits (897), Expect = 4e-95
Identities = 168/329 (51%), Positives = 214/329 (64%), Gaps = 19/329 (5%)
Query: 4 IEHRTVEVNGIKMHIAEKGKEG---PVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
I HRTVE NGI +H+AE G EG VLFLHGFPELWYSWRHQ+ L G+R +APDL
Sbjct: 11 IRHRTVEANGISIHVAEAGGEGGDGAAVLFLHGFPELWYSWRHQMEHLAGRGFRCLAPDL 70
Query: 61 RGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIK 120
RGYGDTD P I SY+ FHVVGD+V+L+D LG+ +VF+VGHD GAII WY+C+FRP+R+
Sbjct: 71 RGYGDTDAPPEIESYSAFHVVGDLVALLDALGLAKVFVVGHDWGAIIAWYMCLFRPDRVT 130
Query: 121 AYVCLSVPFLHR------NPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVM 174
A V SV F+ I+T D YG YYICRFQEPG E + A +++
Sbjct: 131 ALVNTSVAFMRHVFIRSGADAIKTTDHFHKAYGPTYYICRFQEPGVAEEEFAPAHARHII 190
Query: 175 K----NILTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGL 230
+ N T K G P + P P LP+WLTE+D+ YF + FE+TGFTGG+
Sbjct: 191 RRTLCNRFTVHKAGKPESEESP------PPPPLPLPAWLTEEDIDYFAAAFERTGFTGGI 244
Query: 231 NYYRNFNLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEV 290
NYYRN + NWE+ APW K++VP KFI G+ D+ Y +++Y+H GG K +VP LE+V
Sbjct: 245 NYYRNMDRNWEMAAPWADAKVQVPTKFIVGDGDLTYHYAGIQDYLHKGGLKAEVPLLEDV 304
Query: 291 IIQKGVAHFNNQEAEEEISNYIYEFIMKF 319
++ G HF QE EE+S+ IY FI KF
Sbjct: 305 VVIPGAGHFIQQERAEEVSDLIYNFITKF 333
>UniRef100_Q41416 Epoxide hydrolase [Solanum tuberosum]
Length = 305
Score = 348 bits (892), Expect = 1e-94
Identities = 168/307 (54%), Positives = 221/307 (71%), Gaps = 6/307 (1%)
Query: 17 HIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVP--SSISS 74
HIAE G +GP +LF+HGFPELWYSWRHQ+ L GYRAVAPDLRGYGDT + S
Sbjct: 1 HIAELG-QGPTILFIHGFPELWYSWRHQMVYLAERGYRAVAPDLRGYGDTTGAPINDPSK 59
Query: 75 YTCFHVVGDIVSLIDLLGV--EQVFLVGHDMGAIIGWYLCMFRPERIKAYVCLSVPFLHR 132
++ FH+VGD+V+L++ + ++VF+V HD GA I W+LC+FRP+++KA V LSV +L R
Sbjct: 60 FSIFHLVGDVVALLEAIAPNEDKVFVVAHDWGAFIAWHLCLFRPDKVKALVNLSVHYLPR 119
Query: 133 NPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTTRKTGPPIFPKGE 192
N + V+G++A+YG+DYYICRFQ G++EA+ A +G V+K +LT R P FPKG+
Sbjct: 120 NSNMNPVEGLKALYGEDYYICRFQVQGDIEAEFAPIGAKSVLKKMLTYRDPAPFYFPKGK 179
Query: 193 YGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNWELTAPWTGVKIK 252
G D P L +WL+E++L Y+ SKFE+TGFTG LNYYR +++ ELTAPW G ++K
Sbjct: 180 -GLEAIADAPIALSTWLSEEELDYYASKFEQTGFTGALNYYRALSIDSELTAPWQGAEVK 238
Query: 253 VPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHFNNQEAEEEISNYI 312
VP KFI GE +VY KEYIH GGFK+ VP LEEV++ +G AHF NQE EIS +I
Sbjct: 239 VPTKFIVGEFALVYHMRGAKEYIHNGGFKKYVPLLEEVVVLEGAAHFVNQERPHEISKHI 298
Query: 313 YEFIMKF 319
Y+FI KF
Sbjct: 299 YDFIQKF 305
>UniRef100_Q8SAY7 Putative hydrolase [Oryza sativa]
Length = 333
Score = 347 bits (890), Expect = 2e-94
Identities = 168/319 (52%), Positives = 213/319 (66%), Gaps = 7/319 (2%)
Query: 4 IEHRTVEVNGIKMHIAEKGK---EGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDL 60
+ HRTVE NGI MH+AE G P VLF+HGFPELWYSWRHQ+ L + GYR VAPDL
Sbjct: 7 VRHRTVEANGISMHVAEAGPGSGTAPAVLFVHGFPELWYSWRHQMGHLAARGYRCVAPDL 66
Query: 61 RGYGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIK 120
RGYG T P +SYT FH+VGD+V+L+D L + QVF+VGHD GAI+ W LC+ RP+R++
Sbjct: 67 RGYGGTTAPPEHTSYTIFHLVGDLVALLDALELPQVFVVGHDWGAIVSWNLCLLRPDRVR 126
Query: 121 AYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTT 180
A V LSV F+ R P + +D R YGDDYY+CRFQEPG +E ++A + K L
Sbjct: 127 ALVNLSVAFMPRRPAEKPLDYFRGAYGDDYYVCRFQEPG-VEKELASLDLKRFFKLALIV 185
Query: 181 RKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
+ TG + T LP WL+E+D++Y S + KTGF GG+NYYR F+LNW
Sbjct: 186 QTTGSSAMSIKKMRANNREVT---LPPWLSEEDISYVASVYAKTGFAGGINYYRCFDLNW 242
Query: 241 ELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQKGVAHFN 300
EL APWTG K+ VP KFI G+ D+ Y +K YIH G K+DVP LEEV++ KG HF
Sbjct: 243 ELMAPWTGAKVLVPTKFIVGDGDLAYHLPGVKSYIHKGRLKKDVPMLEEVVVIKGAGHFI 302
Query: 301 NQEAEEEISNYIYEFIMKF 319
QE +EIS++IY +I KF
Sbjct: 303 QQERAQEISDHIYNYIKKF 321
>UniRef100_O23444 Putative epoxide hydrolase [Arabidopsis thaliana]
Length = 536
Score = 342 bits (877), Expect = 8e-93
Identities = 162/309 (52%), Positives = 216/309 (69%), Gaps = 7/309 (2%)
Query: 1 MERIEHRTVEVNGIKMHIAEKGKEG----PVVLFLHGFPELWYSWRHQIAALGSLGYRAV 56
++ +EH+T++VNGI MH+AEK G P++LFLHGFPELWY+WRHQ+ AL SLGYR +
Sbjct: 51 LDGVEHKTLKVNGINMHVAEKPGSGSGEDPIILFLHGFPELWYTWRHQMVALSSLGYRTI 110
Query: 57 APDLRGYGDTDVPSSISSYTCFHVVGDIVSLIDLL--GVEQVFLVGHDMGAIIGWYLCMF 114
APDLRGYGDT+ P + YT +V+LI + G + V +VGHD GA+I W LC +
Sbjct: 111 APDLRGYGDTEAPEKVEDYTLLKRGRSVVALIVAVTGGDKAVSVVGHDWGAMIAWQLCQY 170
Query: 115 RPERIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVM 174
RPE++KA V +SV F RNP V +R V+GDDYY+CRFQ+ GE+E + ++GT V+
Sbjct: 171 RPEKVKALVNMSVLFSPRNPVRVPVPTLRHVFGDDYYVCRFQKAGEIETEFKKLGTENVL 230
Query: 175 KNILTTRKTGPPIFPKGEYGTGFNPDTPDNLPSWLTEDDLAYFVSKFEKTGFTGGLNYYR 234
K LT + GP PK +Y + + LP WLT++DL Y+V+K+E GFTG +NYYR
Sbjct: 231 KEFLTYKTPGPLNLPKDKYFKR-SENAASALPLWLTQEDLDYYVTKYENKGFTGPINYYR 289
Query: 235 NFNLNWELTAPWTGVKIKVPVKFITGELDMVYTSFNLKEYIHGGGFKEDVPNLEEVIIQK 294
N + NWELTAPWTG KI+VPVKFI G+ D+ Y KEYI+GGGFK DVP L+E ++ K
Sbjct: 290 NIDRNWELTAPWTGAKIRVPVKFIIGDQDLTYNFPGAKEYINGGGFKRDVPLLDETVVLK 349
Query: 295 GVAHFNNQE 303
G+ HF ++E
Sbjct: 350 GLGHFLHEE 358
Score = 205 bits (522), Expect = 1e-51
Identities = 97/162 (59%), Positives = 118/162 (71%), Gaps = 10/162 (6%)
Query: 5 EHRTVEVNGIKMHIAEKGKE--------GPVVLFLHGFPELWYSWRHQIAALGSLGYRAV 56
+H V+VNGI MH+AEK PV+LFLHGFPELWY+WRHQ+ AL SLGYR +
Sbjct: 371 DHSFVKVNGITMHVAEKSPSVAGNGAIRPPVILFLHGFPELWYTWRHQMVALSSLGYRTI 430
Query: 57 APDLRGYGDTDVPSSISSYTCFHVVGDIVSLIDLL--GVEQVFLVGHDMGAIIGWYLCMF 114
APDLRGYGDTD P S+ +YT HVVGD++ LID + E+VF+VGHD GAII W+LC+F
Sbjct: 431 APDLRGYGDTDAPESVDAYTSLHVVGDLIGLIDAVVGDREKVFVVGHDWGAIIAWHLCLF 490
Query: 115 RPERIKAYVCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQ 156
RP+R+KA V +SV F NPK + +A YGDDYYICRFQ
Sbjct: 491 RPDRVKALVNMSVVFDPWNPKRKPTSTFKAFYGDDYYICRFQ 532
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.142 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 612,565,105
Number of Sequences: 2790947
Number of extensions: 28573242
Number of successful extensions: 61081
Number of sequences better than 10.0: 2509
Number of HSP's better than 10.0 without gapping: 907
Number of HSP's successfully gapped in prelim test: 1603
Number of HSP's that attempted gapping in prelim test: 58459
Number of HSP's gapped (non-prelim): 2691
length of query: 319
length of database: 848,049,833
effective HSP length: 127
effective length of query: 192
effective length of database: 493,599,564
effective search space: 94771116288
effective search space used: 94771116288
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144513.17


