

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.7 - phase: 0
(329 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
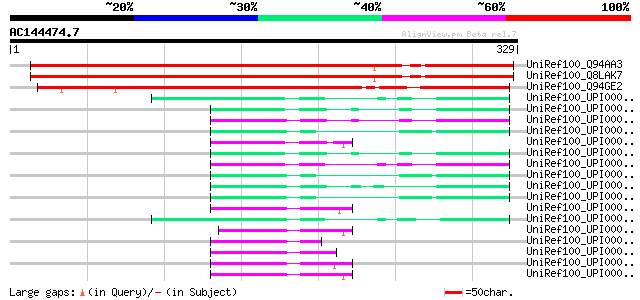
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q94AA3 AT3g09210/F3L24_8 [Arabidopsis thaliana] 342 1e-92
UniRef100_Q8LAK7 Hypothetical protein [Arabidopsis thaliana] 338 1e-91
UniRef100_Q94GE2 Putative transcription factor [Oryza sativa] 243 5e-63
UniRef100_UPI00002690A6 UPI00002690A6 UniRef100 entry 54 4e-06
UniRef100_UPI0000322140 UPI0000322140 UniRef100 entry 54 8e-06
UniRef100_UPI0000335581 UPI0000335581 UniRef100 entry 53 1e-05
UniRef100_UPI000030DD88 UPI000030DD88 UniRef100 entry 53 1e-05
UniRef100_UPI0000288771 UPI0000288771 UniRef100 entry 53 1e-05
UniRef100_UPI0000256719 UPI0000256719 UniRef100 entry 53 1e-05
UniRef100_UPI00002ED7D8 UPI00002ED7D8 UniRef100 entry 51 4e-05
UniRef100_UPI00002CBEC8 UPI00002CBEC8 UniRef100 entry 51 5e-05
UniRef100_UPI00002D97D8 UPI00002D97D8 UniRef100 entry 50 6e-05
UniRef100_UPI00002961B8 UPI00002961B8 UniRef100 entry 50 6e-05
UniRef100_UPI000028EB7C UPI000028EB7C UniRef100 entry 50 6e-05
UniRef100_UPI000025608F UPI000025608F UniRef100 entry 50 6e-05
UniRef100_UPI0000294E72 UPI0000294E72 UniRef100 entry 50 1e-04
UniRef100_UPI00002654CD UPI00002654CD UniRef100 entry 50 1e-04
UniRef100_UPI0000329C4B UPI0000329C4B UniRef100 entry 49 1e-04
UniRef100_UPI000030F19D UPI000030F19D UniRef100 entry 49 1e-04
UniRef100_UPI00002CAE26 UPI00002CAE26 UniRef100 entry 49 1e-04
>UniRef100_Q94AA3 AT3g09210/F3L24_8 [Arabidopsis thaliana]
Length = 333
Score = 342 bits (876), Expect = 1e-92
Identities = 177/319 (55%), Positives = 228/319 (70%), Gaps = 11/319 (3%)
Query: 14 PRFYTPSLSTKPPLSISATSSPELLTAKERRRLRNERRESN-ATNWKEEVENKLIQKTKK 72
P YTP T+ ++ LTAKERR+LRNERRES +W+EEVE KLI+K KK
Sbjct: 18 PSIYTPINKTQFSIAACVIERNHQLTAKERRQLRNERRESKLGYSWREEVEEKLIKKPKK 77
Query: 73 VNKSWKDELNLDNLMKLGPQWWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHE 132
+W +ELNLD L + GPQWW VRVSR++G TA+ LAR+LA+ FP++EF VYAP++
Sbjct: 78 RYATWTEELNLDTLAESGPQWWAVRVSRLRGHETAQILARALARQFPEMEFTVYAPSVQV 137
Query: 133 KKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPR 192
K++LK+GSISVK KP+FPGCIF+RC LNK +HD I++ +GVGGFIGSKVGNTK+QINKPR
Sbjct: 138 KRKLKNGSISVKPKPVFPGCIFIRCILNKEIHDSIRDVDGVGGFIGSKVGNTKRQINKPR 197
Query: 193 PVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKEL----ESDVSKAIVDSKP 248
PV + D+EAIFKQAK QE AD FEE ++ S ++EL SDV + + +SKP
Sbjct: 198 PVDDSDLEAIFKQAKEAQEKADSEFEEADRAEEEASILASQELLALSNSDVIETVAESKP 257
Query: 249 KRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHF 308
KR RK T+ E + KK KL GSTV+++SG+F F G LKKLN KT ATV F
Sbjct: 258 KRAPRKA----TLATETKA--KKKKLAAGSTVRVLSGTFAEFVGNLKKLNRKTAKATVGF 311
Query: 309 TMFGKENIVDLDVSEIVPE 327
T+FGKE +V++D++E+VPE
Sbjct: 312 TLFGKETLVEIDINELVPE 330
>UniRef100_Q8LAK7 Hypothetical protein [Arabidopsis thaliana]
Length = 333
Score = 338 bits (867), Expect = 1e-91
Identities = 176/319 (55%), Positives = 227/319 (70%), Gaps = 11/319 (3%)
Query: 14 PRFYTPSLSTKPPLSISATSSPELLTAKERRRLRNERRESN-ATNWKEEVENKLIQKTKK 72
P YTP T+ ++ LTAKERR+LRNERRES +W+EEVE KLI+K KK
Sbjct: 18 PSIYTPINKTQFSIAACVIERNHQLTAKERRQLRNERRESKLGYSWREEVEEKLIKKPKK 77
Query: 73 VNKSWKDELNLDNLMKLGPQWWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHE 132
+W +ELNLD L + GPQWW VRVSR++G TA+ LAR+LA+ FP++EF VYAP++
Sbjct: 78 RYATWTEELNLDTLAESGPQWWAVRVSRLRGHETAQILARALARQFPEMEFTVYAPSVQV 137
Query: 133 KKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPR 192
K++LK+GSISVK KP+FPGCIF+RC LNK +HD I++ +GVGGFI SKVGNTK+QINKPR
Sbjct: 138 KRKLKNGSISVKPKPVFPGCIFIRCILNKEIHDSIRDVDGVGGFIVSKVGNTKRQINKPR 197
Query: 193 PVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKEL----ESDVSKAIVDSKP 248
PV + D+EAIFKQAK QE AD FEE ++ S ++EL SDV + + +SKP
Sbjct: 198 PVDDSDLEAIFKQAKEAQEKADSEFEEADRAEEEASILASQELLALSNSDVIETVAESKP 257
Query: 249 KRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHF 308
KR RK T+ E + KK KL GSTV+++SG+F F G LKKLN KT ATV F
Sbjct: 258 KRAPRKA----TLATETKA--KKKKLAAGSTVRVLSGTFAEFVGNLKKLNRKTAKATVGF 311
Query: 309 TMFGKENIVDLDVSEIVPE 327
T+FGKE +V++D++E+VPE
Sbjct: 312 TLFGKETLVEIDINELVPE 330
>UniRef100_Q94GE2 Putative transcription factor [Oryza sativa]
Length = 330
Score = 243 bits (620), Expect = 5e-63
Identities = 131/313 (41%), Positives = 194/313 (61%), Gaps = 19/313 (6%)
Query: 19 PSLSTKPPLSISAT--SSPELLTAKERRRLRNERRESNATNWKEEVENKLI-----QKTK 71
P S P +S+S + LT +ERR+ R ERRE A +WKEEV+ +LI ++ K
Sbjct: 25 PRASPGPTISVSMSVDGGEGELTGRERRKQRGERRELRARDWKEEVQERLIHEPARRRKK 84
Query: 72 KVNKSWKDELNLDNLMKLGPQWWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIH 131
++W++ LNLD L + GPQWW VRVS G + L +++++ +P++ FK+Y P+I
Sbjct: 85 PPKRTWRENLNLDFLAEHGPQWWLVRVSMAPGTDYVDLLTKAISRRYPELSFKIYNPSIQ 144
Query: 132 EKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKP 191
KKRLK+GSIS KSKPL PG +FL C LNK +HD+I++ EG GFIG+ VG+ K+QI KP
Sbjct: 145 VKKRLKNGSISTKSKPLHPGLVFLYCTLNKEVHDFIRDTEGCYGFIGATVGSIKRQIKKP 204
Query: 192 RPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRG 251
+P+ +++E+I ++ K EQE D+ FEE E V S NK +E S+ ++ +K KR
Sbjct: 205 KPIPVEEVESIIREEKEEQERVDREFEEMENGGIVESF--NKPVED--SELMLMNKIKRQ 260
Query: 252 SRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMF 311
+K ++ G++V ++SG F GF G+L ++N K K AT+ T+F
Sbjct: 261 FKKPISK--------GGSNHNAFTPGASVHVLSGPFEGFTGSLLEVNRKNKKATLQLTLF 312
Query: 312 GKENIVDLDVSEI 324
GKE+ VDLD +I
Sbjct: 313 GKESFVDLDFDQI 325
>UniRef100_UPI00002690A6 UPI00002690A6 UniRef100 entry
Length = 174
Score = 54.3 bits (129), Expect = 4e-06
Identities = 46/232 (19%), Positives = 90/232 (37%), Gaps = 63/232 (27%)
Query: 93 WWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSKPLFPGC 152
W+ V+ + EA+ L K+ + H+ +K G + K K FPG
Sbjct: 3 WYIVQAYSGFEKKVVEAIKEELKKYKLSSNLEEILVPTHQVTEVKKGKRTKKEKKFFPGY 62
Query: 153 IFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQEN 212
+ ++ +L K ++ IK + V GF+GS +KP P+++++++ I Q
Sbjct: 63 VLIKIDLTKQIYHLIKNLQKVSGFLGS--------ADKPTPISDNEIKRILGQ------- 107
Query: 213 ADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKP 272
VS++ V ++KT I
Sbjct: 108 --------------------------VSESAV-------TQKTGQSFEI----------- 123
Query: 273 KLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
G V++ G F F G +++++ + V ++FG+ VDL+ +++
Sbjct: 124 ----GEKVKVCDGPFASFNGLIEEIDEEKSRLKVSVSIFGRATPVDLEFNQV 171
>UniRef100_UPI0000322140 UPI0000322140 UniRef100 entry
Length = 174
Score = 53.5 bits (127), Expect = 8e-06
Identities = 41/194 (21%), Positives = 77/194 (39%), Gaps = 63/194 (32%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G S K K FPG + ++ EL K ++ IK + V GF+GS +K
Sbjct: 41 HQVTEVKKGKRSKKEKKYFPGYVLIKIELTKQIYHLIKNLQKVSGFLGS--------ADK 92
Query: 191 PRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKR 250
P P+++++++ I Q ++A+N
Sbjct: 93 PTPISDNEIKRILGQV---------------SESAIN----------------------- 114
Query: 251 GSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTM 310
+KT I G V++ G F F G +++++ + V ++
Sbjct: 115 --QKTGMNFEI---------------GEKVKVCDGPFASFTGLIEEIDEEKSRLKVSVSI 157
Query: 311 FGKENIVDLDVSEI 324
FG+ VDL+ +++
Sbjct: 158 FGRPTPVDLEFNQV 171
>UniRef100_UPI0000335581 UPI0000335581 UniRef100 entry
Length = 174
Score = 53.1 bits (126), Expect = 1e-05
Identities = 41/194 (21%), Positives = 78/194 (40%), Gaps = 63/194 (32%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K K FPG + ++ EL K ++ IK + V GF+GS +K
Sbjct: 41 HQVTEVKKGKRTKKDKKFFPGYVLIKIELTKQIYHLIKNLQKVSGFLGSS--------DK 92
Query: 191 PRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKR 250
P P+++++++ I Q ++AVN
Sbjct: 93 PTPISDNEIKRILGQV---------------TESAVN----------------------- 114
Query: 251 GSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTM 310
+KT I G V++ G F F+G +++++ + V ++
Sbjct: 115 --QKTGINFEI---------------GEKVKVCDGPFASFSGLIEEIDEEKSRLKVSVSI 157
Query: 311 FGKENIVDLDVSEI 324
FG+ VDL+ +++
Sbjct: 158 FGRATPVDLEFNQV 171
>UniRef100_UPI000030DD88 UPI000030DD88 UniRef100 entry
Length = 140
Score = 53.1 bits (126), Expect = 1e-05
Identities = 43/194 (22%), Positives = 77/194 (39%), Gaps = 63/194 (32%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K K FPG I ++ +LNK ++ IK + V GF+G + K
Sbjct: 7 HKVTEVKKGKRTQKQKKYFPGYILVKLDLNKQIYHKIKNIQKVSGFLGPE--------GK 58
Query: 191 PRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKR 250
P PV+E+++
Sbjct: 59 PVPVSENEV--------------------------------------------------- 67
Query: 251 GSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTM 310
+K NQ+T TE SA ++ G V++ G F F+G +++++ V ++
Sbjct: 68 --KKIMNQITETETNPSAGISFEI--GEKVRVCDGPFASFSGLVEEIDEDKSRLKVSVSI 123
Query: 311 FGKENIVDLDVSEI 324
FG+ VDL+ +++
Sbjct: 124 FGRPTPVDLEYNQV 137
>UniRef100_UPI0000288771 UPI0000288771 UniRef100 entry
Length = 182
Score = 52.8 bits (125), Expect = 1e-05
Identities = 32/97 (32%), Positives = 51/97 (51%), Gaps = 16/97 (16%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K K FPG + ++ EL K ++ IK + V GF+GS +K
Sbjct: 41 HQVTEVKKGKRTKKEKKYFPGYVLVKIELTKQIYHLIKNLQKVSGFLGS--------ADK 92
Query: 191 PRPVAEDDMEAIFKQAKVEQENADK-----AFEEEEK 222
P P+++D+++ I Q E+ADK +FE EK
Sbjct: 93 PTPISDDEIKRILGQV---SESADKQKSGISFEIGEK 126
Score = 33.9 bits (76), Expect = 6.3
Identities = 15/64 (23%), Positives = 30/64 (46%)
Query: 261 ITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLD 320
++E A K G V++ G F F G +++++ + V ++FG+ VDL+
Sbjct: 108 VSESADKQKSGISFEIGEKVKVCDGPFASFNGLIEEIDEEKSRLKVSVSIFGRATPVDLE 167
Query: 321 VSEI 324
+
Sbjct: 168 FKSV 171
>UniRef100_UPI0000256719 UPI0000256719 UniRef100 entry
Length = 174
Score = 52.8 bits (125), Expect = 1e-05
Identities = 42/194 (21%), Positives = 76/194 (38%), Gaps = 63/194 (32%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K K FPG + ++ EL K ++ IK + V GF+GS K
Sbjct: 41 HQVTEVKKGKRTKKEKKYFPGYVLVKVELTKQIYHLIKNLQKVSGFLGS--------AEK 92
Query: 191 PRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKR 250
P P+++D+++ I Q ++AVN
Sbjct: 93 PTPISDDEIKRILGQV---------------SESAVN----------------------- 114
Query: 251 GSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTM 310
+KT I G V++ G F F G +++++ + V ++
Sbjct: 115 --QKTGISFEI---------------GEKVKVCDGPFASFNGLIEEIDEEKSRLKVSVSI 157
Query: 311 FGKENIVDLDVSEI 324
FG+ VDL+ +++
Sbjct: 158 FGRATPVDLEFNQV 171
>UniRef100_UPI00002ED7D8 UPI00002ED7D8 UniRef100 entry
Length = 174
Score = 51.2 bits (121), Expect = 4e-05
Identities = 41/194 (21%), Positives = 78/194 (40%), Gaps = 63/194 (32%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K K FPG + ++ EL K ++ IK + V GF+GS +K
Sbjct: 41 HQVTEVKKGKRTKKEKKFFPGYVLIKIELTKQIYHMIKNLQKVSGFLGS--------TDK 92
Query: 191 PRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKR 250
P P+++++++ I Q VS++ V
Sbjct: 93 PTPISDNEIKRILGQ---------------------------------VSESAV------ 113
Query: 251 GSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTM 310
++KT I G V++ G F F G +++++ + V ++
Sbjct: 114 -TQKTGISFEI---------------GEKVKVCDGPFASFNGLIEEIDEEKSRLKVSVSI 157
Query: 311 FGKENIVDLDVSEI 324
FG+ VDL+ +++
Sbjct: 158 FGRATPVDLEFNQV 171
>UniRef100_UPI00002CBEC8 UPI00002CBEC8 UniRef100 entry
Length = 152
Score = 50.8 bits (120), Expect = 5e-05
Identities = 42/194 (21%), Positives = 76/194 (38%), Gaps = 63/194 (32%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H +K G + K K FPG + ++ +L+K ++ IK + V GF+G + K
Sbjct: 19 HNVTEVKKGKRTKKQKKYFPGYVLVKLDLSKDIYHKIKNIQKVSGFLGPE--------GK 70
Query: 191 PRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKR 250
P PV+ED++
Sbjct: 71 PVPVSEDEV--------------------------------------------------- 79
Query: 251 GSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTM 310
+K NQLT +E SA ++ G V++ G F F G +++++ + V ++
Sbjct: 80 --KKIMNQLTQSETNPSAGITFEI--GEKVRVCDGPFASFNGLVEEIDEEKSRLKVSVSI 135
Query: 311 FGKENIVDLDVSEI 324
FG+ VDL+ +++
Sbjct: 136 FGRPTPVDLEYNQV 149
>UniRef100_UPI00002D97D8 UPI00002D97D8 UniRef100 entry
Length = 178
Score = 50.4 bits (119), Expect = 6e-05
Identities = 40/194 (20%), Positives = 76/194 (38%), Gaps = 63/194 (32%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K K FPG + ++ EL K ++ IK + V GF+GS +K
Sbjct: 41 HQVTEVKKGKRTKKEKKYFPGYVLIKIELTKQIYHLIKNLQKVSGFLGSS--------DK 92
Query: 191 PRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKR 250
P P+++++++ I Q ++A+N +S +S I
Sbjct: 93 PTPISDNEIKRILGQV---------------SESAINQ-------KSGISFEI------- 123
Query: 251 GSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTM 310
G V++ G F F G ++ ++ + V ++
Sbjct: 124 --------------------------GEKVKVCDGPFASFTGLIEPIDEEKSRLKVSVSI 157
Query: 311 FGKENIVDLDVSEI 324
FG+ VDL+ ++
Sbjct: 158 FGRATPVDLEFYQV 171
>UniRef100_UPI00002961B8 UPI00002961B8 UniRef100 entry
Length = 169
Score = 50.4 bits (119), Expect = 6e-05
Identities = 41/194 (21%), Positives = 76/194 (39%), Gaps = 63/194 (32%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K+K FPG + ++ +LNK ++ IK + V GF+G + K
Sbjct: 36 HKVTEVKKGKRTQKNKKYFPGYVLVKLDLNKQIYHKIKNIQKVSGFLGPE--------GK 87
Query: 191 PRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKR 250
P PV+E+++
Sbjct: 88 PVPVSENEV--------------------------------------------------- 96
Query: 251 GSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTM 310
+K NQL E SA ++ G V++ G F F G +++++ + V ++
Sbjct: 97 --KKIMNQLVEVETNPSAGISFEI--GEKVRVCDGPFASFTGLVEEIDEEKSRLKVSVSI 152
Query: 311 FGKENIVDLDVSEI 324
FG+ VDL+ +++
Sbjct: 153 FGRPTPVDLEYNQV 166
>UniRef100_UPI000028EB7C UPI000028EB7C UniRef100 entry
Length = 145
Score = 50.4 bits (119), Expect = 6e-05
Identities = 31/94 (32%), Positives = 48/94 (50%), Gaps = 10/94 (10%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K K FPG I ++ +LNK ++ IK + V GF+G + K
Sbjct: 41 HKVTEVKKGKRTQKQKKYFPGYILVKLDLNKQIYHKIKNIQKVSGFLGPE--------GK 92
Query: 191 PRPVAEDDMEAIFKQAKVEQEN--ADKAFEEEEK 222
P PV+E++++ I Q + N A FE EK
Sbjct: 93 PVPVSENEVKKIMNQITETEANPSAGITFEIGEK 126
>UniRef100_UPI000025608F UPI000025608F UniRef100 entry
Length = 174
Score = 50.4 bits (119), Expect = 6e-05
Identities = 46/232 (19%), Positives = 89/232 (37%), Gaps = 63/232 (27%)
Query: 93 WWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSKPLFPGC 152
W+ V+ + EA+ L K + H+ +K G + K K FPG
Sbjct: 3 WYIVQAYSGFEKKVVEAIKEELKKHKLSDNLQEILVPTHQVTEVKKGKRTKKEKKFFPGY 62
Query: 153 IFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQEN 212
+ ++ EL K ++ IK + V GF+GS +KP P+++++++ I Q
Sbjct: 63 VLIKIELTKQIYHLIKNLQKVSGFLGSS--------DKPTPISDNEIKRILGQ------- 107
Query: 213 ADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKP 272
VS++ V ++K I E+
Sbjct: 108 --------------------------VSESAV-------TQKVGINFEIGEK-------- 126
Query: 273 KLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
V++ G F F G +++++ + V ++FG+ VDL+ +++
Sbjct: 127 -------VKVCDGPFASFNGLIEEIDEEKSRLKVSVSIFGRATPVDLEFNQV 171
>UniRef100_UPI0000294E72 UPI0000294E72 UniRef100 entry
Length = 174
Score = 49.7 bits (117), Expect = 1e-04
Identities = 29/89 (32%), Positives = 46/89 (51%), Gaps = 10/89 (11%)
Query: 136 LKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVA 195
+K G + K K FPG + ++ +LNK ++ IK + V GF+G + KP PV+
Sbjct: 46 VKKGKRTQKQKKYFPGYVLVKLDLNKQIYHKIKNIQKVSGFLGPE--------GKPVPVS 97
Query: 196 EDDMEAIFKQAKVEQENADK--AFEEEEK 222
E++++ I QA Q + FE EK
Sbjct: 98 ENEIKKIINQANESQTSTTSGITFEVGEK 126
>UniRef100_UPI00002654CD UPI00002654CD UniRef100 entry
Length = 106
Score = 49.7 bits (117), Expect = 1e-04
Identities = 23/72 (31%), Positives = 40/72 (54%), Gaps = 8/72 (11%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K K FPG + ++ EL K ++ IK + V GF+GS +K
Sbjct: 41 HQVTEVKKGKRTKKEKKFFPGYVLIKIELTKQIYHLIKNLQKVSGFLGSS--------DK 92
Query: 191 PRPVAEDDMEAI 202
P P+++D+++ I
Sbjct: 93 PTPISDDEIKRI 104
>UniRef100_UPI0000329C4B UPI0000329C4B UniRef100 entry
Length = 139
Score = 49.3 bits (116), Expect = 1e-04
Identities = 25/82 (30%), Positives = 43/82 (51%), Gaps = 8/82 (9%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G K K +PG + ++ +LNK ++ IK + V GF+G + K
Sbjct: 41 HKITEVKQGKRKQKQKKYYPGYVLVKLDLNKQIYHKIKSIQKVSGFLGPE--------GK 92
Query: 191 PRPVAEDDMEAIFKQAKVEQEN 212
P PV+E+ ++ I QA ++N
Sbjct: 93 PIPVSENQIKKIIDQANSSKDN 114
>UniRef100_UPI000030F19D UPI000030F19D UniRef100 entry
Length = 174
Score = 49.3 bits (116), Expect = 1e-04
Identities = 28/94 (29%), Positives = 47/94 (49%), Gaps = 10/94 (10%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K + FPG + ++ EL K ++ IK + V GF+GS +K
Sbjct: 41 HQITEVKKGKRTKKERKYFPGYVLVKVELTKQIYHLIKNLQKVSGFLGSS--------DK 92
Query: 191 PRPVAEDDMEAIFKQAKVE--QENADKAFEEEEK 222
P P+++D+++ I Q E + FE EK
Sbjct: 93 PTPISDDEIKRILGQVSESAINEKSGLNFEIGEK 126
Score = 34.7 bits (78), Expect = 3.7
Identities = 15/64 (23%), Positives = 33/64 (51%)
Query: 261 ITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLD 320
++E A + K G V++ G F F G +++++ + V ++FG+ VDL+
Sbjct: 108 VSESAINEKSGLNFEIGEKVKVCDGPFASFNGLIEEIDEEKSRLKVSVSIFGRATPVDLE 167
Query: 321 VSEI 324
+++
Sbjct: 168 FNQV 171
>UniRef100_UPI00002CAE26 UPI00002CAE26 UniRef100 entry
Length = 157
Score = 49.3 bits (116), Expect = 1e-04
Identities = 28/94 (29%), Positives = 47/94 (49%), Gaps = 10/94 (10%)
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
H+ +K G + K K FPG + ++ EL K ++ IK + V GF+GS +K
Sbjct: 41 HQVTEVKKGKRTKKEKKYFPGYVLIKIELTKQIYHLIKNLQKVSGFLGSS--------DK 92
Query: 191 PRPVAEDDMEAIFKQAKVEQENADK--AFEEEEK 222
P P+++++++ I Q N +FE EK
Sbjct: 93 PTPISDNEIKRILGQVSESAINQKSGISFEIGEK 126
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.129 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 543,221,761
Number of Sequences: 2790947
Number of extensions: 22637896
Number of successful extensions: 95812
Number of sequences better than 10.0: 768
Number of HSP's better than 10.0 without gapping: 351
Number of HSP's successfully gapped in prelim test: 433
Number of HSP's that attempted gapping in prelim test: 94471
Number of HSP's gapped (non-prelim): 1591
length of query: 329
length of database: 848,049,833
effective HSP length: 127
effective length of query: 202
effective length of database: 493,599,564
effective search space: 99707111928
effective search space used: 99707111928
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144474.7


