

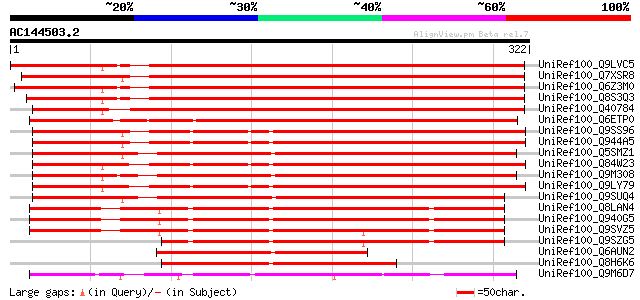
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.2 + phase: 0
(322 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LVC5 Apospory-associated protein C [Arabidopsis thal... 429 e-119
UniRef100_Q7XSR8 OSJNBa0041A02.19 protein [Oryza sativa] 425 e-118
UniRef100_Q6Z3M0 Putative Aldose 1-epimerase [Oryza sativa] 418 e-115
UniRef100_Q8S3Q3 Putative apospory-associated protein C [Oryza s... 411 e-113
UniRef100_Q40784 Possible apospory-associated protein C [Pennise... 403 e-111
UniRef100_Q6ETP0 Apospory-associated protein C-like [Oryza sativa] 363 4e-99
UniRef100_Q9SS96 F4P13.13 protein [Arabidopsis thaliana] 351 1e-95
UniRef100_Q944A5 AT3g01590/F4P13_13 [Arabidopsis thaliana] 350 4e-95
UniRef100_Q5SMZ1 Aldose 1-epimerase-like [Oryza sativa] 347 3e-94
UniRef100_Q84W23 Hypothetical protein At5g14500 [Arabidopsis tha... 343 4e-93
UniRef100_Q9M308 Hypothetical protein F2A19.210 [Arabidopsis tha... 341 1e-92
UniRef100_Q9LY79 Hypothetical protein F18O22_290 [Arabidopsis th... 339 7e-92
UniRef100_Q9SUQ4 Hypothetical protein F9D16.200 [Arabidopsis tha... 311 2e-83
UniRef100_Q8LAN4 Possible apospory-associated like protein [Arab... 290 3e-77
UniRef100_Q940G5 Possible apospory-associated like protein [Arab... 287 2e-76
UniRef100_Q9SVZ5 Possible apospory-associated like protein [Arab... 278 1e-73
UniRef100_Q9SZG5 Possible apospory-associated like protein [Arab... 226 8e-58
UniRef100_Q6AUN2 Hypothetical protein OSJNBa0040E06.8 [Oryza sat... 169 9e-41
UniRef100_Q8H6K6 Apospory-associated-like protein [Brassica rapa... 162 9e-39
UniRef100_Q9M6D7 Apospory-associated protein C [Chlamydomonas re... 127 4e-28
>UniRef100_Q9LVC5 Apospory-associated protein C [Arabidopsis thaliana]
Length = 312
Score = 429 bits (1102), Expect = e-119
Identities = 214/323 (66%), Positives = 252/323 (77%), Gaps = 16/323 (4%)
Query: 1 MSRETMYVKHCMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK---- 56
M+ E + + G+NGL+KI+LRE+RG SAEVYLYG VTSWKNE GEELL +SSK
Sbjct: 1 MATERHHFELAKGINGLDKIVLRESRGRSAEVYLYGSHVTSWKNENGEELLHLSSKAIFK 60
Query: 57 PEKEVKKTNQYNCFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNS 116
P K ++ CF +N + + F +N++W ++ NPPP P NS S
Sbjct: 61 PPKPIRGGIPL-CFPQFSN-----------FGTLESHGFARNRIWEVEANPPPLPLNSCS 108
Query: 117 RAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHT 176
AFVDL+L+ +EDD K WP+ +EFRLRIALG G+L LTSRIRNTN DGK FTFTFAYHT
Sbjct: 109 SAFVDLILRPTEDDLKIWPNNFEFRLRIALGTEGELTLTSRIRNTNSDGKPFTFTFAYHT 168
Query: 177 YLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHE 236
Y V+DISEVR+EGLETLDYLD+LK+RERFTEQGDAITFE EVDK+YLSTPTKIAI+DHE
Sbjct: 169 YFSVSDISEVRVEGLETLDYLDNLKDRERFTEQGDAITFESEVDKIYLSTPTKIAILDHE 228
Query: 237 RKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWK 296
+KRTF IRKDGL DAVVWNPWDKKSK ISDLGD++YKHMLCV+ A +E+ ITLKPGEEWK
Sbjct: 229 KKRTFVIRKDGLADAVVWNPWDKKSKTISDLGDEDYKHMLCVEAAAIERPITLKPGEEWK 288
Query: 297 GRQEISAVPSSYCSGQLDPRKVL 319
GR E+SAVPSSY SGQLDP+KVL
Sbjct: 289 GRLELSAVPSSYSSGQLDPKKVL 311
>UniRef100_Q7XSR8 OSJNBa0041A02.19 protein [Oryza sativa]
Length = 325
Score = 425 bits (1092), Expect = e-118
Identities = 204/315 (64%), Positives = 246/315 (77%), Gaps = 14/315 (4%)
Query: 8 VKHCMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQY 67
V+ G +GL+K++LRE RGFSAEV+LYGGQVTSWKN+RG+ELLFVSSK + K +
Sbjct: 20 VERAKGPSGLDKVVLREARGFSAEVHLYGGQVTSWKNDRGDELLFVSSKATFKPPKAIRG 79
Query: 68 N---CFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVL 124
CF + ++ F +N+ W +D NPPPFPT++ ++AFVDL+L
Sbjct: 80 GIPICFPQFGT-----------HGILEKHGFARNRFWAIDDNPPPFPTSTAAKAFVDLIL 128
Query: 125 KNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDIS 184
K SE+D K WPH +EFRLR+ALGP GDL LTSRIRNTN DG+ F+FTFAYHTY V+DIS
Sbjct: 129 KTSEEDLKIWPHSFEFRLRVALGPGGDLALTSRIRNTNTDGRPFSFTFAYHTYFSVSDIS 188
Query: 185 EVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIR 244
EVRIEGLETLDY+D L+ +ER TEQGDAI FE EVDK+YL P KIAIIDHE+KRTF +R
Sbjct: 189 EVRIEGLETLDYIDCLRGKERSTEQGDAIVFESEVDKIYLDAPAKIAIIDHEKKRTFVLR 248
Query: 245 KDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAV 304
KDGLPDAV+WNPWDK++K + D GD+EYKHMLCV+PA VEK ITLKPGEEWKG+ ++SAV
Sbjct: 249 KDGLPDAVLWNPWDKRTKNMQDFGDEEYKHMLCVEPAAVEKPITLKPGEEWKGKMDLSAV 308
Query: 305 PSSYCSGQLDPRKVL 319
PSSYCSGQLDP KVL
Sbjct: 309 PSSYCSGQLDPNKVL 323
>UniRef100_Q6Z3M0 Putative Aldose 1-epimerase [Oryza sativa]
Length = 337
Score = 418 bits (1074), Expect = e-115
Identities = 203/320 (63%), Positives = 244/320 (75%), Gaps = 16/320 (5%)
Query: 4 ETMYVKHCMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEK 59
+T + + G +G+EK++LR RGFSAEVYLYG QVTSWKN+ EELLFVSSK P K
Sbjct: 28 QTPFYELVKGNSGIEKVLLRGTRGFSAEVYLYGAQVTSWKNDHAEELLFVSSKAIFKPPK 87
Query: 60 EVKKTNQYNCFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAF 119
++ CF + Q+ F +N+LW +D NPPP P N ++F
Sbjct: 88 AIRGGIPI-CFPQFGT-----------HGNLEQHGFARNRLWTIDDNPPPLPVNPAIKSF 135
Query: 120 VDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLY 179
VDL+L+ S++D K WPH +EFRLR+ALGP GDL LTSRIRNTN DG+SF++TFAYHTY
Sbjct: 136 VDLILRPSDEDLKIWPHSFEFRLRVALGPNGDLSLTSRIRNTNTDGRSFSYTFAYHTYFS 195
Query: 180 VTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKR 239
V+DISEVR+EGLET+DYLD+LK +ERFTEQGDAI FE E+DKVYL+ P+KIAIIDHE+KR
Sbjct: 196 VSDISEVRVEGLETMDYLDNLKGKERFTEQGDAIVFESEIDKVYLAAPSKIAIIDHEKKR 255
Query: 240 TFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQ 299
TF + K+GLPDAVVWNPWDKK+K + D GD EYKHMLCV+PA VEK ITLKPGEEWKGR
Sbjct: 256 TFVLTKEGLPDAVVWNPWDKKAKAMQDFGDGEYKHMLCVEPAAVEKPITLKPGEEWKGRL 315
Query: 300 EISAVPSSYCSGQLDPRKVL 319
+SAVPSSYCSGQLDP KVL
Sbjct: 316 ALSAVPSSYCSGQLDPLKVL 335
>UniRef100_Q8S3Q3 Putative apospory-associated protein C [Oryza sativa]
Length = 344
Score = 411 bits (1057), Expect = e-113
Identities = 198/313 (63%), Positives = 244/313 (77%), Gaps = 16/313 (5%)
Query: 11 CMGVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQ 66
C GVNGL+K++LRE RG SAEVYLYGG VTSWK+E GEELLFVS+K P K ++
Sbjct: 33 CKGVNGLDKVVLREVRGSSAEVYLYGGHVTSWKDEHGEELLFVSNKAIFKPPKAIRGGIP 92
Query: 67 YNCFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKN 126
CF +N + + F +N+ W ++ +PPPFP ++++A+VDL+L +
Sbjct: 93 I-CFPQFSN-----------FGNLDPHGFARNRTWSVENDPPPFPVPTSNKAYVDLILTH 140
Query: 127 SEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEV 186
+E+D K WPH YEFRLR+ALGP GDLMLTSRIRNTN DGK F+FTFAYHTY ++DISEV
Sbjct: 141 TEEDLKIWPHSYEFRLRVALGPGGDLMLTSRIRNTNADGKPFSFTFAYHTYFSISDISEV 200
Query: 187 RIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKD 246
R+EGLETLDYLD+L+ R R+TEQGDAI FE E+D++YL TP+KIAIIDHE+KRTF +RK
Sbjct: 201 RVEGLETLDYLDNLQERNRYTEQGDAIVFESELDRIYLGTPSKIAIIDHEKKRTFVVRKG 260
Query: 247 GLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPS 306
GLPDAVVWNPWDKK+K +SD GDDEYK M+CV+ A +EK ITLKPGEEW G+ E+SAVPS
Sbjct: 261 GLPDAVVWNPWDKKAKAMSDFGDDEYKRMVCVEAAAIEKPITLKPGEEWTGKLELSAVPS 320
Query: 307 SYCSGQLDPRKVL 319
SY SGQLDP +VL
Sbjct: 321 SYYSGQLDPDRVL 333
>UniRef100_Q40784 Possible apospory-associated protein C [Pennisetum ciliare]
Length = 329
Score = 403 bits (1035), Expect = e-111
Identities = 194/309 (62%), Positives = 238/309 (76%), Gaps = 16/309 (5%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYNCF 70
+GLEK++LR R AE+YLYGGQVTSWKN+ GEELLF+SSK P K ++
Sbjct: 31 SGLEKVVLRGARNCCAEIYLYGGQVTSWKNDNGEELLFLSSKAIFKPPKAIR-------- 82
Query: 71 DMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDD 130
+ L + Q+ F +N+ W +D +PPP P N +AFVDL+L+ +E+D
Sbjct: 83 ----GGIPICLPQFGTHGNLEQHGFARNRFWSIDNDPPPLPVNPAIKAFVDLILRPAEED 138
Query: 131 TKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEG 190
K WPH +EFRLR+ALGP+GDL LTSRIRNTN DG+ F++TFAYHTY +V+DISEVR+EG
Sbjct: 139 LKIWPHSFEFRLRVALGPSGDLSLTSRIRNTNTDGRPFSYTFAYHTYFFVSDISEVRVEG 198
Query: 191 LETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPD 250
LET+DYLD+LK +ERFTEQGDAI FE EVDKVYL+ P+KIAIIDHE+K+TF + K+GLPD
Sbjct: 199 LETMDYLDNLKAKERFTEQGDAIVFESEVDKVYLAAPSKIAIIDHEKKKTFVVTKEGLPD 258
Query: 251 AVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCS 310
AVVWNPWDKK+K + D GD EYK+MLCV+PA VEK ITLKPGEEW+GR +SAVPSSYCS
Sbjct: 259 AVVWNPWDKKAKAMQDFGDAEYKNMLCVEPAAVEKPITLKPGEEWRGRIALSAVPSSYCS 318
Query: 311 GQLDPRKVL 319
GQLDP KVL
Sbjct: 319 GQLDPLKVL 327
>UniRef100_Q6ETP0 Apospory-associated protein C-like [Oryza sativa]
Length = 319
Score = 363 bits (931), Expect = 4e-99
Identities = 179/303 (59%), Positives = 221/303 (72%), Gaps = 6/303 (1%)
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYNCFDM 72
G GLEK++LRE G S +V+LYGGQV W+NE E+LLFVS K ++ C
Sbjct: 14 GATGLEKLVLREAHGCSVQVFLYGGQVIFWENEYREQLLFVSRKSSYVFQEF----CLPC 69
Query: 73 NNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDTK 132
N R I+ + + Q+ F +N+LW +D PPPFP + S +DL+LK S +D K
Sbjct: 70 NTQNGRSIMQFGN-HGVLEQHGFARNRLWSVDETPPPFPATT-SNCHIDLILKQSPEDLK 127
Query: 133 NWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGLE 192
WPH +EFRLR+AL PTGDLMLTSRI+NTN DGK F F F+Y+TY V+DISEVR+EGLE
Sbjct: 128 IWPHSFEFRLRVALSPTGDLMLTSRIKNTNADGKPFKFGFSYNTYFSVSDISEVRVEGLE 187
Query: 193 TLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAV 252
TLDY+D+L+ +R TEQGDA+ FE E +KVYLS P KI IIDH++KRTFE+RK+GLPD V
Sbjct: 188 TLDYIDNLQCGKRCTEQGDAVVFESEAEKVYLSAPPKIVIIDHDKKRTFELRKEGLPDVV 247
Query: 253 VWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCSGQ 312
VWNPWD+K+K I D G++EYK MLCV A EK ITL+PGEEW+GRQEIS VPSSY SGQ
Sbjct: 248 VWNPWDRKAKTILDFGEEEYKCMLCVGAANAEKPITLRPGEEWQGRQEISVVPSSYSSGQ 307
Query: 313 LDP 315
DP
Sbjct: 308 WDP 310
>UniRef100_Q9SS96 F4P13.13 protein [Arabidopsis thaliana]
Length = 306
Score = 351 bits (901), Expect = 1e-95
Identities = 177/309 (57%), Positives = 225/309 (72%), Gaps = 18/309 (5%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYN---CFD 71
+G +IIL E RG +AEV L+GGQV SWKNER EELL++SSK + + K + CF
Sbjct: 10 DGSSRIILTEPRGSTAEVLLFGGQVISWKNERREELLYMSSKAQYKPPKAIRGGIPVCFP 69
Query: 72 MNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDT 131
N + ++ F +NK W D +P P P +N ++ VDL+LK++EDD
Sbjct: 70 QFGN-----------FGGLERHGFARNKFWSHDEDPSPLPP-ANKQSSVDLILKSTEDDL 117
Query: 132 KNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGL 191
K WPH +E R+RI++ P G L L R+RN D K+F+F FA YLYV+DISEVR+EGL
Sbjct: 118 KTWPHSFELRIRISISP-GKLTLIPRVRNI--DSKAFSFMFALRNYLYVSDISEVRVEGL 174
Query: 192 ETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDA 251
ETLDYLD+L +ERFTEQ DAITF+GEVD+VYL+TPTKIA+IDHERKRT E+RK+G+P+A
Sbjct: 175 ETLDYLDNLIGKERFTEQADAITFDGEVDRVYLNTPTKIAVIDHERKRTIELRKEGMPNA 234
Query: 252 VVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCSG 311
VVWNPWDKK+K I+D+GD++YK MLCV +E + LKP EEWKGRQE+S V SSYCSG
Sbjct: 235 VVWNPWDKKAKTIADMGDEDYKTMLCVDSGVIEPLVLLKPREEWKGRQELSIVSSSYCSG 294
Query: 312 QLDPRKVLF 320
QLDPRKVL+
Sbjct: 295 QLDPRKVLY 303
>UniRef100_Q944A5 AT3g01590/F4P13_13 [Arabidopsis thaliana]
Length = 306
Score = 350 bits (897), Expect = 4e-95
Identities = 176/309 (56%), Positives = 225/309 (71%), Gaps = 18/309 (5%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYN---CFD 71
+G +IIL E RG +AEV L+GGQV SWKNER EELL++SSK + + K + CF
Sbjct: 10 DGSSRIILTEPRGSTAEVLLFGGQVISWKNERREELLYMSSKAQYKPPKAIRGGIPVCFP 69
Query: 72 MNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDT 131
N + ++ F +NK W D +P P P +N ++ VDL+LK++EDD
Sbjct: 70 QFGN-----------FGGLERHGFARNKFWSHDEDPSPLPP-ANKQSSVDLILKSTEDDL 117
Query: 132 KNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGL 191
K WPH +E R+RI++ P G L L R+RN D K+F+F FA YLYV+DISEVR+EGL
Sbjct: 118 KTWPHSFELRIRISISP-GKLTLIPRVRNI--DSKAFSFMFALRNYLYVSDISEVRVEGL 174
Query: 192 ETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDA 251
ETLDYLD+L +ERFTEQ DAITF+GEVD+VYL+TPTKIA+IDHERKRT E+RK+G+P+A
Sbjct: 175 ETLDYLDNLIGKERFTEQADAITFDGEVDRVYLNTPTKIAVIDHERKRTIELRKEGMPNA 234
Query: 252 VVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCSG 311
VVWNPWDKK+K I+D+GD++YK MLCV +E + LKP E+WKGRQE+S V SSYCSG
Sbjct: 235 VVWNPWDKKAKTITDMGDEDYKTMLCVDSGVIEPLVLLKPREKWKGRQELSIVSSSYCSG 294
Query: 312 QLDPRKVLF 320
QLDPRKVL+
Sbjct: 295 QLDPRKVLY 303
>UniRef100_Q5SMZ1 Aldose 1-epimerase-like [Oryza sativa]
Length = 312
Score = 347 bits (889), Expect = 3e-94
Identities = 177/304 (58%), Positives = 218/304 (71%), Gaps = 17/304 (5%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYN---CFD 71
NG +++++R RG S V L+GGQV SW+N+RGEELLF SSK + K + CF
Sbjct: 18 NGADQVMIRSPRGASVLVSLHGGQVVSWRNDRGEELLFTSSKAIFKPPKAMRGGIPICFP 77
Query: 72 MNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNS-RAFVDLVLKNSEDD 130
N L Q+ F +N+LW +D PP N N+ + VDL+LK SEDD
Sbjct: 78 QFGNCGTLE-----------QHGFARNRLWAIDDEAPPLNHNDNNGKVSVDLLLKPSEDD 126
Query: 131 TKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEG 190
K WPH +EFRLR++L GDL L SR+RN N GK F+F+FAYHTYL V+DISEVRIEG
Sbjct: 127 LKCWPHCFEFRLRVSLSTDGDLSLVSRVRNVN--GKPFSFSFAYHTYLSVSDISEVRIEG 184
Query: 191 LETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPD 250
LETLDYLD+L RERFTEQGDAITFE EVD+VY+ +P+ IA++DHE+KRTF +RK+GLPD
Sbjct: 185 LETLDYLDNLSQRERFTEQGDAITFESEVDRVYVGSPSVIAVLDHEKKRTFIVRKEGLPD 244
Query: 251 AVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCS 310
VVWNPWDKKSK ++D GD+EYK MLCV A VE+AITLKPGEEW G+ E+SAV S+ CS
Sbjct: 245 VVVWNPWDKKSKTMADFGDEEYKQMLCVDAAAVERAITLKPGEEWTGKLELSAVASTNCS 304
Query: 311 GQLD 314
LD
Sbjct: 305 DHLD 308
>UniRef100_Q84W23 Hypothetical protein At5g14500 [Arabidopsis thaliana]
Length = 306
Score = 343 bits (880), Expect = 4e-93
Identities = 174/310 (56%), Positives = 223/310 (71%), Gaps = 20/310 (6%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYNCF 70
+G +I+L + G +AEV LYGGQV SWKNER E+LL++S+K P K ++ +
Sbjct: 10 DGSSRILLTDPAGSTAEVLLYGGQVVSWKNERREKLLYMSTKAQLKPPKAIRGGLPISFP 69
Query: 71 DMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDD 130
N + ++ F +N+ W LD +P P P +N ++ VDLVLK++EDD
Sbjct: 70 QFGN------------FGALERHGFARNRFWSLDNDPSPLPP-ANQQSTVDLVLKSTEDD 116
Query: 131 TKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEG 190
K WPH +E R+RI++ P G L + R+RNT D K+F+F F+ YLYV+DISEVR+EG
Sbjct: 117 LKIWPHSFELRVRISISP-GKLTIIPRVRNT--DTKAFSFMFSLRNYLYVSDISEVRVEG 173
Query: 191 LETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPD 250
LETLDYLD+L RERFTEQ DAITF+GEVDKVYL+TPTKIAIIDHERKRT E+RK+G+P+
Sbjct: 174 LETLDYLDNLVRRERFTEQADAITFDGEVDKVYLNTPTKIAIIDHERKRTIELRKEGMPN 233
Query: 251 AVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCS 310
A VWNPWDKK+K I+D+GD++Y MLCV +E I LKP EEWKGRQE+S V SSYCS
Sbjct: 234 AAVWNPWDKKAKSIADMGDEDYTTMLCVDSGAIESPIVLKPHEEWKGRQELSIVSSSYCS 293
Query: 311 GQLDPRKVLF 320
GQLDPRKVL+
Sbjct: 294 GQLDPRKVLY 303
>UniRef100_Q9M308 Hypothetical protein F2A19.210 [Arabidopsis thaliana]
Length = 317
Score = 341 bits (875), Expect = 1e-92
Identities = 172/305 (56%), Positives = 221/305 (72%), Gaps = 19/305 (6%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYNCF 70
NG+++++LR G SA++ L+GGQV SW+NE GEELLF S+K P K ++ Q C+
Sbjct: 21 NGVDQVLLRNPHGASAKISLHGGQVISWRNELGEELLFTSNKAIFKPPKSMRGGIQI-CY 79
Query: 71 DMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSN-SRAFVDLVLKNSED 129
+ L Q+ F +NK+W++D NPPP +N + ++FVDL+LK SED
Sbjct: 80 PQFGDCGSLD-----------QHGFARNKIWVIDENPPPLNSNESLGKSFVDLLLKPSED 128
Query: 130 DTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIE 189
D K WPH +EFRLR++L GDL LTSRIRN N GK F+F+FAYHTYL V+DISEVRIE
Sbjct: 129 DLKQWPHSFEFRLRVSLAVDGDLTLTSRIRNIN--GKPFSFSFAYHTYLSVSDISEVRIE 186
Query: 190 GLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLP 249
GLETLDYLD+L R+ TEQGDAITFE E+D+ YL +P +A++DHERKRT+ I K+GLP
Sbjct: 187 GLETLDYLDNLSQRQLLTEQGDAITFESEMDRTYLRSPKVVAVLDHERKRTYVIGKEGLP 246
Query: 250 DAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYC 309
D VVWNPW+KKSK ++D GD+EYK MLCV A VE+ ITLKPGEEW GR ++AV SS+C
Sbjct: 247 DTVVWNPWEKKSKTMADFGDEEYKSMLCVDGAAVERPITLKPGEEWTGRLILTAVKSSFC 306
Query: 310 SGQLD 314
QL+
Sbjct: 307 FDQLE 311
>UniRef100_Q9LY79 Hypothetical protein F18O22_290 [Arabidopsis thaliana]
Length = 381
Score = 339 bits (869), Expect = 7e-92
Identities = 174/311 (55%), Positives = 223/311 (70%), Gaps = 21/311 (6%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYNCF 70
+G +I+L + G +AEV LYGGQV SWKNER E+LL++S+K P K ++ +
Sbjct: 84 DGSSRILLTDPAGSTAEVLLYGGQVVSWKNERREKLLYMSTKAQLKPPKAIRGGLPISFP 143
Query: 71 DMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDD 130
N + ++ F +N+ W LD +P P P +N ++ VDLVLK++EDD
Sbjct: 144 QFGN------------FGALERHGFARNRFWSLDNDPSPLPP-ANQQSTVDLVLKSTEDD 190
Query: 131 TKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEG 190
K WPH +E R+RI++ P G L + R+RNT D K+F+F F+ YLYV+DISEVR+EG
Sbjct: 191 LKIWPHSFELRVRISISP-GKLTIIPRVRNT--DTKAFSFMFSLRNYLYVSDISEVRVEG 247
Query: 191 LETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPD 250
LETLDYLD+L RERFTEQ DAITF+GEVDKVYL+TPTKIAIIDHERKRT E+RK+G+P+
Sbjct: 248 LETLDYLDNLMRRERFTEQADAITFDGEVDKVYLNTPTKIAIIDHERKRTIELRKEGMPN 307
Query: 251 -AVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYC 309
A VWNPWDKK+K I+D+GD++Y MLCV +E I LKP EEWKGRQE+S V SSYC
Sbjct: 308 AAAVWNPWDKKAKSIADMGDEDYTTMLCVDSGAIESPIVLKPHEEWKGRQELSIVSSSYC 367
Query: 310 SGQLDPRKVLF 320
SGQLDPRKVL+
Sbjct: 368 SGQLDPRKVLY 378
>UniRef100_Q9SUQ4 Hypothetical protein F9D16.200 [Arabidopsis thaliana]
Length = 306
Score = 311 bits (797), Expect = 2e-83
Identities = 154/298 (51%), Positives = 213/298 (70%), Gaps = 18/298 (6%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYN---CFD 71
NG ++I+L+ RG S ++ L+GGQV SWK ++G+ELLF S+K + + CF
Sbjct: 22 NGTDQILLQNPRGASVKISLHGGQVLSWKTDKGDELLFNSTKANLKPPHPVRGGIPICFP 81
Query: 72 MNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPT-NSNSRAFVDLVLKNSEDD 130
+ L Q+ F +NK+W+++ NPP P+ +S +A+VDLVLK+S++D
Sbjct: 82 QFGTRGSLE-----------QHGFARNKMWLVENNPPALPSFDSTGKAYVDLVLKSSDED 130
Query: 131 TKN-WPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIE 189
T WP+ +EF LR++L G+L L SR+RN N K F+F+ AYHTY ++DISEVR+E
Sbjct: 131 TMRIWPYSFEFHLRVSLALDGNLTLISRVRNINS--KPFSFSIAYHTYFSISDISEVRLE 188
Query: 190 GLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLP 249
GLETLDYLD++ +RERFTEQGDA+TFE E+D+VYL++ +AI DHERKRTF I+K+GLP
Sbjct: 189 GLETLDYLDNMHDRERFTEQGDALTFESEIDRVYLNSKDVVAIFDHERKRTFLIKKEGLP 248
Query: 250 DAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSS 307
D VVWNPW+KK++ ++DLGDDEY+HMLCV A +EK ITLKPGEEW G+ +S V S+
Sbjct: 249 DVVVWNPWEKKARALTDLGDDEYRHMLCVDGAAIEKPITLKPGEEWTGKLHLSLVLST 306
>UniRef100_Q8LAN4 Possible apospory-associated like protein [Arabidopsis thaliana]
Length = 317
Score = 290 bits (743), Expect = 3e-77
Identities = 150/298 (50%), Positives = 200/298 (66%), Gaps = 21/298 (7%)
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYNCFDM 72
GVNGL+KII+R+ RG SAEVYLYGGQV+SWKNE GEELL +SSK F
Sbjct: 35 GVNGLDKIIIRDRRGRSAEVYLYGGQVSSWKNENGEELLVMSSKA-----------IFQP 83
Query: 73 NNNKCRLILLLQDLYAEAF---QYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSED 129
I +L Y+ + F++ + W ++ PPP P S S A VDL++++S +
Sbjct: 84 PTPIRGGIPVLFPQYSNTGPLPSHGFVRQRFWEVETKPPPLP--SLSTAHVDLIVRSSNE 141
Query: 130 DTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIE 189
D K WPH++E+RLR+ALG GDL LTSR++NT D K F FTFA H Y V++ISE+ +E
Sbjct: 142 DLKIWPHKFEYRLRVALGHDGDLTLTSRVKNT--DTKPFNFTFALHPYFAVSNISEIHVE 199
Query: 190 GLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLP 249
GL LDYLD KNR RFT+ ITF ++D++YLSTP ++ I+DH++K+T + K+G
Sbjct: 200 GLHNLDYLDQQKNRTRFTDHEKVITFNAQLDRLYLSTPDQLRIVDHKKKKTIVVHKEGQV 259
Query: 250 DAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSS 307
DAVVWNPWDKK +SDLG ++YKH + V+ A V K IT+ PG+EWKG +S VPS+
Sbjct: 260 DAVVWNPWDKK---VSDLGVEDYKHFVTVESAAVAKPITVNPGKEWKGILHVSVVPSN 314
>UniRef100_Q940G5 Possible apospory-associated like protein [Arabidopsis thaliana]
Length = 318
Score = 287 bits (735), Expect = 2e-76
Identities = 149/298 (50%), Positives = 199/298 (66%), Gaps = 21/298 (7%)
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYNCFDM 72
GVNGL+KII+R+ RG SAEVYLYGGQV+SWKNE GEELL +SSK F
Sbjct: 36 GVNGLDKIIIRDRRGRSAEVYLYGGQVSSWKNENGEELLVMSSKA-----------IFQP 84
Query: 73 NNNKCRLILLLQDLYAEAF---QYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSED 129
I +L Y+ + F++ + W ++ PPP P S S A VDL++++S +
Sbjct: 85 PTPIRGGIPVLFPQYSNTGPLPSHGFVRQRFWEVETKPPPLP--SLSTAHVDLIVRSSNE 142
Query: 130 DTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIE 189
D K WPH++E+RLR+ALG GDL LTSR++NT D K F FTFA H Y V++ISE+ +E
Sbjct: 143 DLKIWPHKFEYRLRVALGHDGDLTLTSRVKNT--DTKPFNFTFALHPYFAVSNISEIHVE 200
Query: 190 GLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLP 249
GL LDYLD KNR RFT+ ITF ++D++YLSTP ++ I+DH++K+T + K+G
Sbjct: 201 GLHNLDYLDQQKNRTRFTDHEKVITFNAQLDRLYLSTPDQLRIVDHKKKKTIVVHKEGQV 260
Query: 250 DAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSS 307
DAVVWNPWDKK +SDLG ++YK + V+ A V K IT+ PG+EWKG +S VPS+
Sbjct: 261 DAVVWNPWDKK---VSDLGVEDYKRFVTVESAAVAKPITVNPGKEWKGILHVSVVPSN 315
>UniRef100_Q9SVZ5 Possible apospory-associated like protein [Arabidopsis thaliana]
Length = 329
Score = 278 bits (712), Expect = 1e-73
Identities = 149/310 (48%), Positives = 199/310 (64%), Gaps = 33/310 (10%)
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQYNCFDM 72
GVNGL+KII+R+ RG SAEVYLYGGQV+SWKNE GEELL +SSK F
Sbjct: 35 GVNGLDKIIIRDRRGRSAEVYLYGGQVSSWKNENGEELLVMSSKA-----------IFQP 83
Query: 73 NNNKCRLILLLQDLYAEAF---QYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSED 129
I +L Y+ + F++ + W ++ PPP P S S A VDL++++S +
Sbjct: 84 PTPIRGGIPVLFPQYSNTGPLPSHGFVRQRFWEVETKPPPLP--SLSTAHVDLIVRSSNE 141
Query: 130 DTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIE 189
D K WPH++E+RLR+ALG GDL LTSR++NT D K F FTFA H Y V++ISE+ +E
Sbjct: 142 DLKIWPHKFEYRLRVALGHDGDLTLTSRVKNT--DTKPFNFTFALHPYFAVSNISEIHVE 199
Query: 190 GLETLDYLDSLKNRERFTEQGDAITFEGE------------VDKVYLSTPTKIAIIDHER 237
GL LDYLD KNR RFT+ ITF + +D++YLSTP ++ I+DH++
Sbjct: 200 GLHNLDYLDQQKNRTRFTDHEKVITFNAQSITKKKHTLQKKLDRLYLSTPDQLRIVDHKK 259
Query: 238 KRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKG 297
K+T + K+G DAVVWNPWDKK +SDLG ++YK + V+ A V K IT+ PG+EWKG
Sbjct: 260 KKTIVVHKEGQVDAVVWNPWDKK---VSDLGVEDYKRFVTVESAAVAKPITVNPGKEWKG 316
Query: 298 RQEISAVPSS 307
+S VPS+
Sbjct: 317 ILHVSVVPSN 326
>UniRef100_Q9SZG5 Possible apospory-associated like protein [Arabidopsis thaliana]
Length = 239
Score = 226 bits (575), Expect = 8e-58
Identities = 110/225 (48%), Positives = 152/225 (66%), Gaps = 19/225 (8%)
Query: 95 FLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLML 154
F++ + W ++ PPP P S S A VDL++++S +D K WPH++E+RLR+ALG GDL L
Sbjct: 19 FVRQRFWEVETKPPPLP--SLSTAHVDLIVRSSNEDLKIWPHKFEYRLRVALGHDGDLTL 76
Query: 155 TSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAIT 214
TSR++NT D K F FTFA H Y V++ISE+ +EGL LDYLD KNR RFT+ IT
Sbjct: 77 TSRVKNT--DTKPFNFTFALHPYFAVSNISEIHVEGLHNLDYLDQQKNRTRFTDHEKVIT 134
Query: 215 FEGE------------VDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAVVWNPWDKKSK 262
F + +D++YLSTP ++ I+DH++K+T + K+G DAVVWNPWDKK
Sbjct: 135 FNAQSITKKKHTLQKKLDRLYLSTPDQLRIVDHKKKKTIVVHKEGQVDAVVWNPWDKK-- 192
Query: 263 IISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSS 307
+SDLG ++YK + V+ A V K IT+ PG+EWKG +S VPS+
Sbjct: 193 -VSDLGVEDYKRFVTVESAAVAKPITVNPGKEWKGILHVSVVPSN 236
>UniRef100_Q6AUN2 Hypothetical protein OSJNBa0040E06.8 [Oryza sativa]
Length = 149
Score = 169 bits (428), Expect = 9e-41
Identities = 86/132 (65%), Positives = 101/132 (76%), Gaps = 3/132 (2%)
Query: 92 QYVFLKNKLWILDPNPPPFPTN-SNSRAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTG 150
Q+ F +N++W LD PP N +NS+A VDL+LK SEDD K WPH +EFRLR++L G
Sbjct: 20 QHGFARNRIWALDEEHPPLNQNDNNSKASVDLILKPSEDDLKCWPHGFEFRLRVSLTKDG 79
Query: 151 DLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQG 210
+L L SRIRN N GK F+F+F YHTYL V+DISEVRIEGLETLDYLD+L RERFTEQG
Sbjct: 80 NLSLVSRIRNVN--GKPFSFSFGYHTYLSVSDISEVRIEGLETLDYLDNLSQRERFTEQG 137
Query: 211 DAITFEGEVDKV 222
DAITFE EV V
Sbjct: 138 DAITFESEVKNV 149
>UniRef100_Q8H6K6 Apospory-associated-like protein [Brassica rapa subsp. pekinensis]
Length = 176
Score = 162 bits (411), Expect = 9e-39
Identities = 74/146 (50%), Positives = 107/146 (72%), Gaps = 2/146 (1%)
Query: 95 FLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDTKNWPHRYEFRLRIALGPTGDLML 154
F + + W +D +PPP P++ +S A VDL+LK+SE D+K WP+++ +RLR+ALG GDL+L
Sbjct: 32 FARQRFWEIDAHPPPLPSHPSSSAHVDLILKSSEADSKIWPNKFVYRLRVALGHGGDLIL 91
Query: 155 TSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAIT 214
TSR++N+ D K F FT H Y V+DIS++++E L+ LDYLD KNR RFT+ G IT
Sbjct: 92 TSRVKNS--DVKPFNFTVGLHPYFSVSDISKIQVEELQNLDYLDRQKNRTRFTDHGKFIT 149
Query: 215 FEGEVDKVYLSTPTKIAIIDHERKRT 240
F ++D++YL TP KI I+DH +K+T
Sbjct: 150 FSSQLDRLYLRTPKKIRIVDHNKKKT 175
>UniRef100_Q9M6D7 Apospory-associated protein C [Chlamydomonas reinhardtii]
Length = 338
Score = 127 bits (319), Expect = 4e-28
Identities = 98/315 (31%), Positives = 143/315 (45%), Gaps = 45/315 (14%)
Query: 13 GVNGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQY----- 67
G +G ++L+ G SAE+YL+G VTSWK G+E+L+V +P+ K+
Sbjct: 48 GKSGSPVVVLKHACGASAEMYLFGACVTSWKQPSGDEVLYV--RPDAVFDKSKPISGGVP 105
Query: 68 NCFDMNNNKCRLILLLQDLYAEAFQYVFLKNKLWIL-----DPNPPPFPTNSNSRAFVDL 122
+CF Q Q+ F +N W + DPNP V++
Sbjct: 106 HCFP------------QFGPGAMQQHGFARNLDWAVSTTSADPNP------DEKDPSVEV 147
Query: 123 VLKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTD 182
VL S+ K WPH+++ ++L L ++R N D K F FT A HTY+ V +
Sbjct: 148 VLTESDYTLKMWPHKFKAVYTVSLHGE---QLNLQLRVINTDDKPFDFTAALHTYIEVLE 204
Query: 183 ISEVRIEGLETLDYLDSL---KNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKR 239
I++ ++ GL+ L YL K E E DA+TF G D VYL++P + ++
Sbjct: 205 IAKAKVTGLKGLTYLCKAVDPKVPETKKEDRDAVTFTGYTDSVYLNSPAHVE-LEVGTGA 263
Query: 240 TFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQ 299
+ G D VVWNP D Y+H CV+ A KA T+KPGE W
Sbjct: 264 AVALDSTGWEDTVVWNPHLTMK--------DCYQHFCCVENAKFGKAATVKPGESWTATV 315
Query: 300 EISAVPSSYCSGQLD 314
+S V S L+
Sbjct: 316 TMSVVDVSKLKHDLE 330
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 590,312,014
Number of Sequences: 2790947
Number of extensions: 26138360
Number of successful extensions: 59588
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 81
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 59310
Number of HSP's gapped (non-prelim): 157
length of query: 322
length of database: 848,049,833
effective HSP length: 127
effective length of query: 195
effective length of database: 493,599,564
effective search space: 96251914980
effective search space used: 96251914980
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144503.2


