

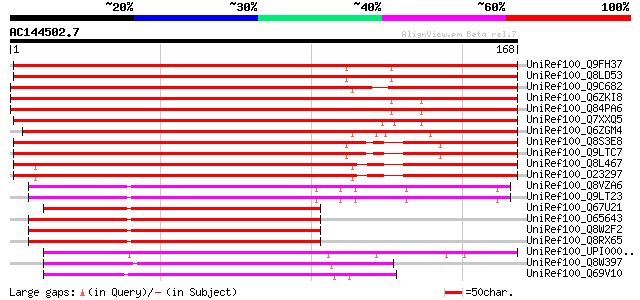
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144502.7 - phase: 2
(168 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FH37 Arabidopsis thaliana genomic DNA, chromosome 5,... 278 4e-74
UniRef100_Q8LD53 BHLH transcription factor, putative [Arabidopsi... 276 1e-73
UniRef100_Q9C682 BHLH transcription factor, putative [Arabidopsi... 234 8e-61
UniRef100_Q6ZKI8 Helix-loop-helix-like protein [Oryza sativa] 214 7e-55
UniRef100_Q84PA6 Helix-loop-helix-like protein [Oryza sativa] 214 7e-55
UniRef100_Q7XXQ5 Hypothetical protein [Oryza sativa] 214 7e-55
UniRef100_Q6ZGM4 Putative bHLH protein [Oryza sativa] 177 7e-44
UniRef100_Q8S3E8 Putative bHLH transcription factor [Arabidopsis... 170 1e-41
UniRef100_Q9LTC7 Emb|CAB10220.1 [Arabidopsis thaliana] 170 1e-41
UniRef100_Q8L467 AT4g14410/dl3245w [Arabidopsis thaliana] 166 2e-40
UniRef100_O23297 Hypothetical protein dl3245w [Arabidopsis thali... 166 2e-40
UniRef100_Q8VZA6 Hypothetical protein At3g19860; MPN9.10 [Arabid... 102 5e-21
UniRef100_Q9LT23 Emb|CAA18500.1 [Arabidopsis thaliana] 102 5e-21
UniRef100_Q67U21 Basic helix-loop-helix-like protein [Oryza sativa] 92 5e-18
UniRef100_O65643 Putative Myc-type transcription factor [Arabido... 84 1e-15
UniRef100_Q8W2F2 Putative transcription factor BHLH11 [Arabidops... 84 1e-15
UniRef100_Q8RX65 AT4g36060/T19K4_190 [Arabidopsis thaliana] 84 1e-15
UniRef100_UPI00002C6501 UPI00002C6501 UniRef100 entry 84 2e-15
UniRef100_Q8W397 Putative amelogenin [Oryza sativa] 69 4e-11
UniRef100_Q69V10 Amelogenin-like protein [Oryza sativa] 68 1e-10
>UniRef100_Q9FH37 Arabidopsis thaliana genomic DNA, chromosome 5, TAC clone:K5F14
[Arabidopsis thaliana]
Length = 234
Score = 278 bits (711), Expect = 4e-74
Identities = 142/169 (84%), Positives = 152/169 (89%), Gaps = 2/169 (1%)
Query: 2 RSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGE 61
R ES +ATSSKACREK RRDRLNDKF+ELG+ILEPG P KTDKAAIL+DAVRMVTQLRGE
Sbjct: 66 RCESSSATSSKACREKQRRDRLNDKFMELGAILEPGNPPKTDKAAILVDAVRMVTQLRGE 125
Query: 62 AQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAP-PSFLPTPTAL 120
AQKLKD+NS LQ+KIKELK EKNELRDEKQRLK EKEKLEQQLK+MNAP PSF P P +
Sbjct: 126 AQKLKDSNSSLQDKIKELKTEKNELRDEKQRLKTEKEKLEQQLKAMNAPQPSFFPAPPMM 185
Query: 121 PAAFA-AQGQAHGNKLVPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
P AFA AQGQA GNK+VP ISYPGVAMWQFMPPA+VDTSQDHVLRPPVA
Sbjct: 186 PTAFASAQGQAPGNKMVPIISYPGVAMWQFMPPASVDTSQDHVLRPPVA 234
>UniRef100_Q8LD53 BHLH transcription factor, putative [Arabidopsis thaliana]
Length = 234
Score = 276 bits (707), Expect = 1e-73
Identities = 141/169 (83%), Positives = 152/169 (89%), Gaps = 2/169 (1%)
Query: 2 RSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGE 61
R ES +ATSSKACREK RRDRLNDKF+ELG+ILEPG P KTDKAAIL+DAVRMVTQLRGE
Sbjct: 66 RCESSSATSSKACREKQRRDRLNDKFMELGAILEPGNPPKTDKAAILVDAVRMVTQLRGE 125
Query: 62 AQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAP-PSFLPTPTAL 120
AQKLKD+NS LQ+KIKELK EKNELRDEKQRLK EKEKLEQQLK++NAP PSF P P +
Sbjct: 126 AQKLKDSNSSLQDKIKELKTEKNELRDEKQRLKTEKEKLEQQLKAINAPQPSFFPAPPMM 185
Query: 121 PAAFA-AQGQAHGNKLVPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
P AFA AQGQA GNK+VP ISYPGVAMWQFMPPA+VDTSQDHVLRPPVA
Sbjct: 186 PTAFASAQGQAPGNKMVPIISYPGVAMWQFMPPASVDTSQDHVLRPPVA 234
>UniRef100_Q9C682 BHLH transcription factor, putative [Arabidopsis thaliana]
Length = 226
Score = 234 bits (596), Expect = 8e-61
Identities = 119/172 (69%), Positives = 136/172 (78%), Gaps = 9/172 (5%)
Query: 1 VRSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRG 60
+++ESC ++SKACREK RRDRLNDKF EL S+LEPGR KTDK AI+ DA+RMV Q R
Sbjct: 60 IKTESCTGSNSKACREKQRRDRLNDKFTELSSVLEPGRTPKTDKVAIINDAIRMVNQARD 119
Query: 61 EAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPS----FLPT 116
EAQKLKD NS LQEKIKELK EKNELRDEKQ+LK EKE+++QQLK++ P FLP
Sbjct: 120 EAQKLKDLNSSLQEKIKELKDEKNELRDEKQKLKVEKERIDQQLKAIKTQPQPQPCFLPN 179
Query: 117 PTALPAAFAAQGQAHGNKLVPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
P L +Q QA G+KLVPF +YPG AMWQFMPPAAVDTSQDHVLRPPVA
Sbjct: 180 PQTL-----SQAQAPGSKLVPFTTYPGFAMWQFMPPAAVDTSQDHVLRPPVA 226
>UniRef100_Q6ZKI8 Helix-loop-helix-like protein [Oryza sativa]
Length = 253
Score = 214 bits (545), Expect = 7e-55
Identities = 108/178 (60%), Positives = 134/178 (74%), Gaps = 10/178 (5%)
Query: 1 VRSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRG 60
VRS SC +SKA REK+RRD++ND+F+ELG+ LEPG+P K+DKAAIL DA RMV QLR
Sbjct: 76 VRSGSCGRPTSKASREKIRRDKMNDRFLELGTTLEPGKPVKSDKAAILSDATRMVIQLRA 135
Query: 61 EAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPSFLPTPTAL 120
EA++LKD N L++KIKELK EK+ELRDEKQ+LK EKE LEQQ+K + A P+++P PT +
Sbjct: 136 EAKQLKDTNESLEDKIKELKAEKDELRDEKQKLKVEKETLEQQVKILTATPAYMPHPTLM 195
Query: 121 PAAFA---------AQGQAHGNKL-VPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
PA + AQGQA G KL +PF+ YPG MWQFMPP+ VDTS+D PPVA
Sbjct: 196 PAPYPQAPLAPFHHAQGQAAGQKLMMPFVGYPGYPMWQFMPPSEVDTSKDSEACPPVA 253
>UniRef100_Q84PA6 Helix-loop-helix-like protein [Oryza sativa]
Length = 216
Score = 214 bits (545), Expect = 7e-55
Identities = 108/178 (60%), Positives = 134/178 (74%), Gaps = 10/178 (5%)
Query: 1 VRSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRG 60
VRS SC +SKA REK+RRD++ND+F+ELG+ LEPG+P K+DKAAIL DA RMV QLR
Sbjct: 39 VRSGSCGRPTSKASREKIRRDKMNDRFLELGTTLEPGKPVKSDKAAILSDATRMVIQLRA 98
Query: 61 EAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPSFLPTPTAL 120
EA++LKD N L++KIKELK EK+ELRDEKQ+LK EKE LEQQ+K + A P+++P PT +
Sbjct: 99 EAKQLKDTNESLEDKIKELKAEKDELRDEKQKLKVEKETLEQQVKILTATPAYMPHPTLM 158
Query: 121 PAAFA---------AQGQAHGNKL-VPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
PA + AQGQA G KL +PF+ YPG MWQFMPP+ VDTS+D PPVA
Sbjct: 159 PAPYPQAPLAPFHHAQGQAAGQKLMMPFVGYPGYPMWQFMPPSEVDTSKDSEACPPVA 216
>UniRef100_Q7XXQ5 Hypothetical protein [Oryza sativa]
Length = 256
Score = 214 bits (545), Expect = 7e-55
Identities = 111/170 (65%), Positives = 135/170 (79%), Gaps = 3/170 (1%)
Query: 2 RSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGE 61
RSES SSKACREK+RRD+LN++F+ELG++LEPG+ K DK++IL DA+R++ +LR E
Sbjct: 87 RSESGTRPSSKACREKVRRDKLNERFLELGAVLEPGKTPKMDKSSILNDAIRVMAELRSE 146
Query: 62 AQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPSFLPTPTALP 121
AQKLK++N LQEKIKELK EKNELRDEKQ+LKAEKE LEQQ+K +NA PSF+P P +P
Sbjct: 147 AQKLKESNESLQEKIKELKAEKNELRDEKQKLKAEKESLEQQIKFLNARPSFVPHPPVIP 206
Query: 122 A-AFAA-QGQAHGNKL-VPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
A AF A QGQA G KL +P I YPG MWQFMPP+ VDT+ D PPVA
Sbjct: 207 ASAFTAPQGQAAGQKLMMPVIGYPGFPMWQFMPPSDVDTTDDTKSCPPVA 256
>UniRef100_Q6ZGM4 Putative bHLH protein [Oryza sativa]
Length = 236
Score = 177 bits (450), Expect = 7e-44
Identities = 98/173 (56%), Positives = 125/173 (71%), Gaps = 9/173 (5%)
Query: 5 SCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQK 64
S + SKACREK+RRDRLND+F+EL S++ P + AK DKA IL DA R++ +LRGEA+K
Sbjct: 64 SSSGPKSKACREKIRRDRLNDRFLELSSVINPDKQAKLDKANILSDAARLLAELRGEAEK 123
Query: 65 LKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPS----FLPTPTAL 120
LK++N L+E IK+LKVEKNELRDEK LKAEKE+LEQQ+K+++ P+ LP P A
Sbjct: 124 LKESNEKLRETIKDLKVEKNELRDEKVTLKAEKERLEQQVKALSVAPTGFVPHLPHPAAF 183
Query: 121 -PAA---FAAQGQAHGNKLVPF-ISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
PAA F QA GNK P ++ G+AMWQ++PP AVDT+QD L PP A
Sbjct: 184 HPAAFPPFIPPYQALGNKNAPTPAAFQGMAMWQWLPPTAVDTTQDPKLWPPNA 236
>UniRef100_Q8S3E8 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 291
Score = 170 bits (430), Expect = 1e-41
Identities = 95/172 (55%), Positives = 120/172 (69%), Gaps = 13/172 (7%)
Query: 2 RSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGE 61
R+ SC+ +KACREKLRR++LNDKF++L S+LEPGR KTDK+AIL DA+R+V QLRGE
Sbjct: 128 RTGSCSKPGTKACREKLRREKLNDKFMDLSSVLEPGRTPKTDKSAILDDAIRVVNQLRGE 187
Query: 62 AQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAP-PSFLPTPTAL 120
A +L++ N L E+IK LK +KNELR+EK LKAEKEK+EQQLKSM P P F+P+
Sbjct: 188 AHELQETNQKLLEEIKSLKADKNELREEKLVLKAEKEKMEQQLKSMVVPSPGFMPSQH-- 245
Query: 121 PAAFAAQGQAHGNKLVPFISY----PGVAMWQFMPPAAVDTSQDHVLRPPVA 168
PAAF H +K+ Y P + MW +PPA DTS+D PPVA
Sbjct: 246 PAAF------HSHKMAVAYPYGYYPPNMPMWSPLPPADRDTSRDLKNLPPVA 291
>UniRef100_Q9LTC7 Emb|CAB10220.1 [Arabidopsis thaliana]
Length = 320
Score = 170 bits (430), Expect = 1e-41
Identities = 95/172 (55%), Positives = 120/172 (69%), Gaps = 13/172 (7%)
Query: 2 RSESCAATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGE 61
R+ SC+ +KACREKLRR++LNDKF++L S+LEPGR KTDK+AIL DA+R+V QLRGE
Sbjct: 157 RTGSCSKPGTKACREKLRREKLNDKFMDLSSVLEPGRTPKTDKSAILDDAIRVVNQLRGE 216
Query: 62 AQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAP-PSFLPTPTAL 120
A +L++ N L E+IK LK +KNELR+EK LKAEKEK+EQQLKSM P P F+P+
Sbjct: 217 AHELQETNQKLLEEIKSLKADKNELREEKLVLKAEKEKMEQQLKSMVVPSPGFMPSQH-- 274
Query: 121 PAAFAAQGQAHGNKLVPFISY----PGVAMWQFMPPAAVDTSQDHVLRPPVA 168
PAAF H +K+ Y P + MW +PPA DTS+D PPVA
Sbjct: 275 PAAF------HSHKMAVAYPYGYYPPNMPMWSPLPPADRDTSRDLKNLPPVA 320
>UniRef100_Q8L467 AT4g14410/dl3245w [Arabidopsis thaliana]
Length = 283
Score = 166 bits (420), Expect = 2e-40
Identities = 92/169 (54%), Positives = 120/169 (70%), Gaps = 11/169 (6%)
Query: 2 RSESCA-ATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRG 60
R+ SC+ +KACRE+LRR++LN++F++L S+LEPGR KTDK AIL DA+R++ QLR
Sbjct: 124 RTGSCSRGGGTKACRERLRREKLNERFMDLSSVLEPGRTPKTDKPAILDDAIRILNQLRD 183
Query: 61 EAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPS-FLPTPTA 119
EA KL++ N L E+IK LK EKNELR+EK LKA+KEK EQQLKSM AP S F+P
Sbjct: 184 EALKLEETNQKLLEEIKSLKAEKNELREEKLVLKADKEKTEQQLKSMTAPSSGFIP---H 240
Query: 120 LPAAFAAQGQAHGNKLVPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
+PAAF + NK+ + SY + MW +MP + DTS+D LRPP A
Sbjct: 241 IPAAF------NHNKMAVYPSYGYMPMWHYMPQSVRDTSRDQELRPPAA 283
>UniRef100_O23297 Hypothetical protein dl3245w [Arabidopsis thaliana]
Length = 277
Score = 166 bits (420), Expect = 2e-40
Identities = 92/169 (54%), Positives = 120/169 (70%), Gaps = 11/169 (6%)
Query: 2 RSESCA-ATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRG 60
R+ SC+ +KACRE+LRR++LN++F++L S+LEPGR KTDK AIL DA+R++ QLR
Sbjct: 118 RTGSCSRGGGTKACRERLRREKLNERFMDLSSVLEPGRTPKTDKPAILDDAIRILNQLRD 177
Query: 61 EAQKLKDANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKSMNAPPS-FLPTPTA 119
EA KL++ N L E+IK LK EKNELR+EK LKA+KEK EQQLKSM AP S F+P
Sbjct: 178 EALKLEETNQKLLEEIKSLKAEKNELREEKLVLKADKEKTEQQLKSMTAPSSGFIP---H 234
Query: 120 LPAAFAAQGQAHGNKLVPFISYPGVAMWQFMPPAAVDTSQDHVLRPPVA 168
+PAAF + NK+ + SY + MW +MP + DTS+D LRPP A
Sbjct: 235 IPAAF------NHNKMAVYPSYGYMPMWHYMPQSVRDTSRDQELRPPAA 277
>UniRef100_Q8VZA6 Hypothetical protein At3g19860; MPN9.10 [Arabidopsis thaliana]
Length = 284
Score = 102 bits (253), Expect = 5e-21
Identities = 70/185 (37%), Positives = 99/185 (52%), Gaps = 26/185 (14%)
Query: 7 AATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLK 66
A S KA REKLRR++LN+ F+ELG++L+P RP K DKA IL D V+++ +L E KLK
Sbjct: 5 ARKSQKAGREKLRREKLNEHFVELGNVLDPERP-KNDKATILTDTVQLLKELTSEVNKLK 63
Query: 67 DANSGLQEKIKELKVEKNELRDEKQRLKAEKEKL----EQQLKSMN-------------A 109
+ L ++ +EL EKN+LR+EK LK++ E L +Q+L+SM+
Sbjct: 64 SEYTALTDESRELTQEKNDLREEKTSLKSDIENLNLQYQQRLRSMSPWGAAMDHTVMMAP 123
Query: 110 PPSF-LPTPTALPAAFAAQGQA------HGNKLVPFISYPGVAMWQFMPPAAVDTSQD-H 161
PPSF P P A+P + GN+ I P +MPP V Q H
Sbjct: 124 PPSFPYPMPIAMPPGSIPMHPSMPSYTYFGNQNPSMIPAPCPTYMPYMPPNTVVEQQSVH 183
Query: 162 VLRPP 166
+ + P
Sbjct: 184 IPQNP 188
>UniRef100_Q9LT23 Emb|CAA18500.1 [Arabidopsis thaliana]
Length = 337
Score = 102 bits (253), Expect = 5e-21
Identities = 70/185 (37%), Positives = 99/185 (52%), Gaps = 26/185 (14%)
Query: 7 AATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLK 66
A S KA REKLRR++LN+ F+ELG++L+P RP K DKA IL D V+++ +L E KLK
Sbjct: 58 ARKSQKAGREKLRREKLNEHFVELGNVLDPERP-KNDKATILTDTVQLLKELTSEVNKLK 116
Query: 67 DANSGLQEKIKELKVEKNELRDEKQRLKAEKEKL----EQQLKSMN-------------A 109
+ L ++ +EL EKN+LR+EK LK++ E L +Q+L+SM+
Sbjct: 117 SEYTALTDESRELTQEKNDLREEKTSLKSDIENLNLQYQQRLRSMSPWGAAMDHTVMMAP 176
Query: 110 PPSF-LPTPTALPAAFAAQGQA------HGNKLVPFISYPGVAMWQFMPPAAVDTSQD-H 161
PPSF P P A+P + GN+ I P +MPP V Q H
Sbjct: 177 PPSFPYPMPIAMPPGSIPMHPSMPSYTYFGNQNPSMIPAPCPTYMPYMPPNTVVEQQSVH 236
Query: 162 VLRPP 166
+ + P
Sbjct: 237 IPQNP 241
>UniRef100_Q67U21 Basic helix-loop-helix-like protein [Oryza sativa]
Length = 343
Score = 92.0 bits (227), Expect = 5e-18
Identities = 50/92 (54%), Positives = 63/92 (68%), Gaps = 1/92 (1%)
Query: 12 KACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSG 71
KA REK+RRDRLN++F ELGS L+P RP + DKA IL DA++M+ L + KLK +
Sbjct: 44 KADREKMRRDRLNEQFQELGSTLDPDRP-RNDKATILSDAIQMLKDLTSQVNKLKAEYTS 102
Query: 72 LQEKIKELKVEKNELRDEKQRLKAEKEKLEQQ 103
L E+ +EL EKNELRDEK LK E + L Q
Sbjct: 103 LSEEARELTQEKNELRDEKVSLKFEVDNLNTQ 134
>UniRef100_O65643 Putative Myc-type transcription factor [Arabidopsis thaliana]
Length = 272
Score = 84.0 bits (206), Expect = 1e-15
Identities = 44/97 (45%), Positives = 66/97 (67%), Gaps = 1/97 (1%)
Query: 7 AATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLK 66
A S KA REKLRRD+L ++F+ELG+ L+P RP K+DKA++L D ++M+ + + +LK
Sbjct: 44 AVCSQKAEREKLRRDKLKEQFLELGNALDPNRP-KSDKASVLTDTIQMLKDVMNQVDRLK 102
Query: 67 DANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQ 103
L ++ +EL EK+ELR+EK LK++ E L Q
Sbjct: 103 AEYETLSQESRELIQEKSELREEKATLKSDIEILNAQ 139
>UniRef100_Q8W2F2 Putative transcription factor BHLH11 [Arabidopsis thaliana]
Length = 286
Score = 84.0 bits (206), Expect = 1e-15
Identities = 44/97 (45%), Positives = 66/97 (67%), Gaps = 1/97 (1%)
Query: 7 AATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLK 66
A S KA REKLRRD+L ++F+ELG+ L+P RP K+DKA++L D ++M+ + + +LK
Sbjct: 44 AVCSQKAEREKLRRDKLKEQFLELGNALDPNRP-KSDKASVLTDTIQMLKDVMNQVDRLK 102
Query: 67 DANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQ 103
L ++ +EL EK+ELR+EK LK++ E L Q
Sbjct: 103 AEYETLSQESRELIQEKSELREEKATLKSDIEILNAQ 139
>UniRef100_Q8RX65 AT4g36060/T19K4_190 [Arabidopsis thaliana]
Length = 268
Score = 84.0 bits (206), Expect = 1e-15
Identities = 44/97 (45%), Positives = 66/97 (67%), Gaps = 1/97 (1%)
Query: 7 AATSSKACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLK 66
A S KA REKLRRD+L ++F+ELG+ L+P RP K+DKA++L D ++M+ + + +LK
Sbjct: 26 AVCSQKAEREKLRRDKLKEQFLELGNALDPNRP-KSDKASVLTDTIQMLKDVMNQVDRLK 84
Query: 67 DANSGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQ 103
L ++ +EL EK+ELR+EK LK++ E L Q
Sbjct: 85 AEYETLSQESRELIQEKSELREEKATLKSDIEILNAQ 121
>UniRef100_UPI00002C6501 UPI00002C6501 UniRef100 entry
Length = 177
Score = 83.6 bits (205), Expect = 2e-15
Identities = 64/177 (36%), Positives = 90/177 (50%), Gaps = 20/177 (11%)
Query: 12 KACREKLRRDRLNDKFIELGSILEPGR--PAKTDKAAILIDAVRMVTQLRGEAQKLKDAN 69
K+ REKLRR+ LNDKF+EL ++L+P P KT+KA I+ +A R++ QLR E KL
Sbjct: 1 KSRREKLRREALNDKFMELSALLDPTAVGPFKTEKATIVTEAARVIKQLRAELAKLSATL 60
Query: 70 SGLQEKIKELKVEKNELRDEKQRLKAEKEKLEQQL-KSMNAPPSFLPTPTAL-------- 120
+QE LK E + + +K L+ +K KL+ QL M++ P P P A+
Sbjct: 61 ESMQESNLALKKETSCVAADKAALQQDKAKLQHQLYYFMSSMPFASPPPGAVFASITGAY 120
Query: 121 -----PAAFAAQGQAHGNKLVPFISYPG--VAMWQF--MPPAAVDTSQDHVLRPPVA 168
+A AQ + KL P G AMW F + + +D LR PVA
Sbjct: 121 HLPRGTSALLAQQNDNAEKLAPNHLAAGSTPAMWNFPRLIVQSTTAEEDAKLRAPVA 177
>UniRef100_Q8W397 Putative amelogenin [Oryza sativa]
Length = 252
Score = 69.3 bits (168), Expect = 4e-11
Identities = 45/125 (36%), Positives = 67/125 (53%), Gaps = 10/125 (8%)
Query: 12 KACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSG 71
K+ REKL+R LND F ELG++LE R + KA IL D R++ L + + L+ NS
Sbjct: 41 KSEREKLKRGHLNDLFGELGNMLEADRQSN-GKACILTDTTRILRDLLSQVKSLRQENST 99
Query: 72 LQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLK---------SMNAPPSFLPTPTALPA 122
LQ + + +E+NEL+DE L++E L+ +L+ A S LP P +
Sbjct: 100 LQNESNYVTMERNELQDENGALRSEISDLQNELRMRATGSPGWGHGATGSPLPVPPSPGT 159
Query: 123 AFAAQ 127
F +Q
Sbjct: 160 VFPSQ 164
>UniRef100_Q69V10 Amelogenin-like protein [Oryza sativa]
Length = 265
Score = 67.8 bits (164), Expect = 1e-10
Identities = 42/120 (35%), Positives = 68/120 (56%), Gaps = 4/120 (3%)
Query: 12 KACREKLRRDRLNDKFIELGSILEPGRPAKTDKAAILIDAVRMVTQLRGEAQKLKDANSG 71
K+ REKL+RD+ ND F ELG++LEP R KA +L + R++ L + + L+ NS
Sbjct: 40 KSEREKLKRDKQNDLFNELGNLLEPDR-QNNGKACVLGETTRILKDLLSQVESLRKENSS 98
Query: 72 LQEKIKELKVEKNELRDEKQRLKAEKEKLEQQLKS-MNAPP--SFLPTPTALPAAFAAQG 128
L+ + + +E+NEL D+ L+ E +L+ +L++ M P S + T AL + G
Sbjct: 99 LKNESHYVALERNELHDDNSMLRTEILELQNELRTRMEGNPVWSHVNTRPALRVPYPTTG 158
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.130 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 266,185,399
Number of Sequences: 2790947
Number of extensions: 10425971
Number of successful extensions: 110076
Number of sequences better than 10.0: 2910
Number of HSP's better than 10.0 without gapping: 1169
Number of HSP's successfully gapped in prelim test: 1783
Number of HSP's that attempted gapping in prelim test: 98163
Number of HSP's gapped (non-prelim): 12064
length of query: 168
length of database: 848,049,833
effective HSP length: 118
effective length of query: 50
effective length of database: 518,718,087
effective search space: 25935904350
effective search space used: 25935904350
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 70 (31.6 bits)
Medicago: description of AC144502.7


