

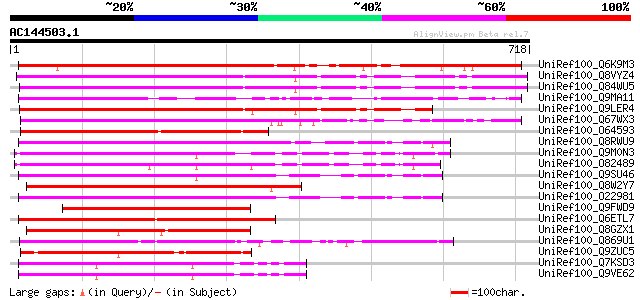
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.1 + phase: 0 /pseudo
(718 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6K9M3 Putative MAP4K alpha1 [Oryza sativa] 638 0.0
UniRef100_Q8VYZ4 Hypothetical protein At5g14720 [Arabidopsis tha... 551 e-155
UniRef100_Q84WU5 Hypothetical protein At5g14720 [Arabidopsis tha... 550 e-155
UniRef100_Q9MA11 F20B17.7 [Arabidopsis thaliana] 533 e-150
UniRef100_Q9LER4 Protein kinase-like protein [Arabidopsis thaliana] 532 e-149
UniRef100_Q67WX3 Putative oxidative-stress responsive [Oryza sat... 516 e-145
UniRef100_O64593 F17O7.3 protein [Arabidopsis thaliana] 486 e-135
UniRef100_Q8RWU9 Hypothetical protein At4g24100 [Arabidopsis tha... 483 e-135
UniRef100_Q9M0N3 Hypothetical protein AT4g10730 [Arabidopsis tha... 481 e-134
UniRef100_O82489 T12H20.4 protein [Arabidopsis thaliana] 480 e-134
UniRef100_Q9SU46 Hypothetical protein AT4g24100 [Arabidopsis tha... 478 e-133
UniRef100_Q8W2Y7 Putative Ste-20 related kinase, 3'-partial [Ory... 478 e-133
UniRef100_O22981 Ser/Thr protein kinase isolog [Arabidopsis thal... 446 e-124
UniRef100_Q9FWD9 Putative Ste20-related protein kinase [Oryza sa... 390 e-107
UniRef100_Q6ETL7 Putative oxidative-stress responsive 1 [Oryza s... 385 e-105
UniRef100_Q8GZX1 Putative Ste20-related protein kinase [Oryza sa... 382 e-104
UniRef100_Q869U1 Similar to Arabidopsis thaliana (Mouse-ear cres... 346 2e-93
UniRef100_Q9ZUC5 F5O8.25 protein [Arabidopsis thaliana] 321 4e-86
UniRef100_Q7KSD3 CG7693-PA, isoform A [Drosophila melanogaster] 313 9e-84
UniRef100_Q9VE62 Ste20-like protein kinase [Drosophila melanogas... 313 9e-84
>UniRef100_Q6K9M3 Putative MAP4K alpha1 [Oryza sativa]
Length = 761
Score = 638 bits (1645), Expect = 0.0
Identities = 366/763 (47%), Positives = 478/763 (61%), Gaps = 106/763 (13%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDL-------- 64
E++KYPI E Y+LYEEIGQGVSA V+R+LC +EIVA+K+LDFER N DL
Sbjct: 19 ERRKYPIRVEDYELYEEIGQGVSAIVYRSLCKPLDEIVAVKVLDFERTNSDLWLVVMQVG 78
Query: 65 --------------------------NNISREAQTMVLVDHPNVLKSHCSFVSDHNLWVV 98
NNI REAQTM+L+D PNV+K+HCSF ++H+LWVV
Sbjct: 79 YTRIVAIYVPPLDLSKMIVTRICLTQNNIMREAQTMILIDQPNVMKAHCSFTNNHSLWVV 138
Query: 99 MPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGHIHRDVKAGNVLIDSR 158
MP+M+GGSCLHI+K+ +PDGFEE VIATVLREVLKGLEYLHHHGHIHRDVKAGN+L+DSR
Sbjct: 139 MPYMAGGSCLHIMKSVYPDGFEEAVIATVLREVLKGLEYLHHHGHIHRDVKAGNILVDSR 198
Query: 159 GAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHGYNFKADIWSFGITALE 218
G VKLGDFGVSACLFDSGDRQR+RNTFVGTPCWMAPEVMEQLHGY+FKADIWSFGITALE
Sbjct: 199 GVVKLGDFGVSACLFDSGDRQRARNTFVGTPCWMAPEVMEQLHGYDFKADIWSFGITALE 258
Query: 219 LAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQMIACCLVKDPSKRPSASK 278
LAHGHAPFSK+PP+KVLLMTLQNAPPGLDYERDKKFS+ FKQM+A CLVKDPSKRP+A K
Sbjct: 259 LAHGHAPFSKFPPMKVLLMTLQNAPPGLDYERDKKFSRHFKQMVAMCLVKDPSKRPTAKK 318
Query: 279 LLKHSFFKQARSSDYITRTLLEGLPALGDRMEILKRKED-MLAQKKMPDGQMEELSQNEY 337
LLK FFKQARSSD+I+R LLEGLP LG R LK K++ +L+QKKMPDGQ EE+SQ+EY
Sbjct: 319 LLKQPFFKQARSSDFISRKLLEGLPGLGARYLALKEKDEVLLSQKKMPDGQKEEISQDEY 378
Query: 338 KRGISGWNFNLEDMKAQASLINDFDDAMSDISHVSSACSLTNLDAQDKQLPSSSH----- 392
KRGIS WNF+++D+K+QASLI + DD++S S A +LD + + H
Sbjct: 379 KRGISSWNFDMDDLKSQASLITECDDSIS--CKDSDASCFYDLDTILPERATGPHMSRVF 436
Query: 393 -----SRSQTADMSDDDSSITSSSHEPQTSSCLDDHVDHSLGDMENVGRAAEVVVATHPP 447
+ ++ MS+D S+++S P+ CL + ++ T
Sbjct: 437 SIKYDTDTENDVMSNDKSAVSS----PEHPICLARNTS---------------MLRTTNG 477
Query: 448 LHRRGCSSSILPEVTLPPIRAESLRNDREYFKH*CLSNFHH---SHSICSDWFFFNDKYS 504
+H G + + + D + S+FH S S CS F + K S
Sbjct: 478 VHANG---QVRKHSSTESSELDLQEKDSDAIPTSSFSSFHERKFSFSSCSSDGFLSSKES 534
Query: 505 NFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLTELPSRASKTS-ANSDDTDDK 563
+ + + NI++ +K V+ D+ E + K+S AN +D DD+
Sbjct: 535 SKHQI---NIHNRDKCNGGPLQVA-----------DEPSPEAVPKVPKSSAANVEDHDDR 580
Query: 564 AKVPVVQQRGRFKVTSENVDPEKATPSPVLQK------SHSMQVGCLEVCTPTNILQRVC 617
+K P++QQRGRFKVT +V+ +K +Q+ S+ + L + + ++
Sbjct: 581 SKPPLIQQRGRFKVTPGHVELDKDFQYRSIQELMPSVGSNIQAISHLPSLSIPSSIEAAS 640
Query: 618 *VLYGSILSP-----SANVQD*PSFVH--------DILTGNHSNFNEANYCG*IC*NIGY 664
++ GS+ N+ +H D+ + + A+ +
Sbjct: 641 TIIGGSLYMQLYNVLQTNMLQREQILHAMKQLSGCDMAMTSPACIAPASRASSPSSALSI 700
Query: 665 TLLQLESAHEREKELLHEITDLQWRLICTQEELQKLKTDNAQV 707
LE+AHE+EKEL++EIT+LQWRL+C+Q+E+Q+LK AQV
Sbjct: 701 DRSLLEAAHEKEKELVNEITELQWRLVCSQDEIQRLKAKAAQV 743
>UniRef100_Q8VYZ4 Hypothetical protein At5g14720 [Arabidopsis thaliana]
Length = 674
Score = 551 bits (1419), Expect = e-155
Identities = 330/717 (46%), Positives = 424/717 (59%), Gaps = 55/717 (7%)
Query: 10 KETEKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISR 69
+ + +KK+P+ A+ Y+LYEEIG GVSA+VHRALC+ N +VAIK+LD E+ N DL+ I R
Sbjct: 2 ESSSEKKFPLNAKDYKLYEEIGDGVSATVHRALCIPLNVVVAIKVLDLEKCNNDLDGIRR 61
Query: 70 EAQTMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLR 129
E QTM L++HPNVL++HCSF + H LWVVMP+M+GGSCLHI+K+++PDGFEE VIAT+LR
Sbjct: 62 EVQTMSLINHPNVLQAHCSFTTGHQLWVVMPYMAGGSCLHIIKSSYPDGFEEPVIATLLR 121
Query: 130 EVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTP 189
E LK L YLH HGHIHRDVKAGN+L+DS GAVKL DFGVSAC+FD+GDRQRSRNTFVGTP
Sbjct: 122 ETLKALVYLHAHGHIHRDVKAGNILLDSNGAVKLADFGVSACMFDTGDRQRSRNTFVGTP 181
Query: 190 CWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYE 249
CWMAPEVM+QLHGY+FKAD+WSFGITALELAHGHAPFSKYPP+KVLLMTLQNAPPGLDYE
Sbjct: 182 CWMAPEVMQQLHGYDFKADVWSFGITALELAHGHAPFSKYPPMKVLLMTLQNAPPGLDYE 241
Query: 250 RDKKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRM 309
RDK+FSK+FK+M+ CLVKDP KRP++ KLLKH FFK AR +DY+ +T+L GLP LGDR
Sbjct: 242 RDKRFSKAFKEMVGTCLVKDPKKRPTSEKLLKHPFFKHARPADYLVKTILNGLPPLGDRY 301
Query: 310 EILKRKE-DMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSDI 368
+K KE D+L Q K LSQ EY RGIS WNFNLED+K QA+LI+D D + ++
Sbjct: 302 RQIKSKEADLLMQNK--SEYEAHLSQQEYIRGISAWNFNLEDLKTQAALISDDDTSHAEE 359
Query: 369 SHVS-SACSLTNLDAQDKQLPSSSHSR----SQTADMSDDDSSITSSSHEP--QTSSCLD 421
+ C + A + SSS + + D+ D +SS S +P C D
Sbjct: 360 PDFNQKQCERQDESALSPERASSSATAPSQDDELNDIHDLESSFASFPIKPLQALKGCFD 419
Query: 422 DHVDHSLGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFKH* 481
D D + V + L + S+ A++ R
Sbjct: 420 ISEDE---DNATTPDWKDANVNSGQQLLTKASIGSLAETTKEEDTAAQNTSLPRHVISE- 475
Query: 482 CLSNFHHSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDD 541
+ S SI + S F+P ++ D E Q S + L D
Sbjct: 476 --QKKYLSGSIIPE--------STFSPKRITSDADREFQQRRYQTERSYSGSLYRTKRDS 525
Query: 542 VLTELPSRASKTSANSDDTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPVLQKSHSMQV 601
V D+ ++VP V+ +GRFKVTS ++ P+ +T S S
Sbjct: 526 V------------------DETSEVPHVEHKGRFKVTSADLSPKGSTNSTFTPFSGGTSS 567
Query: 602 GCLEVCTPTNILQRVC*VLYGSILSPSANVQD*PSFVHDILTGNHSNFNEANYCG*IC*N 661
T +IL + SIL +A ++ + + A G N
Sbjct: 568 PSCLNATTASILPSI-----QSILQQNAMQRE-----EILRLIKYLEQTSAKQPGSPETN 617
Query: 662 IGYTLLQLESAHEREKELLHEITDLQWRLICTQEELQKLKTDNAQVLS--NALASNN 716
+ LLQ A RE+EL ++ LQ EEL+K K N Q+ + NAL N
Sbjct: 618 VD-DLLQTPPATSRERELQSQVMLLQQSFSSLTEELKKQKQKNGQLENQLNALTHRN 673
>UniRef100_Q84WU5 Hypothetical protein At5g14720 [Arabidopsis thaliana]
Length = 674
Score = 550 bits (1417), Expect = e-155
Identities = 330/713 (46%), Positives = 422/713 (58%), Gaps = 55/713 (7%)
Query: 14 KKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQT 73
+KK+P+ A+ Y+LYEEIG GVSA+VHRALC+ N +VAIK+LD E+ N DL+ I RE QT
Sbjct: 6 EKKFPLNAKDYKLYEEIGDGVSATVHRALCIPLNVVVAIKVLDLEKCNNDLDGIRREVQT 65
Query: 74 MVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLK 133
M L++HPNVL++HCSF + H LWVVMP+M+GGSCLHI+K+++PDGFEE VIAT+LRE LK
Sbjct: 66 MSLINHPNVLQAHCSFTTGHQLWVVMPYMAGGSCLHIIKSSYPDGFEEPVIATLLRETLK 125
Query: 134 GLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMA 193
L YLH HGHIHRDVKAGN+L+DS GAVKL DFGVSAC+FD+GDRQRSRNTFVGTPCWMA
Sbjct: 126 ALVYLHAHGHIHRDVKAGNILLDSNGAVKLADFGVSACMFDTGDRQRSRNTFVGTPCWMA 185
Query: 194 PEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKK 253
PEVM+QLHGY+FKAD+WSFGITALELAHGHAPFSKYPP+KVLLMTLQNAPPGLDYERDK+
Sbjct: 186 PEVMQQLHGYDFKADVWSFGITALELAHGHAPFSKYPPMKVLLMTLQNAPPGLDYERDKR 245
Query: 254 FSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILK 313
FSK+FK+M+ CLVKDP KRP++ KLLKH FFK AR +DY+ +T+L GLP LGDR +K
Sbjct: 246 FSKAFKEMVGTCLVKDPKKRPTSEKLLKHPFFKHARPADYLVKTILNGLPPLGDRYRQIK 305
Query: 314 RKE-DMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSDISHVS 372
KE D+L Q K LSQ EY RGIS WNFNLED+K QA+LI+D D + ++ +
Sbjct: 306 SKEADLLMQNK--SEYEAHLSQQEYIRGISAWNFNLEDLKTQAALISDDDTSHAEEPDFN 363
Query: 373 -SACSLTNLDAQDKQLPSSSHSR----SQTADMSDDDSSITSSSHEP--QTSSCLDDHVD 425
C + A + SSS + + D+ D +SS S +P C D D
Sbjct: 364 QKQCERQDESALSPERASSSATAPSQDDELNDIHDLESSFASFPIKPLQALKGCFDISED 423
Query: 426 HSLGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFKH*CLSN 485
D + V + L + S+ A++ R
Sbjct: 424 E---DNATTPDWKDANVNSGQQLLTKASIGSLAETTKEEDTAAQNTSLPRHVISE---QK 477
Query: 486 FHHSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLTE 545
+ S SI + S F+P ++ D E Q S + L D V
Sbjct: 478 KYLSGSIIPE--------STFSPKRITSDADREFQQRRYQTERSYSGSLYRTKRDSV--- 526
Query: 546 LPSRASKTSANSDDTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPVLQKSHSMQVGCLE 605
D+ ++VP V+ +GRFKVTS ++ P+ +T S S
Sbjct: 527 ---------------DETSEVPHVEHKGRFKVTSADLSPKGSTNSTFTPFSGGTSSPSCL 571
Query: 606 VCTPTNILQRVC*VLYGSILSPSANVQD*PSFVHDILTGNHSNFNEANYCG*IC*NIGYT 665
T +IL + SIL +A ++ + + A G N+
Sbjct: 572 NATTASILPSI-----QSILQQNAMQRE-----EILRLIKYLEQTSAKQPGSPETNVD-D 620
Query: 666 LLQLESAHEREKELLHEITDLQWRLICTQEELQKLKTDNAQVLS--NALASNN 716
LLQ A RE+EL ++ LQ EEL+K K N Q+ + NAL N
Sbjct: 621 LLQTPPATSRERELQSQVMLLQQSFSSLTEELKKQKQKNGQLENQLNALTHRN 673
>UniRef100_Q9MA11 F20B17.7 [Arabidopsis thaliana]
Length = 547
Score = 533 bits (1372), Expect = e-150
Identities = 337/695 (48%), Positives = 397/695 (56%), Gaps = 153/695 (22%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQ 72
EKKKYPIG EHY LYE IGQGVSA VHRALC+ F+E+VAIKILDFERDNCDLNNISREAQ
Sbjct: 2 EKKKYPIGPEHYTLYEFIGQGVSALVHRALCIPFDEVVAIKILDFERDNCDLNNISREAQ 61
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVL 132
TM+LVDHPNVLKSHCSFVSDHNLWV+MP+MSGGSCLHILKAA+PDGFEE +IAT+LRE L
Sbjct: 62 TMMLVDHPNVLKSHCSFVSDHNLWVIMPYMSGGSCLHILKAAYPDGFEEAIIATILREAL 121
Query: 133 KGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWM 192
KGL+YLH HGHIHRDVKAGN+L+ +RGAVKLGDFGVSACLFDSGDRQR+RNTFVGTPCW
Sbjct: 122 KGLDYLHQHGHIHRDVKAGNILLGARGAVKLGDFGVSACLFDSGDRQRTRNTFVGTPCW- 180
Query: 193 APEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDK 252
ADIWSFGIT LELAHGHAPFSKYPP+
Sbjct: 181 --------------ADIWSFGITGLELAHGHAPFSKYPPM-------------------- 206
Query: 253 KFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEIL 312
KSFKQMIA CLVKDPSKRPSA KLLKHSFFKQARSSDYI R LL+GLP L +R++ +
Sbjct: 207 ---KSFKQMIASCLVKDPSKRPSAKKLLKHSFFKQARSSDYIARKLLDGLPDLVNRVQAI 263
Query: 313 KRKEDMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSDISHVS 372
K NEYKRGISG + + ++A D D S+I +
Sbjct: 264 K---------------------NEYKRGISGLSGSATSLQAL-----DSQDTQSEIQEDT 297
Query: 373 SACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSSCLDDHVDHSLGDME 432
+TN Q P S S D SDDDSS+ S S++ S H D SL +
Sbjct: 298 G--QITNKYLQ----PLIHRSLSIARDKSDDDSSLASPSYDSYVYSS-PRHEDLSLNNTH 350
Query: 433 NVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFKH*CLSNFHHSHSI 492
V +TH + ++SI I S+ D
Sbjct: 351 --------VGSTHANNGKPTDATSIPTNQPTEIIAGSSVLAD------------------ 384
Query: 493 CSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLTELPSRASK 552
N P N +S+K Q N S+ N T GDDV TE+ + K
Sbjct: 385 -----------GNGAP----NKGESDKTQEQLQNGSNCNGTHPTVGGDDVPTEMAVKPPK 429
Query: 553 TSANSDDTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPVLQKSHSMQVGCLEVCTPTNI 612
+++ D++DDK+K PVVQQRGRFKVTSEN+D EK VL + S + +V P
Sbjct: 430 AASSLDESDDKSKPPVVQQRGRFKVTSENLDIEK-----VLCQHSSASLPHSDVTLPNLT 484
Query: 613 LQRVC*VLYGSILSPSANVQD*PSFVHDILTGNHSNFNEANYCG*IC*NIGYTLLQLESA 672
V ++Y P +IL N+ Y +C QL +
Sbjct: 485 SSYVYPLVY-------------PVLQTNILERMDVQLNKEVYNNLLC-------PQLRN- 523
Query: 673 HEREKELLHEITDLQWRLICTQEELQKLKTDNAQV 707
W LIC +EELQK KT++AQV
Sbjct: 524 --------------PW-LICAEEELQKYKTEHAQV 543
>UniRef100_Q9LER4 Protein kinase-like protein [Arabidopsis thaliana]
Length = 561
Score = 532 bits (1371), Expect = e-149
Identities = 296/589 (50%), Positives = 373/589 (63%), Gaps = 51/589 (8%)
Query: 14 KKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQT 73
+KK+P+ A+ Y+LYEEIG GVSA+VHRALC+ N +VAIK+LD E+ N DL+ I RE QT
Sbjct: 6 EKKFPLNAKDYKLYEEIGDGVSATVHRALCIPLNVVVAIKVLDLEKCNNDLDGIRREVQT 65
Query: 74 MVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLK 133
M L++HPNVL++HCSF + H LWVVMP+M+GGSCLHI+K+++PDGFEE VIAT+LRE LK
Sbjct: 66 MSLINHPNVLQAHCSFTTGHQLWVVMPYMAGGSCLHIIKSSYPDGFEEPVIATLLRETLK 125
Query: 134 GLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMA 193
L YLH HGHIHRDVKAGN+L+DS GAVKL DFGVSAC+FD+GDRQRSRNTFVGTPCWMA
Sbjct: 126 ALVYLHAHGHIHRDVKAGNILLDSNGAVKLADFGVSACMFDTGDRQRSRNTFVGTPCWMA 185
Query: 194 PEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKK 253
PEVM+QLHGY+FKAD+WSFGITALELAHGHAPFSKYPP+KVLLMTLQNAPPGLDYERDK+
Sbjct: 186 PEVMQQLHGYDFKADVWSFGITALELAHGHAPFSKYPPMKVLLMTLQNAPPGLDYERDKR 245
Query: 254 FSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILK 313
FSK+FK+M+ CLVKDP KRP++ KLLKH FFK AR +DY+ +T+L GLP LGDR +K
Sbjct: 246 FSKAFKEMVGTCLVKDPKKRPTSEKLLKHPFFKHARPADYLVKTILNGLPPLGDRYRQIK 305
Query: 314 RKE-DMLAQKKMPDGQMEELSQ---------NEYKRGISGWNFNLEDMKAQASLINDFDD 363
KE D+L Q K LSQ EY RGIS WNFNLED+K QA+LI+D D
Sbjct: 306 SKEADLLMQNK--SEYEAHLSQARISTFCLIQEYIRGISAWNFNLEDLKTQAALISDDDT 363
Query: 364 AMSDISHVS-SACSLTNLDAQDKQLPSSSHSR----SQTADMSDDDSSITSSSHEP--QT 416
+ ++ + C + A + SSS + + D+ D +SS S +P
Sbjct: 364 SHAEEPDFNQKQCERQDESALSPERASSSATAPSQDDELNDIHDLESSFASFPIKPLQAL 423
Query: 417 SSCLDDHVDHSLGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDRE 476
C D D D + V + L + S+ A++ R
Sbjct: 424 KGCFDISEDE---DNATTPDWKDANVNSGQQLLTKASIGSLAETTKEEDTAAQNTSLPRH 480
Query: 477 YFKH*CLSNFHHSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVT 536
+ S SI + S F+P ++ D E Q S + L
Sbjct: 481 VISE---QKKYLSGSIIPE--------STFSPKRITSDADREFQQRRYQTERSYSGSLYR 529
Query: 537 HTGDDVLTELPSRASKTSANSDDTDDKAKVPVVQQRGRFKVTSENVDPE 585
D V D+ ++VP V+ +GRFKVTS ++ P+
Sbjct: 530 TKRDSV------------------DETSEVPHVEHKGRFKVTSADLSPK 560
>UniRef100_Q67WX3 Putative oxidative-stress responsive [Oryza sativa]
Length = 693
Score = 516 bits (1330), Expect = e-145
Identities = 315/737 (42%), Positives = 426/737 (57%), Gaps = 114/737 (15%)
Query: 15 KKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTM 74
+++P + Y+L EE+G GVSA+V++ALC+ N VAIK+LD E+ + DL+ I RE QTM
Sbjct: 5 RRFPTDPKEYKLCEEVGDGVSATVYKALCIPLNIEVAIKVLDLEKCSNDLDGIRREVQTM 64
Query: 75 VLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKG 134
L+DHPN+L+++CSF + H LWV+MP+M+ GS LHI+K + PDGFEE VIAT+LREVLK
Sbjct: 65 SLIDHPNLLRAYCSFTNGHQLWVIMPYMAAGSALHIMKTSFPDGFEEPVIATLLREVLKA 124
Query: 135 LEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAP 194
L YLH GHIHRDVKAGN+LID+ GAVKLGDFGVSAC+FD+G+RQR+RNTFVGTPCWMAP
Sbjct: 125 LVYLHSQGHIHRDVKAGNILIDTNGAVKLGDFGVSACMFDTGNRQRARNTFVGTPCWMAP 184
Query: 195 EVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKF 254
EVM+QLHGY++KADIWSFGITALELAHGHAPFSKYPP+KVLLMTLQNAPPGLDYERDK+F
Sbjct: 185 EVMQQLHGYDYKADIWSFGITALELAHGHAPFSKYPPMKVLLMTLQNAPPGLDYERDKRF 244
Query: 255 SKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILKR 314
SKSFK ++A CLVKDP KRPS+ KLLKHSFFK AR+++++ R++L+GLP LG+R LK
Sbjct: 245 SKSFKDLVATCLVKDPRKRPSSEKLLKHSFFKHARTAEFLARSILDGLPPLGERFRTLKG 304
Query: 315 KE-DMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLIND------------- 360
KE D+L K+ E+LSQ EY RGISGWNFNLED+K A+LI++
Sbjct: 305 KEADLLLSNKLGSESKEQLSQKEYIRGISGWNFNLEDLKNAAALIDNTNGTCHLDGVNSK 364
Query: 361 FDDAMSDISH-----------VSSA---CSLTNLDAQDKQLPSSSHSRSQTADMS----- 401
F D + + + V+SA + ++ D L SS SR A S
Sbjct: 365 FKDGLQEANEPENIYQGRANLVASARPEDEIQEVEDLDGALASSFPSRPLEALKSCFDVC 424
Query: 402 --DDDSSITSSSHEPQTSSC--------LDDHVDHSLGDMENVGRAAEVVVATHPPLHRR 451
DD + T +P S +++H + E++ R+A V +
Sbjct: 425 GDDDPPTATDLREQPNMESTSPMQQFQQIENHKSANCNG-ESLERSASVPSNLVNSGSHK 483
Query: 452 GCSSSILPEVTLPPIRAESLRNDREYFKH*CLSNFHHSHSICSDWFFFNDKYSNFNPLIF 511
S S++PE L P R ++ ND N H + C+ N + +F
Sbjct: 484 FLSGSLIPEHVLSPYR--NVGNDP-------ARNECHQKNTCN---------RNRSGPLF 525
Query: 512 SNINDSEKLQNLSTNVSSANAILVTHTGDDVLTELPSRASKTSANSDDTDDKAKVPVVQQ 571
+ D LP + S V+Q+
Sbjct: 526 RQMKDPR-------------------------AHLPVEPEEQSEGK----------VIQR 550
Query: 572 RGRFKVTSENVDPEKATPSPVLQKSHSMQVGCL-EVCTPTNILQRVC*VLYGSILSPSAN 630
RGRF+VTS+++ +K S + ++ +G P+ IL + ++ + +
Sbjct: 551 RGRFQVTSDSI-AQKVASSASSSRCSNLPIGVTRSTVHPSTILPTLQFMIQQNTMQKEV- 608
Query: 631 VQD*PSFVHDILTGNHSNFNEANYCG*IC*NIGYTLLQLESAHEREKELLHEITDLQWRL 690
+ S + +I S+ +A+ G Q H REKEL I +LQ +
Sbjct: 609 ISRLISSIEEI-----SDAADASTTG---------SSQPSGVHFREKELQSYIANLQQSV 654
Query: 691 ICTQEELQKLKTDNAQV 707
EE+Q+LK N Q+
Sbjct: 655 TELAEEVQRLKLKNTQL 671
>UniRef100_O64593 F17O7.3 protein [Arabidopsis thaliana]
Length = 553
Score = 486 bits (1250), Expect = e-135
Identities = 227/344 (65%), Positives = 286/344 (82%), Gaps = 4/344 (1%)
Query: 15 KKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTM 74
K++P+ A+ Y+L+EE+G+GVSA+V+RA C++ NEIVA+KILD E+ DL I +E M
Sbjct: 7 KRFPLYAKDYELFEEVGEGVSATVYRARCIALNEIVAVKILDLEKCRNDLETIRKEVHIM 66
Query: 75 VLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKG 134
L+DHPN+LK+HCSF+ +LW+VMP+MSGGSC H++K+ +P+G E+ +IAT+LREVLK
Sbjct: 67 SLIDHPNLLKAHCSFIDSSSLWIVMPYMSGGSCFHLMKSVYPEGLEQPIIATLLREVLKA 126
Query: 135 LEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAP 194
L YLH GHIHRDVKAGN+LI S+G VKLGDFGVSAC+FDSG+R ++RNTFVGTPCWMAP
Sbjct: 127 LVYLHRQGHIHRDVKAGNILIHSKGVVKLGDFGVSACMFDSGERMQTRNTFVGTPCWMAP 186
Query: 195 EVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKF 254
EVM+QL GY+FK +WSFGITALELAHGHAPFSKYPP+KVLLMTLQNAPP LDY+RDKKF
Sbjct: 187 EVMQQLDGYDFK--LWSFGITALELAHGHAPFSKYPPMKVLLMTLQNAPPRLDYDRDKKF 244
Query: 255 SKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILKR 314
SKSF+++IA CLVKDP KRP+A+KLLKH FFK ARS+DY++R +L GL LG+R + LK
Sbjct: 245 SKSFRELIAACLVKDPKKRPTAAKLLKHPFFKHARSTDYLSRKILHGLSPLGERFKKLKE 304
Query: 315 KEDMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLI 358
E L K +G E+LSQ+EY RGIS WNF+LE ++ QASL+
Sbjct: 305 AEAELF--KGINGDKEQLSQHEYMRGISAWNFDLEALRRQASLV 346
>UniRef100_Q8RWU9 Hypothetical protein At4g24100 [Arabidopsis thaliana]
Length = 709
Score = 483 bits (1244), Expect = e-135
Identities = 280/610 (45%), Positives = 366/610 (59%), Gaps = 55/610 (9%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQ 72
+++ + + + Y+L EEIG G SA V+RA+ + NE+VAIK LD +R N +L++I RE+Q
Sbjct: 22 QQRGFSMNPKDYKLMEEIGHGASAVVYRAIYLPTNEVVAIKCLDLDRCNSNLDDIRRESQ 81
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVL 132
TM L+DHPNV+KS CSF DH+LWVVMPFM+ GSCLH++K A+ DGFEE I VL+E L
Sbjct: 82 TMSLIDHPNVIKSFCSFSVDHSLWVVMPFMAQGSCLHLMKTAYSDGFEESAICCVLKETL 141
Query: 133 KGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWM 192
K L+YLH GHIHRDVKAGN+L+D G +KLGDFGVSACLFD+GDRQR+RNTFVGTPCWM
Sbjct: 142 KALDYLHRQGHIHRDVKAGNILLDDNGEIKLGDFGVSACLFDNGDRQRARNTFVGTPCWM 201
Query: 193 APEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDK 252
APEV++ +GYN KADIWSFGITALELAHGHAPFSKYPP+KVLLMT+QNAPPGLDY+RDK
Sbjct: 202 APEVLQPGNGYNSKADIWSFGITALELAHGHAPFSKYPPMKVLLMTIQNAPPGLDYDRDK 261
Query: 253 KFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEIL 312
KFSKSFK+M+A CLVKD +KRP+A KLLKHS FK + + + L LP L R++ L
Sbjct: 262 KFSKSFKEMVAMCLVKDQTKRPTAEKLLKHSCFKHTKPPEQTVKILFSDLPPLWTRVKSL 321
Query: 313 KRKE-DMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSDISHV 371
+ K+ LA K+M E +SQ+EY+RG+S WNF++ D+K QASL+ D DD
Sbjct: 322 QDKDAQQLALKRMATADEEAISQSEYQRGVSAWNFDVRDLKTQASLLIDDDDLEESKEDE 381
Query: 372 SSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSSCLDDHVDHSLGDM 431
C+ N +Q+ S Q + + ++++ E T
Sbjct: 382 EILCAQFNKVNDREQVFDS----LQLYENMNGKEKVSNTEVEEPTC-------------- 423
Query: 432 ENVGRAAEVVVATHPPLHRRGCSSS-ILPEVTLPPIRAESLRNDREYFKH*CLSNFHHSH 490
+ V T L R +S +PE + P+R +S + HS
Sbjct: 424 ----KEKFTFVTTTSSLERMSPNSEHDIPEAKVKPLRRQSQSGPLT-----SRTVLSHSA 474
Query: 491 SICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLTELPSRA 550
S S F ++ P + + S L NLST SS L +
Sbjct: 475 SEKSHIFERSESEPQTAPTVRRAPSFSGPL-NLSTRASS--------------NSLSAPI 519
Query: 551 SKTSANSDDTDDKAKVPVVQQRGRFKVTSENVD----------PEKATPSPVLQKSHSMQ 600
+ D DDK+K +V Q+GRF VTS NVD P ++ + L+KS S+
Sbjct: 520 KYSGGFRDSLDDKSKANLV-QKGRFSVTSGNVDLAKDVPLSIVPRRSPQATPLRKSASVG 578
Query: 601 VGCLEVCTPT 610
LE PT
Sbjct: 579 NWILEPKMPT 588
>UniRef100_Q9M0N3 Hypothetical protein AT4g10730 [Arabidopsis thaliana]
Length = 693
Score = 481 bits (1238), Expect = e-134
Identities = 280/620 (45%), Positives = 371/620 (59%), Gaps = 92/620 (14%)
Query: 7 DKEKETEKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNN 66
DK+K KK + + + Y+L EE+G G SA VHRA+ + NE+VAIK LD +R N +L++
Sbjct: 13 DKKK---KKGFSVNPKDYKLMEEVGYGASAVVHRAIYLPTNEVVAIKSLDLDRCNSNLDD 69
Query: 67 ISREAQTMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIAT 126
I REAQTM L+DHPNV+KS CSF DH+LWVVMPFM+ GSCLH++KAA+PDGFEE I +
Sbjct: 70 IRREAQTMTLIDHPNVIKSFCSFAVDHHLWVVMPFMAQGSCLHLMKAAYPDGFEEAAICS 129
Query: 127 VLREVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFV 186
+L+E LK L+YLH GHIHRDVKAGN+L+D G +KLGDFGVSACLFD+GDRQR+RNTFV
Sbjct: 130 MLKETLKALDYLHRQGHIHRDVKAGNILLDDTGEIKLGDFGVSACLFDNGDRQRARNTFV 189
Query: 187 GTPCWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGL 246
GTPCWMAPEV++ GYN KADIWSFGITALELAHGHAPFSKYPP+KVLLMT+QNAPPGL
Sbjct: 190 GTPCWMAPEVLQPGSGYNSKADIWSFGITALELAHGHAPFSKYPPMKVLLMTIQNAPPGL 249
Query: 247 DYERDKKFSK-------SFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLL 299
DY+RDKKFSK SFK+++A CLVKD +KRP+A KLLKHSFFK + + + L
Sbjct: 250 DYDRDKKFSKKMSSCAQSFKELVALCLVKDQTKRPTAEKLLKHSFFKNVKPPEICVKKLF 309
Query: 300 EGLPALGDRMEILKRKEDMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLIN 359
LP L R++ L Q+EY+RG+S WNFN+ED+K QASL++
Sbjct: 310 VDLPPLWTRVKAL---------------------QSEYQRGVSAWNFNIEDLKEQASLLD 348
Query: 360 DFDDAMSDISHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSSC 419
D DD +++ S ++ +QL + + R Q + S + S + + +
Sbjct: 349 D-DDILTE--------SREEEESFGEQLHNKVNDRGQVS-----GSQLLSENMNGKEKAS 394
Query: 420 LDDHVDHSLGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFK 479
+ V+ + + V S +P+ P+R ++
Sbjct: 395 DTEVVEPICEEKSTLNSTTSSVE------QPASSSEQDVPQAKGKPVRLQT--------- 439
Query: 480 H*CLSNFHHSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTG 539
HS + S N + SE + L ++V A + +G
Sbjct: 440 --------HSGPLSSGVVLINSDSEKVH-----GYERSESERQLKSSVRRAPSF----SG 482
Query: 540 DDVLTELPSRASKTSANS---------DDTDDKAKVPVVQQRGRFKVTSENVDPEKATPS 590
LP+RAS S ++ D DDK+K VVQ +GRF VTSEN+D +A+P
Sbjct: 483 P---LNLPNRASANSLSAPIKSSGGFRDSIDDKSKANVVQIKGRFSVTSENLDLARASP- 538
Query: 591 PVLQKSHSMQVGCLEVCTPT 610
L+KS S+ L+ PT
Sbjct: 539 --LRKSASVGNWILDSKMPT 556
>UniRef100_O82489 T12H20.4 protein [Arabidopsis thaliana]
Length = 607
Score = 480 bits (1236), Expect = e-134
Identities = 279/621 (44%), Positives = 372/621 (58%), Gaps = 83/621 (13%)
Query: 7 DKEKETEKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNN 66
DK+K KK + + + Y+L EE+G G SA VHRA+ + NE+VAIK LD +R N +L++
Sbjct: 13 DKKK---KKGFSVNPKDYKLMEEVGYGASAVVHRAIYLPTNEVVAIKSLDLDRCNSNLDD 69
Query: 67 ISREAQTMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIAT 126
I REAQTM L+DHPNV+KS CSF DH+LWVVMPFM+ GSCLH++KAA+PDGFEE I +
Sbjct: 70 IRREAQTMTLIDHPNVIKSFCSFAVDHHLWVVMPFMAQGSCLHLMKAAYPDGFEEAAICS 129
Query: 127 VLREVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFV 186
+L+E LK L+YLH GHIHRDVKAGN+L+D G +KLGDFGVSACLFD+GDRQR+RNTFV
Sbjct: 130 MLKETLKALDYLHRQGHIHRDVKAGNILLDDTGEIKLGDFGVSACLFDNGDRQRARNTFV 189
Query: 187 GTPCW---------MAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLM 237
GTPCW MAPEV++ GYN KADIWSFGITALELAHGHAPFSKYPP+KVLLM
Sbjct: 190 GTPCWLVTVVFSFRMAPEVLQPGSGYNSKADIWSFGITALELAHGHAPFSKYPPMKVLLM 249
Query: 238 TLQNAPPGLDYERDKKFSK-------SFKQMIACCLVKDPSKRPSASKLLKHSFFKQARS 290
T+QNAPPGLDY+RDKKFSK SFK+++A CLVKD +KRP+A KLLKHSFFK +
Sbjct: 250 TIQNAPPGLDYDRDKKFSKKMSSCAQSFKELVALCLVKDQTKRPTAEKLLKHSFFKNVKP 309
Query: 291 SDYITRTLLEGLPALGDRMEILKRKE-DMLAQKKMPDGQMEELS-----QNEYKRGISGW 344
+ + L LP L R++ L+ K+ LA K M + +S Q+EY+RG+S W
Sbjct: 310 PEICVKKLFVDLPPLWTRVKALQAKDAAQLALKGMASADQDAISQVTNAQSEYQRGVSAW 369
Query: 345 NFNLEDMKAQASLINDFDDAMSDISHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSDDD 404
NFN+ED+K QASL++D DD +++ S ++ +QL + + R Q +
Sbjct: 370 NFNIEDLKEQASLLDD-DDILTE--------SREEEESFGEQLHNKVNDRGQVS-----G 415
Query: 405 SSITSSSHEPQTSSCLDDHVDHSLGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLP 464
S + S + + + + V+ + + V S +P+
Sbjct: 416 SQLLSENMNGKEKASDTEVVEPICEEKSTLNSTTSSVE------QPASSSEQDVPQAKGK 469
Query: 465 PIRAESLRNDREYFKH*CLSNFHHSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLS 524
P+R ++ HS + S N + SE + L
Sbjct: 470 PVRLQT-----------------HSGPLSSGVVLINSDSEKVH-----GYERSESERQLK 507
Query: 525 TNVSSANAILVTHTGDDVLTELPSRASKTSANS---------DDTDDKAKVPVVQQRGRF 575
++V A + +G LP+RAS S ++ D DDK+K VVQ +GRF
Sbjct: 508 SSVRRAPSF----SGP---LNLPNRASANSLSAPIKSSGGFRDSIDDKSKANVVQIKGRF 560
Query: 576 KVTSENVDPEKATPSPVLQKS 596
VTSEN+D + T + + S
Sbjct: 561 SVTSENLDLARPTGQAIKESS 581
>UniRef100_Q9SU46 Hypothetical protein AT4g24100 [Arabidopsis thaliana]
Length = 710
Score = 478 bits (1231), Expect = e-133
Identities = 271/594 (45%), Positives = 355/594 (59%), Gaps = 73/594 (12%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQ 72
+++ + + + Y+L EEIG G SA V+RA+ + NE+VAIK LD +R N +L++I RE+Q
Sbjct: 22 QQRGFSMNPKDYKLMEEIGHGASAVVYRAIYLPTNEVVAIKCLDLDRCNSNLDDIRRESQ 81
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVL 132
TM L+DHPNV+KS CSF DH+LWVVMPFM+ GSCLH++K A+ DGFEE I VL+E L
Sbjct: 82 TMSLIDHPNVIKSFCSFSVDHSLWVVMPFMAQGSCLHLMKTAYSDGFEESAICCVLKETL 141
Query: 133 KGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWM 192
K L+YLH GHIHRDVKAGN+L+D G +KLGDFGVSACLFD+GDRQR+RNTFVGTPCWM
Sbjct: 142 KALDYLHRQGHIHRDVKAGNILLDDNGEIKLGDFGVSACLFDNGDRQRARNTFVGTPCWM 201
Query: 193 APEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDK 252
APEV++ +GYN KADIWSFGITALELAHGHAPFSKYPP+KVLLMT+QNAPPGLDY+RDK
Sbjct: 202 APEVLQPGNGYNSKADIWSFGITALELAHGHAPFSKYPPMKVLLMTIQNAPPGLDYDRDK 261
Query: 253 KFSK-------SFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPAL 305
KFSK SFK+M+A CLVKD +KRP+A KLLKHS FK + + + L LP L
Sbjct: 262 KFSKRTFSCAQSFKEMVAMCLVKDQTKRPTAEKLLKHSCFKHTKPPEQTVKILFSDLPPL 321
Query: 306 GDRMEILKRKE-DMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDA 364
R++ L+ K+ LA K+M E +SQ+EY+RG+S WNF++ D+K QASL+ND +
Sbjct: 322 WTRVKSLQDKDAQQLALKRMATADEEAISQSEYQRGVSAWNFDVRDLKTQASLVNDREQV 381
Query: 365 MSDISHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSSCLDDHV 424
+ QL + + + + ++ ++ + T+S L+
Sbjct: 382 FDSL-----------------QLYENMNGKEKVSNTEVEEPTCKEKFTFVTTTSSLERMS 424
Query: 425 DHSLGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFKH*CLS 484
+S D +PE + P+R +S +
Sbjct: 425 PNSEHD---------------------------IPEAKVKPLRRQSQSGPLT-----SRT 452
Query: 485 NFHHSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLT 544
HS S S F ++ P + + S L NLST SS
Sbjct: 453 VLSHSASEKSHIFERSESEPQTAPTVRRAPSFSGPL-NLSTRASS--------------N 497
Query: 545 ELPSRASKTSANSDDTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPVLQKSHS 598
L + + D DDK+K +V Q+GRF VTS NVD K T P K HS
Sbjct: 498 SLSAPIKYSGGFRDSLDDKSKANLV-QKGRFSVTSGNVDLAKPTAQPQTIKEHS 550
>UniRef100_Q8W2Y7 Putative Ste-20 related kinase, 3'-partial [Oryza sativa]
Length = 541
Score = 478 bits (1231), Expect = e-133
Identities = 233/389 (59%), Positives = 293/389 (74%), Gaps = 9/389 (2%)
Query: 24 YQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLVDHPNVL 83
YQL EE+G G A V+RAL V N++VA+K LD ++ N +++ I REAQ M L++HPNV+
Sbjct: 26 YQLMEEVGYGAHAVVYRALFVPRNDVVAVKCLDLDQLNNNIDEIQREAQIMSLIEHPNVI 85
Query: 84 KSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGH 143
+++CSFV +H+LWVVMPFM+ GSCLH++K A+PDGFEE VI ++L+E LK LEYLH G
Sbjct: 86 RAYCSFVVEHSLWVVMPFMTEGSCLHLMKIAYPDGFEEPVIGSILKETLKALEYLHRQGQ 145
Query: 144 IHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVMEQLHGY 203
IHRDVKAGN+L+D+ G VKLGDFGVSAC+FD GDRQRSRNTFVGTPCWMAPEV++ GY
Sbjct: 146 IHRDVKAGNILVDNAGIVKLGDFGVSACMFDRGDRQRSRNTFVGTPCWMAPEVLQPGTGY 205
Query: 204 NFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKKFSKSFKQMIA 263
NFKADIWSFGITALELAHGHAPFSKYPP+KVLLMTLQNAPPGLDY+RD++FSKSFK+M+A
Sbjct: 206 NFKADIWSFGITALELAHGHAPFSKYPPMKVLLMTLQNAPPGLDYDRDRRFSKSFKEMVA 265
Query: 264 CCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILKRKE-DMLAQK 322
CLVKD +KRP+A KLLKHSFFK A+ + + +L LP L DR++ L+ K+ LA K
Sbjct: 266 MCLVKDQTKRPTAEKLLKHSFFKNAKPPELTMKGILTDLPPLWDRVKALQLKDAAQLALK 325
Query: 323 KMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLIND--------FDDAMSDISHVSSA 374
KMP + E LS +EY+RG+S WNF++ED+KAQASLI D DD I +
Sbjct: 326 KMPSSEQEALSMSEYQRGVSAWNFDVEDLKAQASLIRDDEPPEIKEDDDTARTIEVEKDS 385
Query: 375 CSLTNLDAQDKQLPSSSHSRSQTADMSDD 403
S +L + + R+ T + D
Sbjct: 386 FSRNHLGKSSSTIENFFSGRTSTTAANSD 414
>UniRef100_O22981 Ser/Thr protein kinase isolog [Arabidopsis thaliana]
Length = 572
Score = 446 bits (1148), Expect = e-124
Identities = 260/587 (44%), Positives = 342/587 (57%), Gaps = 66/587 (11%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQ 72
+++ + + + Y+L EEIG G SA V+RA+ + NE+VAIK LD +R N +L++I RE+Q
Sbjct: 22 QQRGFSMNPKDYKLMEEIGHGASAVVYRAIYLPTNEVVAIKCLDLDRCNSNLDDIRRESQ 81
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVL 132
TM L+DHPNV+KS CSF DH+LWVVMPFM+ GSCLH++K A+ DGFEE I VL+E L
Sbjct: 82 TMSLIDHPNVIKSFCSFSVDHSLWVVMPFMAQGSCLHLMKTAYSDGFEESAICCVLKETL 141
Query: 133 KGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWM 192
K L+YLH GHIHRDVKAGN+L+D G +KLGDFGVSACLFD+GDRQR+RNTFVGTPCWM
Sbjct: 142 KALDYLHRQGHIHRDVKAGNILLDDNGEIKLGDFGVSACLFDNGDRQRARNTFVGTPCWM 201
Query: 193 APEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDK 252
APEV++ +GYN KADIWSFGITALELAHGHAPFSKYPP+KVLLMT+QNAPPGLDY+RDK
Sbjct: 202 APEVLQPGNGYNSKADIWSFGITALELAHGHAPFSKYPPMKVLLMTIQNAPPGLDYDRDK 261
Query: 253 KFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEIL 312
KFSKSFK+M+A CLVKD +KRP+A KLLKHS FK + + + L LP L R++ L
Sbjct: 262 KFSKSFKEMVAMCLVKDQTKRPTAEKLLKHSCFKHTKPPEQTVKILFSDLPPLWTRVKSL 321
Query: 313 KRKE-DMLAQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSDISHV 371
+ K+ LA K+M E +SQ + + E + AQ + +ND + +
Sbjct: 322 QDKDAQQLALKRMATADEEAISQLIDDDDLEESKEDEEILCAQFNKVNDREQVFDSL--- 378
Query: 372 SSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSSSHEPQTSSCLDDHVDHSLGDM 431
QL + + + + ++ ++ + T+S L+ +S D
Sbjct: 379 --------------QLYENMNGKEKVSNTEVEEPTCKEKFTFVTTTSSLERMSPNSEHD- 423
Query: 432 ENVGRAAEVVVATHPPLHRRGCSSSILPEVTLPPIRAESLRNDREYFKH*CLSNFHHSHS 491
+PE + P+R +S + HS S
Sbjct: 424 --------------------------IPEAKVKPLRRQSQSGPLT-----SRTVLSHSAS 452
Query: 492 ICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGDDVLTELPSRAS 551
S F ++ P + + S L NLST SS L +
Sbjct: 453 EKSHIFERSESEPQTAPTVRRAPSFSGPL-NLSTRASS--------------NSLSAPIK 497
Query: 552 KTSANSDDTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPVLQKSHS 598
+ D DDK+K +V Q+GRF VTS NVD K T P K HS
Sbjct: 498 YSGGFRDSLDDKSKANLV-QKGRFSVTSGNVDLAKPTAQPQTIKEHS 543
>UniRef100_Q9FWD9 Putative Ste20-related protein kinase [Oryza sativa]
Length = 643
Score = 390 bits (1003), Expect = e-107
Identities = 182/261 (69%), Positives = 220/261 (83%), Gaps = 1/261 (0%)
Query: 74 MVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLK 133
M L++HPNV++++CSFV +H+LWVVMPFM+ GSCLH++K A+PDGFEE VI ++L+E LK
Sbjct: 1 MSLIEHPNVIRAYCSFVVEHSLWVVMPFMTEGSCLHLMKIAYPDGFEEPVIGSILKETLK 60
Query: 134 GLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMA 193
LEYLH G IHRDVKAGN+L+D+ G VKLGDFGVSAC+FD GDRQRSRNTFVGTPCWMA
Sbjct: 61 ALEYLHRQGQIHRDVKAGNILVDNAGIVKLGDFGVSACMFDRGDRQRSRNTFVGTPCWMA 120
Query: 194 PEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDKK 253
PEV++ GYNFKADIWSFGITALELAHGHAPFSKYPP+KVLLMTLQNAPPGLDY+RD++
Sbjct: 121 PEVLQPGTGYNFKADIWSFGITALELAHGHAPFSKYPPMKVLLMTLQNAPPGLDYDRDRR 180
Query: 254 FSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEILK 313
FSKSFK+M+A CLVKD +KRP+A KLLKHSFFK A+ + + +L LP L DR++ L+
Sbjct: 181 FSKSFKEMVAMCLVKDQTKRPTAEKLLKHSFFKNAKPPELTMKGILTDLPPLWDRVKALQ 240
Query: 314 RKE-DMLAQKKMPDGQMEELS 333
K+ LA KKMP + E LS
Sbjct: 241 LKDAAQLALKKMPSSEQEALS 261
Score = 44.3 bits (103), Expect = 0.013
Identities = 58/201 (28%), Positives = 86/201 (41%), Gaps = 34/201 (16%)
Query: 546 LPSRASKTSANS---------DDTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPV---- 592
LP+RAS S ++ D DK+K VV+ +GRF VTSENVD K P+
Sbjct: 426 LPTRASANSLSAPIRSSGGYVDSLGDKSKRNVVEIKGRFSVTSENVDLAKVQEVPLSSLS 485
Query: 593 --------LQKSHSM---QVGCLEVCTPTNILQRVC*VLYGSILSPS-ANVQD*PSFVHD 640
L+KS S+ V + ++ + + SIL P N+ F D
Sbjct: 486 RKSPQASPLKKSASVGDWLVNTKPMSNSHHVKELCNSSVSSSILIPHLENLVKQTMFQQD 545
Query: 641 ILTGNHSNFNEANYCG*IC*NIGYTLLQLES-------AHEREKELLHEITDLQWRLICT 693
++ S+ + + I L +S E+E+ LL +I++LQ R+I
Sbjct: 546 LIMNVLSSLQQNEKVDGVLSGISPQLRNTDSDTMVGSVNSEKERSLLVKISELQSRMITL 605
Query: 694 QEEL--QKLKTDNAQVLSNAL 712
+EL KLK Q NAL
Sbjct: 606 TDELIAAKLKHVQLQQELNAL 626
>UniRef100_Q6ETL7 Putative oxidative-stress responsive 1 [Oryza sativa]
Length = 534
Score = 385 bits (988), Expect = e-105
Identities = 192/359 (53%), Positives = 257/359 (71%), Gaps = 5/359 (1%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFN-EIVAIKILDF-ERDNCDLNNISRE 70
++ ++P A+ Y+L E +G G +A V RA C+ E+VA+KI++ +R D+N+ S E
Sbjct: 22 KRDQFPSRAKDYELLEPVGDGATAVVRRARCLPLGGEVVAVKIMNMSQRSEDDVNHASEE 81
Query: 71 AQTMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLRE 130
+ M +DH N+L ++CSF LW++MP+M+GGSC H++K+++P GF+E IA VLRE
Sbjct: 82 VKMMSTIDHDNLLGAYCSFTEGETLWIIMPYMAGGSCFHLMKSSYPKGFDEKFIAFVLRE 141
Query: 131 VLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSG-DRQRSRNTFVGTP 189
L+GL YLH + +HRDVKAGN+L+D VKL DFG SA L+D +R R T VGTP
Sbjct: 142 TLEGLAYLHRYALVHRDVKAGNILLDQHKGVKLADFGASASLYDPMINRHGKRKTLVGTP 201
Query: 190 CWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYE 249
CWMAPEVMEQ Y+ KADIWSFGITALELAHGHAPFS PP KV L+TLQ+APP L
Sbjct: 202 CWMAPEVMEQKE-YDAKADIWSFGITALELAHGHAPFSTQPPAKVFLLTLQHAPPSLHNT 260
Query: 250 RDKKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRM 309
+DKKFSKSFKQMIA CL+KDPSKRP+A LL+ FFK+ + D + +++L LP+LGDRM
Sbjct: 261 KDKKFSKSFKQMIATCLMKDPSKRPTAQHLLELPFFKKVKFEDNVLKSVLNKLPSLGDRM 320
Query: 310 EILKRKEDML-AQKKMPDGQMEELSQNEYKRGISGWNFNLEDMKAQASLINDFDDAMSD 367
+ ++ E L A+KK D E+ SQ+EY RG+S WNF++E++KAQA+L D +D D
Sbjct: 321 QSIQENEAKLQAEKKPLDKCKEKASQDEYMRGVSEWNFDIEELKAQAALYPDENDGGED 379
>UniRef100_Q8GZX1 Putative Ste20-related protein kinase [Oryza sativa]
Length = 645
Score = 382 bits (982), Expect = e-104
Identities = 194/325 (59%), Positives = 243/325 (74%), Gaps = 18/325 (5%)
Query: 24 YQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMVLVDHPNVL 83
Y+L EE+G G +A V+RA+ + N VA+K LD +R N +L++I +EAQTM L+DHPNV+
Sbjct: 32 YRLLEEVGYGANAVVYRAVFLPSNRTVAVKCLDLDRVNSNLDDIRKEAQTMSLIDHPNVI 91
Query: 84 KSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGLEYLHHHGH 143
+++CSFV DHNLWV+MPFMS GSCLH++K A+PDGFEE VIA++L+E LK LEYLH GH
Sbjct: 92 RAYCSFVVDHNLWVIMPFMSEGSCLHLMKVAYPDGFEEPVIASILKETLKALEYLHRQGH 151
Query: 144 IHRDVK-----AGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWMAPEVME 198
IHRDVK AGN+L+DS G VKLGDFGVSAC+FD GDRQRSRNTFVGTPCWMAPEV++
Sbjct: 152 IHRDVKRNIIQAGNILMDSPGIVKLGDFGVSACMFDRGDRQRSRNTFVGTPCWMAPEVLQ 211
Query: 199 QLHGYNFKAD---------IWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYE 249
GYNFK IW F I+ L ++ + K KVLLMTLQNAPPGLDY+
Sbjct: 212 PGAGYNFKKYVSNHLFTNLIWLFKIS---LRGKNSNYHKNTGNKVLLMTLQNAPPGLDYD 268
Query: 250 RDKKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRM 309
RDK+FSKSFK+M+A CLVKD +KRP+A KLLKHSFFK A+ + +++L LP L DR+
Sbjct: 269 RDKRFSKSFKEMVAMCLVKDQTKRPTAEKLLKHSFFKNAKPPELTVKSILTDLPPLWDRV 328
Query: 310 EILKRKE-DMLAQKKMPDGQMEELS 333
+ L+ K+ LA KKMP + E LS
Sbjct: 329 KALQLKDAAQLALKKMPSSEQEALS 353
Score = 42.0 bits (97), Expect = 0.064
Identities = 51/176 (28%), Positives = 72/176 (39%), Gaps = 34/176 (19%)
Query: 546 LPSRASKTSANSD---------DTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPVLQKS 596
LP+RAS S ++ DK+K VV+ +GRF VTSENVD K P S
Sbjct: 468 LPTRASANSLSAPIRSSGGYVGSLGDKSKRSVVEIKGRFSVTSENVDLAKVQEVPTSGIS 527
Query: 597 HSMQVGCLEVCTPTNILQRVC*VLYGSILSPSANVQD*P--SFVHDILTGNHSNFNEANY 654
+Q GS L SA+V P + D++T S+ +
Sbjct: 528 RKLQ--------------------EGSSLRKSASVGHWPVDAKPMDLITNLLSSLQQNEK 567
Query: 655 CG*IC*NIGYTLLQLE---SAHEREKELLHEITDLQWRLICTQEELQKLKTDNAQV 707
+G E S E E+ LL +I +LQ R+I +EL K + Q+
Sbjct: 568 ADATQYRLGNMDGDTEVETSISEGERSLLVKIFELQSRMISLTDELITTKLQHVQL 623
>UniRef100_Q869U1 Similar to Arabidopsis thaliana (Mouse-ear cress). hypothetical
75.4 kDa protein [Dictyostelium discoideum]
Length = 1028
Score = 346 bits (887), Expect = 2e-93
Identities = 225/614 (36%), Positives = 321/614 (51%), Gaps = 70/614 (11%)
Query: 14 KKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFER-DNCDLNNISREAQ 72
+K YP A+ Y+L E IG+G S V RA+C+ F E VAIKI+D E N L I +E Q
Sbjct: 61 RKNYPNSADQYELKETIGKGGSGLVQRAICLPFQENVAIKIIDLEHCKNVSLEEIRKEIQ 120
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVL 132
M L HPNV+ H SFV + +LWV+M F+S GSC I++ + P GFEE VIAT+L+E L
Sbjct: 121 AMSLCHHPNVVAYHTSFVYNESLWVIMDFLSAGSCSDIMRFSFPQGFEEHVIATILKEAL 180
Query: 133 KGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGDRQRSRNTFVGTPCWM 192
K + Y H G IHRD+K+GN+LIDS G ++L DFGVSA L D+G+ SRNTFVGTPCWM
Sbjct: 181 KAICYFHKTGRIHRDIKSGNILIDSNGNIQLSDFGVSATLIDTGET--SRNTFVGTPCWM 238
Query: 193 APEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPGLDYERDK 252
APE+MEQ++ Y++ DIWSFGITALELA G APF++YPP+KVLL+TLQN PP L+ + +
Sbjct: 239 APEIMEQVN-YDYAVDIWSFGITALELARGKAPFAEYPPMKVLLLTLQNPPPSLEGDGES 297
Query: 253 KFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPALGDRMEIL 312
K+S SFK ++ CL KDPSKRP SKLL+H FFKQA+ DY+ + +L LP LG R ++L
Sbjct: 298 KWSHSFKDLVEKCLQKDPSKRPLPSKLLEHRFFKQAKKPDYLVQHILAKLPPLGQRYQML 357
Query: 313 KRKEDMLAQKKMPDGQMEELSQNEYKRGISG----WNFNLEDMKAQASLINDFDDAMSDI 368
ED A + S ++ G S W F E+ D+ S
Sbjct: 358 --SEDSFA-------MLRNTSSPQFDTGHSNSADEWIFPNENN----------DNNNSST 398
Query: 369 SHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSD-DDSSITSSSHEPQTSSCLDDHVDHS 427
+ ++ + TN PS++++ + + S + SH P TS
Sbjct: 399 TTTTTTTTTTN--------PSNNNNNNNNKTKENLTQSPFETPSHTPSTSP--------- 441
Query: 428 LGDMENVGRAAEVVVATHPPLHRRGCSSSILPEVTLP-------PIRAESLRNDREYFKH 480
G + R + + H L G SS+++P +P P+ A + ++
Sbjct: 442 -GSTPSHSRTS-TPTSNHTAL---GSSSTVVPPPVVPLTLPTAIPVTAAAYHQQQQ---- 492
Query: 481 *CLSNFHHSHSICSDWFFFNDKYSNFNPLIFSNINDSEKLQNLSTNVSSANAILVTHTGD 540
HH S S ++ SN SN++ ++ S+A+ +V +T
Sbjct: 493 ------HHHLSHSSGSIPNHNASSNLGASAHSNVHGLAH-SSIHPTSSAASTTVVNNTQQ 545
Query: 541 DVLTELPSRASKTSANSDDTDDKAKVPVVQQRGRFKVTSENVDPEKATPSPVLQKSHSMQ 600
+ P + + + +V+ +T P+K P S
Sbjct: 546 PQTLQPPQQQHQLQQPIKTPSPINAINIVKLNQSDLITPPKTSPKKEGSIP--SSSSHGN 603
Query: 601 VGCLEVCTPTNILQ 614
+ L +P + LQ
Sbjct: 604 IPSLVTTSPKSPLQ 617
>UniRef100_Q9ZUC5 F5O8.25 protein [Arabidopsis thaliana]
Length = 480
Score = 321 bits (823), Expect = 4e-86
Identities = 172/329 (52%), Positives = 231/329 (69%), Gaps = 23/329 (6%)
Query: 16 KYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQTMV 75
++P+ A+ Y++ EEIG GV +RA C+ +EIVAIKI + E+ DL I +E +
Sbjct: 8 RFPLVAKDYEILEEIGDGV----YRARCILLDEIVAIKIWNLEKCTNDLETIRKEVHRLS 63
Query: 76 LVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDGFEEVVIATVLREVLKGL 135
L+DHPN+L+ HCSF+ +LW+VMPFMS GS L+I+K+ +P+G EE VIA +LRE+LK L
Sbjct: 64 LIDHPNLLRVHCSFIDSSSLWIVMPFMSCGSSLNIMKSVYPNGLEEPVIAILLREILKAL 123
Query: 136 EYLHHHGHIHRDVK-------AGNVLIDSRGAVKLGDFGVSACLFDSGDRQR--SRNTFV 186
YLH GHIHR+VK AGNVL+DS G VKLGDF VSA +FDS +R R S NTFV
Sbjct: 124 VYLHGLGHIHRNVKVLLISIHAGNVLVDSEGTVKLGDFEVSASMFDSVERMRTSSENTFV 183
Query: 187 GTPCWMAPEV-MEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPPG 245
G P MAPE M+Q+ GY+FK DIWSFG+TALELAHGH+P + VL + LQN+ P
Sbjct: 184 GNPRRMAPEKDMQQVDGYDFKVDIWSFGMTALELAHGHSPTT------VLPLNLQNS-PF 236
Query: 246 LDYERDKKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLLEGLPAL 305
+YE D KFSKSF++++A CL++DP KRP+AS+LL++ F +Q S++Y+ T L+GL L
Sbjct: 237 PNYEEDTKFSKSFRELVAACLIEDPEKRPTASQLLEYPFLQQTLSTEYLASTFLDGLSPL 296
Query: 306 GDRMEILKRKEDMLAQKKMPDGQMEELSQ 334
G+R LK ++ L K DG E++SQ
Sbjct: 297 GERYRKLKEEKAKLV--KGVDGNKEKVSQ 323
>UniRef100_Q7KSD3 CG7693-PA, isoform A [Drosophila melanogaster]
Length = 552
Score = 313 bits (803), Expect = 9e-84
Identities = 180/413 (43%), Positives = 245/413 (58%), Gaps = 38/413 (9%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQ 72
EK +P + Y+L + IG G +A VH A C+ NE AIK ++ E+ N ++ + +E Q
Sbjct: 27 EKYTWPNSKDDYELRDVIGVGATAVVHGAYCIPRNEKCAIKRINLEKWNTSMDELLKEIQ 86
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDG------FEEVVIAT 126
M H NV+ H SFV LW+V+ + GGS L I+K F+E IAT
Sbjct: 87 AMSSCFHENVVTYHTSFVVREELWLVLRLLEGGSLLDIIKHKMRTSNCKQGVFDEATIAT 146
Query: 127 VLREVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGD--RQRSRNT 184
VL+EVLKGLEY H +G IHRD+KAGN+LI G +++ DFGVSA L D RQ+ R+T
Sbjct: 147 VLKEVLKGLEYFHSNGQIHRDIKAGNILIGDDGTIQIADFGVSAWLATGRDLSRQKVRHT 206
Query: 185 FVGTPCWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPP 244
FVGTPCWMAPEVMEQ HGY+FKADIWSFGITA+E+A G AP+ KYPP+KVL++TLQN PP
Sbjct: 207 FVGTPCWMAPEVMEQDHGYDFKADIWSFGITAIEMATGTAPYHKYPPMKVLMLTLQNDPP 266
Query: 245 GLDYERD-----KKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLL 299
LD D K + K+F++MI CL K+PSKRP+AS+LLKH+FFK+A+ Y+T+TLL
Sbjct: 267 TLDTGADDKDQYKAYGKTFRKMIVECLQKEPSKRPTASELLKHAFFKKAKDRKYLTQTLL 326
Query: 300 EGLPALGDRMEILKRKEDMLAQKKMP--DGQMEELSQNEYKRGISGWNFNLEDMKAQASL 357
+ P++ R+ A K+ P G++ E+ W+ ED A+
Sbjct: 327 QSGPSMETRVH--------KAAKRQPGASGRLHRTVTGEWV-----WSSEEEDNGGSAT- 372
Query: 358 INDFDDAMSDISHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSS 410
D H SS + D++D+ + + S +D + IT S
Sbjct: 373 ----GSGTGDRKHPSS-----DSDSEDRPMNRLERADSSDSDREEPSPEITHS 416
>UniRef100_Q9VE62 Ste20-like protein kinase [Drosophila melanogaster]
Length = 552
Score = 313 bits (803), Expect = 9e-84
Identities = 180/413 (43%), Positives = 245/413 (58%), Gaps = 38/413 (9%)
Query: 13 EKKKYPIGAEHYQLYEEIGQGVSASVHRALCVSFNEIVAIKILDFERDNCDLNNISREAQ 72
EK +P + Y+L + IG G +A VH A C+ NE AIK ++ E+ N ++ + +E Q
Sbjct: 27 EKYTWPNSKDDYELRDVIGVGATAVVHGAYCIPRNEKCAIKRINLEKWNTSMDELLKEIQ 86
Query: 73 TMVLVDHPNVLKSHCSFVSDHNLWVVMPFMSGGSCLHILKAAHPDG------FEEVVIAT 126
M H NV+ H SFV LW+V+ + GGS L I+K F+E IAT
Sbjct: 87 AMSSCFHENVVTYHTSFVVREELWLVLRLLEGGSLLDIIKHKMRTSNCKQGVFDEATIAT 146
Query: 127 VLREVLKGLEYLHHHGHIHRDVKAGNVLIDSRGAVKLGDFGVSACLFDSGD--RQRSRNT 184
VL+EVLKGLEY H +G IHRD+KAGN+LI G +++ DFGVSA L D RQ+ R+T
Sbjct: 147 VLKEVLKGLEYFHSNGQIHRDIKAGNILIGDDGTIQIADFGVSAWLATGRDLSRQKVRHT 206
Query: 185 FVGTPCWMAPEVMEQLHGYNFKADIWSFGITALELAHGHAPFSKYPPLKVLLMTLQNAPP 244
FVGTPCWMAPEVMEQ HGY+FKADIWSFGITA+E+A G AP+ KYPP+KVL++TLQN PP
Sbjct: 207 FVGTPCWMAPEVMEQDHGYDFKADIWSFGITAIEMATGTAPYHKYPPMKVLMLTLQNDPP 266
Query: 245 GLDYERD-----KKFSKSFKQMIACCLVKDPSKRPSASKLLKHSFFKQARSSDYITRTLL 299
LD D K + K+F++MI CL K+PSKRP+AS+LLKH+FFK+A+ Y+T+TLL
Sbjct: 267 TLDTGADDKDQYKAYGKTFRKMIVECLQKEPSKRPTASELLKHAFFKKAKDRKYLTQTLL 326
Query: 300 EGLPALGDRMEILKRKEDMLAQKKMP--DGQMEELSQNEYKRGISGWNFNLEDMKAQASL 357
+ P++ R+ A K+ P G++ E+ W+ ED A+
Sbjct: 327 QSGPSMETRVH--------KAAKRQPGASGRLHRTVTGEWV-----WSSEEEDNGGSAT- 372
Query: 358 INDFDDAMSDISHVSSACSLTNLDAQDKQLPSSSHSRSQTADMSDDDSSITSS 410
D H SS + D++D+ + + S +D + IT S
Sbjct: 373 ----GSGTGDRKHPSS-----DSDSEDRPMNRLERADSSDSDREEPSPEITHS 416
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,192,534,203
Number of Sequences: 2790947
Number of extensions: 49541926
Number of successful extensions: 223650
Number of sequences better than 10.0: 18683
Number of HSP's better than 10.0 without gapping: 11579
Number of HSP's successfully gapped in prelim test: 7105
Number of HSP's that attempted gapping in prelim test: 173450
Number of HSP's gapped (non-prelim): 23680
length of query: 718
length of database: 848,049,833
effective HSP length: 135
effective length of query: 583
effective length of database: 471,271,988
effective search space: 274751569004
effective search space used: 274751569004
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC144503.1


