

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144477.10 - phase: 0 /pseudo
(200 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
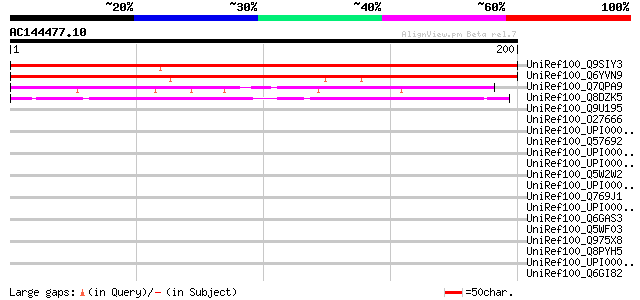
UniRef100_Q9SIY3 Expressed protein [Arabidopsis thaliana] 227 2e-58
UniRef100_Q6YVN9 Hypothetical protein OSJNBa0063H21.119 [Oryza s... 190 2e-47
UniRef100_Q7QPA9 GLP_122_5075_4320 [Giardia lamblia ATCC 50803] 52 7e-06
UniRef100_Q8DZK5 Hypothetical protein SAG1096 [Streptococcus aga... 45 8e-04
UniRef100_Q9U195 Hypothetical protein L2903.03 [Leishmania major] 42 0.012
UniRef100_O27666 Conserved protein [Methanobacterium thermoautot... 38 0.13
UniRef100_UPI000031185C UPI000031185C UniRef100 entry 36 0.51
UniRef100_Q57692 Hypothetical protein MJ0240 [Methanococcus jann... 35 0.87
UniRef100_UPI000025867F UPI000025867F UniRef100 entry 35 1.5
UniRef100_UPI00002782F8 UPI00002782F8 UniRef100 entry 34 1.9
UniRef100_Q5W2W2 Putative TrbE protein [Sulfolobus islandicus] 34 2.5
UniRef100_UPI000021BE31 UPI000021BE31 UniRef100 entry 33 3.3
UniRef100_Q769J1 Polymerase [Hepatitis B virus] 33 3.3
UniRef100_UPI00002C8CCA UPI00002C8CCA UniRef100 entry 33 4.3
UniRef100_Q6GAS3 Hypothetical protein [Staphylococcus aureus] 33 4.3
UniRef100_Q5WF03 Hypothetical protein [Bacillus clausii] 33 4.3
UniRef100_Q975X8 Hypothetical protein ST0300 [Sulfolobus tokodaii] 33 4.3
UniRef100_Q8PYH5 Cobalt import ATP-binding protein cbiO 2 [Metha... 33 4.3
UniRef100_UPI000042E72B UPI000042E72B UniRef100 entry 33 5.7
UniRef100_Q6GI82 Hypothetical protein [Staphylococcus aureus] 33 5.7
>UniRef100_Q9SIY3 Expressed protein [Arabidopsis thaliana]
Length = 210
Score = 227 bits (578), Expect = 2e-58
Identities = 115/209 (55%), Positives = 153/209 (73%), Gaps = 9/209 (4%)
Query: 1 MEVEVKLRLPNADSYHRVTTLLSPFHVITHRQHNLFFDGAGSELSSRRAILRLRFYGD-- 58
MEVEVKLRL A ++ R+TTLL+P+H+ T Q N FFD ++LS RRA+LRLRF +
Sbjct: 1 MEVEVKLRLLTAAAHLRLTTLLTPYHLKTLHQRNTFFDTPKNDLSLRRAVLRLRFLQNAA 60
Query: 59 -------DERCVVSLKAKAVLVDGVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGR 111
RC+VSLKAK L +G+SRVEEDEE+++ IG++CV+ P KL + SR++ R
Sbjct: 61 VSAASPSPPRCIVSLKAKPTLANGISRVEEDEEEIEYWIGKECVESPAKLSDIGSRVLKR 120
Query: 112 VKEEFGVVGENGFVGLGGFKNVRNVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKR 171
VKEE+G GFV LGGF+NVRNVY+W+G+KLEVDET +DFG +EIECE+ +PE K
Sbjct: 121 VKEEYGFNDFLGFVCLGGFENVRNVYEWRGVKLEVDETKYDFGNCYEIECETEEPERVKT 180
Query: 172 ILEEFLKENGIDYSYSVASKFAIFRAGKL 200
++EEFL E I++S S +KFA+FR+GKL
Sbjct: 181 MIEEFLTEEKIEFSNSDMTKFAVFRSGKL 209
>UniRef100_Q6YVN9 Hypothetical protein OSJNBa0063H21.119 [Oryza sativa]
Length = 212
Score = 190 bits (482), Expect = 2e-47
Identities = 107/210 (50%), Positives = 141/210 (66%), Gaps = 10/210 (4%)
Query: 1 MEVEVKLRLPNADSYHRVTTLLSPFHVITHRQHNLFFDGAGSELSSRRAILRLRFYGDDE 60
MEVE+KLRLP+A ++ R+++ L+P T Q NLFFD A L++ A LR+R YG D+
Sbjct: 1 MEVEIKLRLPDAGAHRRLSSFLAPRLRRTDAQRNLFFDAAARPLAAATAALRVRLYGLDD 60
Query: 61 RC----VVSLKAKAVLVDGVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGRVKEEF 116
R V++LK + + GVSRVEE EE LDP I CVD+P LG VES I+ V EE+
Sbjct: 61 RAPSRAVLALKRRPRIDAGVSRVEEVEEPLDPAIALACVDDPASLGGVESPIIRLVSEEY 120
Query: 117 GVVGENG-FVGLGGFKNVRNVY-----DWKGLKLEVDETHFDFGTLFEIECESSDPEEAK 170
GV G+ FV LGGF+N R VY D GL +E+DET FDFGT +E+ECE+++PE+AK
Sbjct: 121 GVGGDAAPFVCLGGFRNTRAVYQLEEGDTLGLVVELDETRFDFGTNYELECETAEPEQAK 180
Query: 171 RILEEFLKENGIDYSYSVASKFAIFRAGKL 200
++LE L G+ Y YS ++KFA F AGKL
Sbjct: 181 QVLERLLTVAGVPYEYSRSNKFACFMAGKL 210
>UniRef100_Q7QPA9 GLP_122_5075_4320 [Giardia lamblia ATCC 50803]
Length = 251
Score = 52.4 bits (124), Expect = 7e-06
Identities = 57/207 (27%), Positives = 95/207 (45%), Gaps = 22/207 (10%)
Query: 1 MEVEVKLRLPNADSYHRVTTLLSPF--HVITHRQHNLFFDGAGSELSSRRAILRLRFY-- 56
+E E+KL L + +SY +V T L ++ + + +FD A EL+ ++A +RLR
Sbjct: 5 IEQEIKLTLCDPNSYAQVLTELDKQAKYIGSVTLTSTYFDTATHELAHQKASMRLRELKT 64
Query: 57 GDDERCVVSLKAKA-VLVDGVSRVEEDE-----EDLDPKIGRDCVDEPGKLGLVESRIMG 110
+LKAK+ + GV R E+E ED +G D G+ L S
Sbjct: 65 ATSISYTATLKAKSSITAAGVFRAIEEEVHISKEDYSALMG----DSTGERVL--SFFKN 118
Query: 111 RVKEEFGVVG---ENGFVGLGGFKNVRNVYDWKGLKLEVDETHFDF---GTLFEIECESS 164
V + V+ E + + F +R Y +K LE+D F G + E+ECE+
Sbjct: 119 NVCDLHTVLDNAIEGALMKIASFTTMRRRYTYKKHTLELDNVIFIVPRPGRMVELECETE 178
Query: 165 DPEEAKRILEEFLKENGIDYSYSVASK 191
+P++A L F + +++S +K
Sbjct: 179 EPQKAMEDLRAFFSTINVMFTHSTTTK 205
>UniRef100_Q8DZK5 Hypothetical protein SAG1096 [Streptococcus agalactiae]
Length = 190
Score = 45.4 bits (106), Expect = 8e-04
Identities = 49/197 (24%), Positives = 84/197 (41%), Gaps = 15/197 (7%)
Query: 1 MEVEVKLRLPNADSYHRVTTLLSPFHVITHRQHNLFFDGAGSELSSRRAILRLRFYGDDE 60
+E+E K L N D ++R+T+L S IT Q N +FD E+ + R LR+R +
Sbjct: 4 LEIEYKTLL-NKDEFNRLTSLFSHVQPIT--QTNYYFDTETFEMKAHRMSLRIRTLPNRA 60
Query: 61 RCVVSLKAKAVLVDGVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGRVKEEFGVVG 120
+ + + ++ + +E K G+ D + I + E+ V
Sbjct: 61 ELTLKIPREVGNLEHNHDLTLEEAKYIVKNGQFPED---------TEIASLILEKG--VD 109
Query: 121 ENGFVGLGGFKNVRNVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKRILEEFLKEN 180
G R + + +D + +E+E E P++ KR ++FLKEN
Sbjct: 110 PTKLAVFGQLTTTRREMETSIGLMALDSNIYADIKDYELELEVKQPKQGKRDFDQFLKEN 169
Query: 181 GIDYSYSVASKFAIFRA 197
I++ Y+ SK A F A
Sbjct: 170 NINFKYA-KSKVARFSA 185
>UniRef100_Q9U195 Hypothetical protein L2903.03 [Leishmania major]
Length = 745
Score = 41.6 bits (96), Expect = 0.012
Identities = 21/54 (38%), Positives = 31/54 (56%)
Query: 1 MEVEVKLRLPNADSYHRVTTLLSPFHVITHRQHNLFFDGAGSELSSRRAILRLR 54
M++EVKL L +A+SY R L+ H+ ++ FFD L R ++LRLR
Sbjct: 1 MQIEVKLSLEDAESYQRTLNTLANHHLKDECYYDFFFDFPYPALQERSSVLRLR 54
Score = 32.0 bits (71), Expect = 9.7
Identities = 23/97 (23%), Positives = 39/97 (39%), Gaps = 7/97 (7%)
Query: 23 SPFHVITHRQHNLFFDGAGSELSSRRAILRLRFYGDDERCVVSLKAKAVLVDGVSRVEED 82
+P + +LFFD L ++ LRLR + +++LKA V G
Sbjct: 485 NPNGYLQETNEDLFFDSPEQTLRRGKSFLRLRKRLHSNKYLLALKAHQVFSGGQQNSLSS 544
Query: 83 EEDLDPKIGRDCVDEPGKLGLVESRIMGRVKEEFGVV 119
+ D+ + R +D P ++ + E F VV
Sbjct: 545 KVDVSEVVARALIDNP-------TQFLHEYAEHFAVV 574
>UniRef100_O27666 Conserved protein [Methanobacterium thermoautotrophicum]
Length = 177
Score = 38.1 bits (87), Expect = 0.13
Identities = 45/202 (22%), Positives = 79/202 (38%), Gaps = 30/202 (14%)
Query: 1 MEVEVKLRLPNADSYHRVTTLLSPFHVITHRQHNLFFDGAGSELSSRRAILRLRFYGDDE 60
+EVEVK ++ + D + HV QH+L+F+ + + LR+R G
Sbjct: 2 IEVEVKAKIHDRDEMVDRILSVGGVHVSDEEQHDLYFNAPHRDFARTDEALRIRRSGG-- 59
Query: 61 RCVVSLKAKAVLVDGVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGRVKEEFGVVG 120
R ++ K + + +R E + E DP + ++ G R++ V +E
Sbjct: 60 RTFITYKGPKIDEESKTRKELETEVADPGTAAEILESLG------FRMVREVVKE----- 108
Query: 121 ENGFVGLGGFKNVRNVYDWKGLKLEVDETHFDFGTLFEIE---CESSDPEEAKRILEEFL 177
R +Y + +D T GT EIE + SD A R + E
Sbjct: 109 -------------RRIYSVGEFTVSID-TVMGLGTYLEIERDLPDGSDYAGALREIFELY 154
Query: 178 KENGIDYSYSVASKFAIFRAGK 199
++ GI+ + S + G+
Sbjct: 155 RKLGIEDGFERRSYLELLELGE 176
>UniRef100_UPI000031185C UPI000031185C UniRef100 entry
Length = 354
Score = 36.2 bits (82), Expect = 0.51
Identities = 29/129 (22%), Positives = 50/129 (38%), Gaps = 15/129 (11%)
Query: 58 DDERCVVSLKAKAVLVDGVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGRVKEEFG 117
DD R + ++ +DG+ E P + + PG + ++R + ++ G
Sbjct: 182 DDVRMALKTNPDSIYIDGM----EGGTGAGPHLATEETGVPGIAAITQAR---KAFDDLG 234
Query: 118 VVGENGFVGLGGFKNVRNVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKRILEEFL 177
GE V GG +N +V K L L D + + C PE ++
Sbjct: 235 KTGEISLVYAGGIRNGADVA--KALALGADAVAIGHSAMMALNCNKDIPE------ADYE 286
Query: 178 KENGIDYSY 186
KE G++ Y
Sbjct: 287 KEMGVEAGY 295
>UniRef100_Q57692 Hypothetical protein MJ0240 [Methanococcus jannaschii]
Length = 175
Score = 35.4 bits (80), Expect = 0.87
Identities = 32/178 (17%), Positives = 75/178 (41%), Gaps = 27/178 (15%)
Query: 1 MEVEVKLRLPNADSYHRVTTLLSPFHVITHRQHNLFFDGAGSELSSRRAILRLRFYGDDE 60
+EVE+K+++ + + L + Q +++F+G + LR+R +D
Sbjct: 2 IEVEIKVKIDDKNKVVEQLKKLGFKFIKKKFQEDIYFNGIDRDFRETDEALRIR--DEDG 59
Query: 61 RCVVSLKAKAVLVDGVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGRVKEEFGVVG 120
V+ K + ++ + E+++ KI ++ K+ + ++
Sbjct: 60 NFFVTYKGPKI-----DKISKTREEIEVKI-----EDKEKMRQIFKKL------------ 97
Query: 121 ENGFVGLGGFKNVRNVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKRILEEFLK 178
GF + + +R +Y + ++ +D+ G E+E SD E ++LEE ++
Sbjct: 98 --GFKEVPPIRKIREIYKKEDIEASIDDVE-GLGLFLELEKSISDINEKDKVLEEMME 152
>UniRef100_UPI000025867F UPI000025867F UniRef100 entry
Length = 131
Score = 34.7 bits (78), Expect = 1.5
Identities = 18/60 (30%), Positives = 32/60 (53%)
Query: 131 KNVRNVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKRILEEFLKENGIDYSYSVAS 190
+N N+ + L LE D+ FDF F + +SDP + + + L++NG+ + Y +S
Sbjct: 43 RNDINIRNGNALSLEFDDESFDFIYSFGVFHHTSDPIKCVKEAKRVLRKNGVIFLYLYSS 102
>UniRef100_UPI00002782F8 UPI00002782F8 UniRef100 entry
Length = 177
Score = 34.3 bits (77), Expect = 1.9
Identities = 18/60 (30%), Positives = 32/60 (53%)
Query: 131 KNVRNVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKRILEEFLKENGIDYSYSVAS 190
+N N+ + L LE D+ FDF F + +SDP + + + L++NG+ + Y +S
Sbjct: 43 RNDINIRNGNALSLEFDDESFDFIYSFGVFHHTSDPIKCIKEAKRVLRKNGVIFLYLYSS 102
>UniRef100_Q5W2W2 Putative TrbE protein [Sulfolobus islandicus]
Length = 624
Score = 33.9 bits (76), Expect = 2.5
Identities = 14/53 (26%), Positives = 30/53 (56%)
Query: 135 NVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKRILEEFLKENGIDYSYS 187
+VY+++G K + ++F T E++ E+++P + ++ EF K D Y+
Sbjct: 64 DVYEFEGTKFPIISSNFQLVTEKELDLETAEPPQRPKVTREFAKYLQTDQGYA 116
>UniRef100_UPI000021BE31 UPI000021BE31 UniRef100 entry
Length = 602
Score = 33.5 bits (75), Expect = 3.3
Identities = 34/120 (28%), Positives = 52/120 (43%), Gaps = 3/120 (2%)
Query: 75 GVSRVEEDEED--LDPKIGRDCVDEPGKLGLVESRIMGRVKEEFGVVGENGFVGLGGFKN 132
GVSRV++ E D L +G DE + + E + + + + E GE +N
Sbjct: 427 GVSRVDDGEADQRLREVLGEILADEDEEPQIAEEQTVAQAENEEVAGGEKTGEKTVHQEN 486
Query: 133 VRNVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKRILEEFLKENGIDYSYSVASKF 192
V V D + + E T + T EIE E + E++ EE L+ DY+ V F
Sbjct: 487 VEEV-DVETPEEEETGTLEELATEAEIEGEKDEEEKSDEDAEEQLEFQSSDYNADVEGYF 545
>UniRef100_Q769J1 Polymerase [Hepatitis B virus]
Length = 840
Score = 33.5 bits (75), Expect = 3.3
Identities = 19/66 (28%), Positives = 33/66 (49%), Gaps = 3/66 (4%)
Query: 56 YGDDERCVVS---LKAKAVLVDGVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGRV 112
+GD+ C S L K V G S+ ++ L P+ G +PG+ G++ +R+
Sbjct: 192 HGDEPFCSQSSGILSRKPVGPSGRSQGKQSRLGLQPQQGAMASGKPGRSGIIRARVHSTT 251
Query: 113 KEEFGV 118
++ FGV
Sbjct: 252 RQSFGV 257
>UniRef100_UPI00002C8CCA UPI00002C8CCA UniRef100 entry
Length = 283
Score = 33.1 bits (74), Expect = 4.3
Identities = 20/61 (32%), Positives = 34/61 (54%), Gaps = 6/61 (9%)
Query: 94 CVDEPGKLGLVESRIMGRVKEEFGVVGENGFVGLGGFKNVRNVYDWKGLKLEVDETHFDF 153
C+D+ G LGLV + I+ ++ E + G FVG+ F+N + LK +++T+F
Sbjct: 10 CIDDYG-LGLVHNEIIRKIVETGVISGVTTFVGMKNFQN-----EATALKALLNKTNFQI 63
Query: 154 G 154
G
Sbjct: 64 G 64
>UniRef100_Q6GAS3 Hypothetical protein [Staphylococcus aureus]
Length = 197
Score = 33.1 bits (74), Expect = 4.3
Identities = 32/157 (20%), Positives = 67/157 (42%), Gaps = 11/157 (7%)
Query: 28 ITHRQHNLFFDGAGSELSSRRAILRLRFYGDDERCVVSLKAKAVLVDGVSRVEEDEEDLD 87
+ +Q N + D +L R+ LR+R + + AK L++ + + + + +++
Sbjct: 30 VLFKQINYYIDTPDFKLKEHRSALRIRVKDNQYEMTLKTPAKVGLLE-YNYIVDIKPEMN 88
Query: 88 PKIGRDCVDEPGKLGLVESRIMGRVKEEFGVVGENGFVGLGGFKNVRNVYDWKGLKLEVD 147
I D + + I + E+FGV + LG R +KG L +D
Sbjct: 89 LIISNDNLPDD---------IRQIIVEQFGVKDTTLSI-LGALTTYRQETKYKGDLLVLD 138
Query: 148 ETHFDFGTLFEIECESSDPEEAKRILEEFLKENGIDY 184
++ + T +E+E E D + + + L E +++
Sbjct: 139 KSEYFDTTDYELEFEVKDYNQGLQKFQSLLNELNLEH 175
>UniRef100_Q5WF03 Hypothetical protein [Bacillus clausii]
Length = 187
Score = 33.1 bits (74), Expect = 4.3
Identities = 43/184 (23%), Positives = 69/184 (37%), Gaps = 17/184 (9%)
Query: 1 MEVEVKLRLPNADSYHRVTTLLSPFHVITHRQHNLFFDGAGSELSSRRAILRLRFYGDDE 60
+E E K L A +Y ++T F +Q N ++D A L S A LR+R
Sbjct: 5 IEYEAKSMLSEA-AYRQLT---EKFGTKAEKQQNDYYDTADFTLKSAGAALRVRRKKGSS 60
Query: 61 RCVVSLKAKAVLVDGVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGRVKEEFGVVG 120
+ A L++ + + +E L L E + +KE G +
Sbjct: 61 VFTLKAPANGGLLETHQPLTKSDEQLLLSGA-----------LPEGEVQSAIKELIGTIP 109
Query: 121 ENGFVGLGGFKNVRNVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKRILEEFLKEN 180
G R +KG L +D + + +EIE E E A+ IL + L E
Sbjct: 110 T--IRHFGSLTTYRMEVPYKGGVLCLDHSVYAGTEDYEIEYEGQSLEHAESILTQLLAEA 167
Query: 181 GIDY 184
+ +
Sbjct: 168 DVPF 171
>UniRef100_Q975X8 Hypothetical protein ST0300 [Sulfolobus tokodaii]
Length = 185
Score = 33.1 bits (74), Expect = 4.3
Identities = 39/182 (21%), Positives = 69/182 (37%), Gaps = 28/182 (15%)
Query: 2 EVEVKLRLPNADSYHRVTTLLSPFHVITHRQHNLFFDGAGSELSSRRAILRLRFYGDDER 61
E+++KL PN + ++ + H Q +++++ + LRLR ++
Sbjct: 7 EIKIKLISPNIEELEKILNIKYKLFNEEH-QIDIYYNSPVRDFRKTDEALRLRLVNNE-- 63
Query: 62 CVVSLKAKAVLVDGVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGRVKEEFGVVGE 121
V L K + S+ E++ KI D + L+ R+
Sbjct: 64 --VELTYKGPKLSSESK---SREEITVKI-----DNLDSMDLILQRL------------- 100
Query: 122 NGFVGLGGFKNVRNVYDWKGLKLEVDETHFDFGTLFEIECESSDPEEAKRILEEFLKENG 181
GF+ + + +R Y + +D FD G EIE EE + FLKE
Sbjct: 101 -GFIKVLKIEKIRKNYKINNFTVSLDRV-FDLGDFVEIEGIDITDEELINFVNNFLKEYN 158
Query: 182 ID 183
I+
Sbjct: 159 IE 160
>UniRef100_Q8PYH5 Cobalt import ATP-binding protein cbiO 2 [Methanosarcina mazei]
Length = 327
Score = 33.1 bits (74), Expect = 4.3
Identities = 17/50 (34%), Positives = 25/50 (50%)
Query: 75 GVSRVEEDEEDLDPKIGRDCVDEPGKLGLVESRIMGRVKEEFGVVGENGF 124
G+ + D++ L P + D P +GL + RVKE +VG NGF
Sbjct: 125 GIVFQDPDDQVLAPSVEEDVAFGPINMGLSREEVKMRVKEALEMVGLNGF 174
>UniRef100_UPI000042E72B UPI000042E72B UniRef100 entry
Length = 964
Score = 32.7 bits (73), Expect = 5.7
Identities = 25/83 (30%), Positives = 41/83 (49%), Gaps = 3/83 (3%)
Query: 41 GSELSSRRAILRLRFYGDDERCVVSLKAKAVLVDGVSRVEEDEEDLDPKIGRDC-VDEPG 99
G+ S L GDDE VV + + +G +VE++ + PK+ + + E G
Sbjct: 402 GTRAKSLALKLEEELLGDDEESVVEGEGEKTKPEGDVKVEDEAAEEKPKVAKSLGLSEKG 461
Query: 100 KLGLVESRIMGRVKEEFGVVGEN 122
+VE IM ++ EE G+ GE+
Sbjct: 462 S-DVVEEIIM-KMVEERGLTGED 482
>UniRef100_Q6GI82 Hypothetical protein [Staphylococcus aureus]
Length = 197
Score = 32.7 bits (73), Expect = 5.7
Identities = 32/157 (20%), Positives = 66/157 (41%), Gaps = 11/157 (7%)
Query: 28 ITHRQHNLFFDGAGSELSSRRAILRLRFYGDDERCVVSLKAKAVLVDGVSRVEEDEEDLD 87
+ +Q N + D +L R+ LR+R + + AK L++ + + + + +++
Sbjct: 30 VLFKQVNYYIDTPDFKLKEHRSALRIRVKDNQYEMTLKTPAKVGLLE-YNYIVDIKPEMN 88
Query: 88 PKIGRDCVDEPGKLGLVESRIMGRVKEEFGVVGENGFVGLGGFKNVRNVYDWKGLKLEVD 147
I D + + I + E+FGV + LG R +KG L +D
Sbjct: 89 LIISNDNLPDD---------IRQIIVEQFGVKDTTLSI-LGALTTYRQETKYKGDLLVLD 138
Query: 148 ETHFDFGTLFEIECESSDPEEAKRILEEFLKENGIDY 184
++ + T +E+E E D + + L E +++
Sbjct: 139 KSEYFDTTDYELEFEVKDYNQGLEKFQSLLNELNLEH 175
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.141 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 343,356,526
Number of Sequences: 2790947
Number of extensions: 14779135
Number of successful extensions: 38011
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 37990
Number of HSP's gapped (non-prelim): 29
length of query: 200
length of database: 848,049,833
effective HSP length: 121
effective length of query: 79
effective length of database: 510,345,246
effective search space: 40317274434
effective search space used: 40317274434
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC144477.10


