

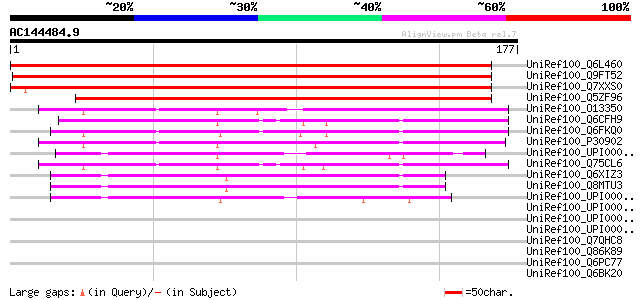
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144484.9 + phase: 0
(177 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6L460 Putative mitochondrial ATP synthase [Solanum de... 297 9e-80
UniRef100_Q9FT52 ATP synthase D chain, mitochondrial [Arabidopsi... 288 6e-77
UniRef100_Q7XXS0 Hypothetical protein [Oryza sativa] 278 4e-74
UniRef100_Q5ZF96 Mitochondrial F0 ATP synthase delta chain [Plan... 227 1e-58
UniRef100_O13350 ATP synthase D chain, mitochondrial [Kluyveromy... 57 2e-07
UniRef100_Q6CFH9 Similar to sp|O13350 Kluyveromyces lactis ATP s... 55 8e-07
UniRef100_Q6FKQ0 Similar to sp|P30902 Saccharomyces cerevisiae Y... 54 1e-06
UniRef100_P30902 ATP synthase D chain, mitochondrial [Saccharomy... 52 7e-06
UniRef100_UPI0000361541 UPI0000361541 UniRef100 entry 47 2e-04
UniRef100_Q75CL6 ACL097Cp [Ashbya gossypii] 47 2e-04
UniRef100_Q6XIZ3 Similar to Drosophila melanogaster ATPsyn-d [Dr... 47 3e-04
UniRef100_Q8MTU3 ATP synthase D chain, mitochondrial [Drosophila... 47 3e-04
UniRef100_UPI00003AD393 UPI00003AD393 UniRef100 entry 46 5e-04
UniRef100_UPI00001807D3 UPI00001807D3 UniRef100 entry 44 0.002
UniRef100_UPI0000430E4C UPI0000430E4C UniRef100 entry 44 0.002
UniRef100_UPI0000361542 UPI0000361542 UniRef100 entry 44 0.002
UniRef100_Q7QHC8 ENSANGP00000012669 [Anopheles gambiae str. PEST] 43 0.003
UniRef100_Q86K89 Hypothetical protein [Dictyostelium discoideum] 43 0.003
UniRef100_Q6PC77 ATP synthase, H+ transporting, mitochondrial F0... 42 0.005
UniRef100_Q6BK20 Similar to CA1907|CaATP7 Candida albicans CaATP... 42 0.005
>UniRef100_Q6L460 Putative mitochondrial ATP synthase [Solanum demissum]
Length = 168
Score = 297 bits (760), Expect = 9e-80
Identities = 141/168 (83%), Positives = 158/168 (93%)
Query: 1 MSGTVKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPE 60
MSGTVKK+ DV FKAG+ IDW+GMAKLLVTDEAR+EF NLRRAFD+VN+QL+TKFSQEPE
Sbjct: 1 MSGTVKKIADVTFKAGRTIDWEGMAKLLVTDEARKEFSNLRRAFDDVNSQLQTKFSQEPE 60
Query: 61 PIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKE 120
PI+WEYYRKGIG+RLVDMYK+ YESIEIPKF DTVTPQYKPKFDALLVELKEAE++SLKE
Sbjct: 61 PINWEYYRKGIGSRLVDMYKEAYESIEIPKFEDTVTPQYKPKFDALLVELKEAEKQSLKE 120
Query: 121 SERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
SERLEKE+ VQ LKK++STMTA+EYFA+HPELKKKFDDEIRND WGY
Sbjct: 121 SERLEKEVAEVQELKKKLSTMTAEEYFAKHPELKKKFDDEIRNDYWGY 168
>UniRef100_Q9FT52 ATP synthase D chain, mitochondrial [Arabidopsis thaliana]
Length = 167
Score = 288 bits (736), Expect = 6e-77
Identities = 136/167 (81%), Positives = 152/167 (90%)
Query: 2 SGTVKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEP 61
SG KK+ DVAFKA + IDWDGMAK+LVTDEARREF NLRRAFDEVNTQL+TKFSQEPEP
Sbjct: 1 SGAGKKIADVAFKASRTIDWDGMAKVLVTDEARREFSNLRRAFDEVNTQLQTKFSQEPEP 60
Query: 62 IDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKES 121
IDW+YYRKGIG +VD YK+ Y+SIEIPK+VD VTP+YKPKFDALLVELKEAE+KSLKES
Sbjct: 61 IDWDYYRKGIGAGIVDKYKEAYDSIEIPKYVDKVTPEYKPKFDALLVELKEAEQKSLKES 120
Query: 122 ERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
ERLEKEI +VQ + K++STMTADEYF +HPELKKKFDDEIRNDNWGY
Sbjct: 121 ERLEKEIADVQEISKKLSTMTADEYFEKHPELKKKFDDEIRNDNWGY 167
>UniRef100_Q7XXS0 Hypothetical protein [Oryza sativa]
Length = 169
Score = 278 bits (711), Expect = 4e-74
Identities = 132/169 (78%), Positives = 155/169 (91%), Gaps = 1/169 (0%)
Query: 1 MSGT-VKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEP 59
MSG VKKV +VA KAGK IDW+GMAK+LV+DEAR+EF LRR F++VN QL+TKFSQEP
Sbjct: 1 MSGNGVKKVAEVAAKAGKAIDWEGMAKMLVSDEARKEFNTLRRTFEDVNHQLQTKFSQEP 60
Query: 60 EPIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLK 119
+PIDWEYYRKGIG+++VDMYK+ YESIEIPK+VDTVTPQYKPKFDALLVELKEAE++SLK
Sbjct: 61 QPIDWEYYRKGIGSKVVDMYKEAYESIEIPKYVDTVTPQYKPKFDALLVELKEAEKESLK 120
Query: 120 ESERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
ESER+EKE+ +Q +KK ISTMTADEYFA+HPE+K+KFDDEIRNDNWGY
Sbjct: 121 ESERIEKELAELQEMKKNISTMTADEYFAKHPEVKQKFDDEIRNDNWGY 169
>UniRef100_Q5ZF96 Mitochondrial F0 ATP synthase delta chain [Plantago major]
Length = 145
Score = 227 bits (578), Expect = 1e-58
Identities = 104/145 (71%), Positives = 132/145 (90%)
Query: 24 MAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTRLVDMYKQHY 83
MAK+LV+DEAR+EFF LRRAFD+VNTQL+TKFSQEP PIDWEYYRKG+G+RLVDMYKQ +
Sbjct: 1 MAKVLVSDEARKEFFALRRAFDDVNTQLQTKFSQEPVPIDWEYYRKGLGSRLVDMYKQAH 60
Query: 84 ESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLKKRISTMTA 143
+ I+IPK+VD VTP+Y KFDAL VE+++AE++SL+ ++RLEKEI +V+ LK ++STMTA
Sbjct: 61 DEIKIPKYVDNVTPEYTVKFDALSVEMQQAEQESLEVTKRLEKEIADVEELKNKVSTMTA 120
Query: 144 DEYFAEHPELKKKFDDEIRNDNWGY 168
DEYFA+HPE+K+KFD+EIRND WGY
Sbjct: 121 DEYFAKHPEVKEKFDEEIRNDYWGY 145
>UniRef100_O13350 ATP synthase D chain, mitochondrial [Kluyveromyces lactis]
Length = 173
Score = 57.4 bits (137), Expect = 2e-07
Identities = 40/171 (23%), Positives = 79/171 (45%), Gaps = 13/171 (7%)
Query: 11 VAFKAGKKIDWDGM-AKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRK 69
+A A K+DW + + L +T + + + ++ DE QL + +P +D+ +YR
Sbjct: 2 LAKSAANKLDWAKVISSLKLTGKTATQLSSFKKRNDEARRQL-LELQSQPTSVDFSHYRS 60
Query: 70 GI-GTRLVDMYKQHYES-----IEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESER 123
+ T +VD +Q Y+S +++ K + T+ F++ +E EK + + +
Sbjct: 61 VLKNTEVVDKIEQFYKSYKPVSVDVSKQLSTIEA-----FESQAIENAAETEKLVAQELK 115
Query: 124 LEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
KE +N + +T DE PE+ K ++ ++ W YKE+
Sbjct: 116 DLKETLNNIESARPFDQLTVDELTKARPEIDAKVEEMVKKGRWDVPGYKEK 166
>UniRef100_Q6CFH9 Similar to sp|O13350 Kluyveromyces lactis ATP synthase D chain
[Yarrowia lipolytica]
Length = 176
Score = 55.1 bits (131), Expect = 8e-07
Identities = 42/161 (26%), Positives = 76/161 (47%), Gaps = 7/161 (4%)
Query: 18 KIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGI-GTRLV 76
K+DW + L A R E + + +P +D+ +YRK + ++V
Sbjct: 12 KVDWGKIVSSLGLTGATVSSLQAFRKRHEEAKKNAYELQNQPTTVDFAHYRKVLKNQKVV 71
Query: 77 DMYKQHYESIEIPKFVDTVTPQYKP--KFDALLVE-LKEAEEKSLKESERLEKEIVNVQS 133
D +QH++S + P D V+ Q K F+A +E K E K +E L+K + N++S
Sbjct: 72 DEIEQHFKSFK-PVTYD-VSKQLKTIDAFEAKAIEDAKATEGKVNQEIGDLQKTLENIES 129
Query: 134 LKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
+ ++ D+ F P+L+KK ++ ++ W Y E+
Sbjct: 130 ARP-FDQLSVDDVFKARPDLEKKIEEMVKKGRWSVPGYNEK 169
>UniRef100_Q6FKQ0 Similar to sp|P30902 Saccharomyces cerevisiae YKL016c ATP7 [Candida
glabrata]
Length = 176
Score = 54.3 bits (129), Expect = 1e-06
Identities = 42/164 (25%), Positives = 80/164 (48%), Gaps = 7/164 (4%)
Query: 15 AGKKIDWDGM-AKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIG- 72
A K+DW + + L +T + + ++ DE QL + Q+P +D+ +YR +
Sbjct: 9 AANKLDWAKVISSLKLTGKTATQLSTFKKRNDEARRQL-FELEQQPVEVDFAHYRSVLNN 67
Query: 73 TRLVDMYKQHYESIEIPKFVDTVTPQYK-PKFDALLVE-LKEAEEKSLKESERLEKEIVN 130
+ +VD +++ +S P VDT F+ L +E KE EE KE + L++ + N
Sbjct: 68 SEVVDKIERYVKSYS-PVTVDTSKQLGAIASFEKLALENAKETEEIVSKELKALKETLAN 126
Query: 131 VQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
++S + +T DE PE+ ++ + ++N W Y ++
Sbjct: 127 IESARP-FDQLTVDELVKAKPEIDERVAELVKNGKWEIPSYYQK 169
>UniRef100_P30902 ATP synthase D chain, mitochondrial [Saccharomyces cerevisiae]
Length = 173
Score = 52.0 bits (123), Expect = 7e-06
Identities = 37/166 (22%), Positives = 74/166 (44%), Gaps = 5/166 (3%)
Query: 11 VAFKAGKKIDWDGM-AKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRK 69
+A A K+DW + + L +T + + ++ DE QL + +P +D+ +YR
Sbjct: 2 LAKSAANKLDWAKVISSLRITGSTATQLSSFKKRNDEARRQL-LELQSQPTEVDFSHYRS 60
Query: 70 GI-GTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDA-LLVELKEAEEKSLKESERLEKE 127
+ T ++D + + + + K + Q F+ + KE E KE + L+
Sbjct: 61 VLKNTSVIDKIESYVKQYKPVKIDASKQLQVIESFEKHAMTNAKETESLVSKELKDLQST 120
Query: 128 IVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKE 173
+ N+QS + +T D+ PE+ K ++ ++ W YK+
Sbjct: 121 LDNIQSARP-FDELTVDDLTKIKPEIDAKVEEMVKKGKWDVPGYKD 165
>UniRef100_UPI0000361541 UPI0000361541 UniRef100 entry
Length = 161
Score = 47.4 bits (111), Expect = 2e-04
Identities = 39/157 (24%), Positives = 72/157 (45%), Gaps = 19/157 (12%)
Query: 17 KKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGI-GTRL 75
K IDW A+L+ ++ R FN + + + + P IDW YYR + + +
Sbjct: 9 KAIDWLAFAELVPPNQ--RGMFNALKTRSDAIAAKLSSLPETPAAIDWSYYRSTVANSGM 66
Query: 76 VDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNV-QSL 134
VD +++ ++++++P+ VDT T A+ ++ EA + + + E + I Q L
Sbjct: 67 VDDFEKKFKALKVPEPVDTQT-------SAIGIQETEANKSAAEYIEASKARIAEYEQEL 119
Query: 135 KK-----RISTMTADEYFAEHPELKKKFDDEIRNDNW 166
K MT ++ A PE K D+++ W
Sbjct: 120 SKFKNMIPFDQMTIEDLNAAFPETKL---DKVKYPYW 153
>UniRef100_Q75CL6 ACL097Cp [Ashbya gossypii]
Length = 174
Score = 47.0 bits (110), Expect = 2e-04
Identities = 42/169 (24%), Positives = 79/169 (45%), Gaps = 9/169 (5%)
Query: 11 VAFKAGKKIDWDGM-AKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRK 69
+A A K+DW + A L +T + + ++ DE +L + +P +D+ +YR
Sbjct: 3 LARSAANKLDWAKVIASLKLTGKTAAQLTQFKKRNDEARRRL-FELESQPAEVDFAHYRA 61
Query: 70 GI-GTRLVDMYKQHYESIEIPKFVDTVTPQYKP--KFDALLV-ELKEAEEKSLKESERLE 125
+ T +VD + + P VD V+ Q F+A V + +E E +E + L+
Sbjct: 62 VLKNTAVVDQIEASCRAYR-PVTVD-VSKQISSLEAFEAQAVRDAQETEAVVGRELKALK 119
Query: 126 KEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
+ + N+++ + +T DE A PE+ K + ++ W YKE+
Sbjct: 120 ETLANIEAARP-FDELTVDELAAARPEIDTKVAEMVKRGKWEVPGYKEK 167
>UniRef100_Q6XIZ3 Similar to Drosophila melanogaster ATPsyn-d [Drosophila yakuba]
Length = 178
Score = 46.6 bits (109), Expect = 3e-04
Identities = 32/139 (23%), Positives = 67/139 (48%), Gaps = 4/139 (2%)
Query: 15 AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTR 74
A I+W +A+ + ++ + F + ++ + + P IDW Y+K +
Sbjct: 7 AQSSINWSALAERVPANQ--KSSFGAFKTKSDIYVRAVLANPECPPQIDWANYKKLVPVA 64
Query: 75 -LVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQS 133
LVD +++ YE++++P D V+ Q + + E+ ++ S + + +KEI +++S
Sbjct: 65 GLVDSFQKQYEALKVPYPQDKVSSQVDAEIKSSQSEIDAYKKASEQRIQNYQKEIAHLKS 124
Query: 134 LKKRISTMTADEYFAEHPE 152
L MT ++Y PE
Sbjct: 125 LLP-YDQMTMEDYRDAFPE 142
>UniRef100_Q8MTU3 ATP synthase D chain, mitochondrial [Drosophila melanogaster]
Length = 190
Score = 46.6 bits (109), Expect = 3e-04
Identities = 32/139 (23%), Positives = 67/139 (48%), Gaps = 4/139 (2%)
Query: 15 AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTR 74
A I+W +A+ + ++ + F + ++ + + P IDW Y+K +
Sbjct: 19 AQSSINWSALAERVPANQ--KSSFGAFKTKSDIYVRAVLANPECPPQIDWANYKKLVPVA 76
Query: 75 -LVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQS 133
LVD +++ YE++++P D V+ Q + A E+ ++ S + + +KEI +++S
Sbjct: 77 GLVDSFQKQYEALKVPYPQDKVSSQVDAEIKASQSEIDAYKKASEQRIQNYQKEIAHLKS 136
Query: 134 LKKRISTMTADEYFAEHPE 152
L MT ++Y P+
Sbjct: 137 LLP-YDQMTMEDYRDAFPD 154
>UniRef100_UPI00003AD393 UPI00003AD393 UniRef100 entry
Length = 161
Score = 45.8 bits (107), Expect = 5e-04
Identities = 36/144 (25%), Positives = 68/144 (47%), Gaps = 10/144 (6%)
Query: 15 AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIG-T 73
A K IDW A+ + ++ R FN + + + ++P IDW YY+ +
Sbjct: 7 AVKAIDWAAFAERVPANQ--RAMFNALKTRSDALSARLAALPEKPPAIDWTYYKTAVAKA 64
Query: 74 RLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESE-RLEKEIVNVQ 132
+VD +++ + ++++P+ VDT T K DA E + + +K S+ R+ + +Q
Sbjct: 65 GMVDEFQKKFSALKVPEPVDTQT----AKIDAQEQEAAKGIVEYVKASKARIAEYEQQLQ 120
Query: 133 SLKKRI--STMTADEYFAEHPELK 154
L+ I MT ++ PE +
Sbjct: 121 KLRSMIPFEQMTFEDLHEAFPETR 144
>UniRef100_UPI00001807D3 UPI00001807D3 UniRef100 entry
Length = 161
Score = 43.9 bits (102), Expect = 0.002
Identities = 40/162 (24%), Positives = 70/162 (42%), Gaps = 25/162 (15%)
Query: 17 KKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIG-TRL 75
K IDW +++ ++ + N +++++ S++P IDW YYR +G L
Sbjct: 9 KTIDWVSFVEIMPQNQ--KAIGNALKSWNKTFHTRLASMSEKPPAIDWAYYRTNVGKAGL 66
Query: 76 VDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEE---------KSLKESERLEK 126
VD +++ Y ++IP D T LV+ +E EE S + EK
Sbjct: 67 VDDFEKKYNVLKIPVPEDKYT---------ALVDAEEKEEVKTCAQFVSGSQARVQEYEK 117
Query: 127 EIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
++ ++S+ MT D+ PE K DE + W +
Sbjct: 118 QLEKIKSMIP-FDQMTIDDLNEIFPETKL---DEKKYPYWSH 155
>UniRef100_UPI0000430E4C UPI0000430E4C UniRef100 entry
Length = 164
Score = 43.9 bits (102), Expect = 0.002
Identities = 42/169 (24%), Positives = 74/169 (42%), Gaps = 18/169 (10%)
Query: 14 KAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGT 73
KA K I+W + + + + E + ++ + Q + ++ IDW YY+K I T
Sbjct: 2 KALKAINWSAITERIPSSE--KAALTAFKSKSDRYLQRMMAYPEDLPKIDWTYYKKTIIT 59
Query: 74 R-LVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQ 132
LVD + + YE+I IP D T A+ E KE +K + + +I +Q
Sbjct: 60 PGLVDKFYKEYEAISIPYPTDKYT-------QAIDSEQKEIADKIQSFIQEVNSQIAELQ 112
Query: 133 SLKKRI------STMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEEQ 175
RI S MT +++ P+ + D+E W ++ +E+
Sbjct: 113 QNLDRIKNMIPFSEMTMEDFSDIQPKGTLRPDEE--PTTWPHTEDSQEK 159
>UniRef100_UPI0000361542 UPI0000361542 UniRef100 entry
Length = 163
Score = 43.9 bits (102), Expect = 0.002
Identities = 23/81 (28%), Positives = 44/81 (53%), Gaps = 3/81 (3%)
Query: 17 KKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGI-GTRL 75
K IDW A+L+ ++ R FN + + + + P IDW YYR + + +
Sbjct: 10 KAIDWLAFAELVPPNQ--RGMFNALKTRSDAIAAKLSSLPETPAAIDWSYYRSTVANSGM 67
Query: 76 VDMYKQHYESIEIPKFVDTVT 96
VD +++ ++++++P+ VDT T
Sbjct: 68 VDDFEKKFKALKVPEPVDTQT 88
>UniRef100_Q7QHC8 ENSANGP00000012669 [Anopheles gambiae str. PEST]
Length = 161
Score = 43.1 bits (100), Expect = 0.003
Identities = 39/128 (30%), Positives = 59/128 (45%), Gaps = 22/128 (17%)
Query: 57 QEPEPIDWEYYRKGIGTR-LVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAE- 114
+ P IDW +Y+K + +VD +++ YE ++IP D VT LVE +E E
Sbjct: 47 ETPPKIDWSFYKKNVPVAGMVDKFQKAYEGLQIPYPADNVTK---------LVEAQEKEV 97
Query: 115 ----EKSLKESE-RLEKEIVNVQSLKKRI--STMTADEYFAEHPELKKKFDDEIRNDNWG 167
K +KESE R+ + +LK + MT +++ PE + D R W
Sbjct: 98 QQDIAKFVKESEVRISDYQTQIATLKALLPFDQMTMEDFKDSFPE--QALDPINRPSFW- 154
Query: 168 YSHYKEEQ 175
H EEQ
Sbjct: 155 -PHTPEEQ 161
>UniRef100_Q86K89 Hypothetical protein [Dictyostelium discoideum]
Length = 234
Score = 43.1 bits (100), Expect = 0.003
Identities = 29/120 (24%), Positives = 58/120 (48%), Gaps = 13/120 (10%)
Query: 56 SQEPEPIDWEYYRKGIGTRLVDMY----KQHYESI-----EIPKFVDTVTPQYKPKFDAL 106
S E I+W++Y+K + V+ + K+ +E+I E KF+D + + K +A
Sbjct: 98 SSEKVVINWDFYKKILPEDTVEFFRALEKKTHETILSWEAEDKKFIDFLLKKIKSDEEAS 157
Query: 107 LVELKEAEEKSLKESERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNW 166
KE+++ + LE + ++ + T T ++ ++PE+ ++ EI ND W
Sbjct: 158 ----KESQKSLYLTIQALEADRKQMEDFLTNLRTTTFEDLHGKYPEIDEQIYQEISNDEW 213
>UniRef100_Q6PC77 ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d
[Brachydanio rerio]
Length = 161
Score = 42.4 bits (98), Expect = 0.005
Identities = 40/159 (25%), Positives = 75/159 (47%), Gaps = 15/159 (9%)
Query: 15 AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIG-T 73
A K IDW A+ + ++ R F +L+ D ++ +L + ++P I+W +YR +
Sbjct: 7 AVKAIDWLAFAERVPPNQ-RAMFNSLKTRSDAISAKLAS-LPEKPSTINWSHYRAVVAKA 64
Query: 74 RLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESE----RLEKEIV 129
+VD +++ + + +P+ VDT T K DA E ++ L+ S+ EKE+
Sbjct: 65 GMVDEFEKKFAGLTVPEPVDTQT----AKIDAQEQEANKSAAAYLEASKARIAEYEKELE 120
Query: 130 NVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
+++ MT ++ PE K D+ + W Y
Sbjct: 121 KFRNMIP-FDQMTIEDLNNTFPETKL---DKQKYPYWPY 155
>UniRef100_Q6BK20 Similar to CA1907|CaATP7 Candida albicans CaATP7 F1F0-ATPase
complex [Debaryomyces hansenii]
Length = 175
Score = 42.4 bits (98), Expect = 0.005
Identities = 34/170 (20%), Positives = 78/170 (45%), Gaps = 5/170 (2%)
Query: 8 VTDVAFKAGKKIDWDGMAKLL-VTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEY 66
++ VA A K++W + L +T ++ +E + SQ+ +D+ +
Sbjct: 1 MSSVAKSATTKVNWAKITSTLGLTGNTASSLTAFKKRNEEARKE-NLSLSQQSTEVDFNH 59
Query: 67 YRKGIG-TRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKS-LKESERL 124
YR + ++++D ++ + + + + T + F+ VE + EKS L E ++L
Sbjct: 60 YRSVLSNSKVIDEIEKAVSAFKPVTYDVSKTLKTIDMFEQKAVENAKLTEKSVLDEVKQL 119
Query: 125 EKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
++ + +++ + +T D+ P+L +K ++N W YKE+
Sbjct: 120 QETLKDIEGARP-FDQLTVDDVAKARPDLDEKVTYMVKNSKWEVPTYKEK 168
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.132 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 295,836,109
Number of Sequences: 2790947
Number of extensions: 12106845
Number of successful extensions: 51163
Number of sequences better than 10.0: 322
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 282
Number of HSP's that attempted gapping in prelim test: 50784
Number of HSP's gapped (non-prelim): 537
length of query: 177
length of database: 848,049,833
effective HSP length: 119
effective length of query: 58
effective length of database: 515,927,140
effective search space: 29923774120
effective search space used: 29923774120
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC144484.9


