

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.2 - phase: 0
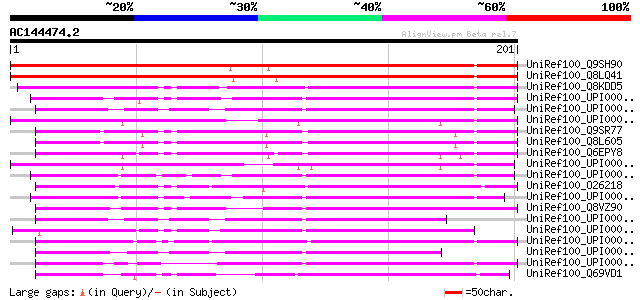
(201 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SH90 Hypothetical protein At2g37970; T8P21.12 [Arabi... 262 3e-69
UniRef100_Q8LQ41 P0529H11.19 protein [Oryza sativa] 233 2e-60
UniRef100_Q8KDD5 Hypothetical protein CT1119 [Chlorobium tepidum] 110 2e-23
UniRef100_UPI000029E73E UPI000029E73E UniRef100 entry 105 5e-22
UniRef100_UPI0000344277 UPI0000344277 UniRef100 entry 104 1e-21
UniRef100_UPI00002EBB25 UPI00002EBB25 UniRef100 entry 103 3e-21
UniRef100_Q9SR77 T22K18.4 protein [Arabidopsis thaliana] 102 5e-21
UniRef100_Q8L605 Hypothetical protein At3g10130 [Arabidopsis tha... 102 5e-21
UniRef100_Q6EPY8 SOUL heme-binding protein-like [Oryza sativa] 100 2e-20
UniRef100_UPI0000298E21 UPI0000298E21 UniRef100 entry 100 3e-20
UniRef100_UPI00002E5468 UPI00002E5468 UniRef100 entry 98 1e-19
UniRef100_O26218 Hypothetical protein MTH115 [Methanobacterium t... 96 6e-19
UniRef100_UPI00002C69CE UPI00002C69CE UniRef100 entry 94 3e-18
UniRef100_Q8VZ90 Hypothetical protein At5g20140 [Arabidopsis tha... 94 3e-18
UniRef100_UPI0000278AFB UPI0000278AFB UniRef100 entry 91 1e-17
UniRef100_UPI000032CEC3 UPI000032CEC3 UniRef100 entry 91 2e-17
UniRef100_UPI000033FBAD UPI000033FBAD UniRef100 entry 90 3e-17
UniRef100_UPI0000278014 UPI0000278014 UniRef100 entry 90 4e-17
UniRef100_UPI00002A2065 UPI00002A2065 UniRef100 entry 88 1e-16
UniRef100_Q69VD1 SOUL heme-binding protein-like [Oryza sativa] 88 1e-16
>UniRef100_Q9SH90 Hypothetical protein At2g37970; T8P21.12 [Arabidopsis thaliana]
Length = 215
Score = 262 bits (670), Expect = 3e-69
Identities = 132/216 (61%), Positives = 160/216 (73%), Gaps = 16/216 (7%)
Query: 1 MGLVFGRFSAETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIG 60
MG+VFG+ + ETPKY + K+ Y IR+Y P++ AE+TYD S FKGDKDGGF++L YIG
Sbjct: 1 MGMVFGKIAVETPKYTVTKSGDGYEIREYPPAVAAEVTYDASEFKGDKDGGFQLLAKYIG 60
Query: 61 IFGKPQNTKTEKISMTTPVITKENKS----------SSEKIAMTVPVVTNE-----KNKM 105
+FGKP+N K EKI+MT PVITKE + SEKI MT PVVT E + K+
Sbjct: 61 VFGKPENEKPEKIAMTAPVITKEGEKIAMTAPVITKESEKIEMTSPVVTKEGGGEGRKKL 120
Query: 106 VTMQFTLPSMYLKVEEVPKPIDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCL 165
VTMQF LPSMY K EE P+P DERVVI+EEGG+KYGV+ F G+AS+ VV EKV+KL L
Sbjct: 121 VTMQFLLPSMYKKAEEAPRPTDERVVIKEEGGRKYGVIKFSGIASESVVSEKVKKLSSHL 180
Query: 166 EKDGFKVIGDFLLGRYNPPAITIPMFRTNEVLIPVE 201
EKDGFK+ GDF+L RYNPP T+P FRTNEV+IPVE
Sbjct: 181 EKDGFKITGDFVLARYNPP-WTLPPFRTNEVMIPVE 215
>UniRef100_Q8LQ41 P0529H11.19 protein [Oryza sativa]
Length = 216
Score = 233 bits (594), Expect = 2e-60
Identities = 120/217 (55%), Positives = 146/217 (66%), Gaps = 17/217 (7%)
Query: 1 MGLVFGRFSAETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIG 60
MG+V G+ + ETPK+E+L T Y +RKY P +VAE+TYDP+ KGD+DGGF VL +YIG
Sbjct: 1 MGMVLGKITVETPKHEVLHTGAGYEVRKYPPCVVAEVTYDPAEMKGDRDGGFTVLGNYIG 60
Query: 61 IFGKPQNTKTEKISMTTPVITKENKSS------------SEKIAMTVPVVTNEKNK---- 104
G PQNTK EKI MT PVIT S E +AMT PV+T E+
Sbjct: 61 ALGNPQNTKPEKIDMTAPVITSGEPESIAMTAPVITSGEPEPVAMTAPVITAEERSQGKG 120
Query: 105 MVTMQFTLPSMYLKVEEVPKPIDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLC 164
+TMQF LPS Y KVEE P+P DERVV+R+ G +KYGVV F G+ D+VVKEK E L+
Sbjct: 121 QMTMQFLLPSKYSKVEEAPRPTDERVVLRQVGERKYGVVRFSGLTGDKVVKEKAEWLKAA 180
Query: 165 LEKDGFKVIGDFLLGRYNPPAITIPMFRTNEVLIPVE 201
LEKDGF V G F+L RYNPP T+P RTNEV++PVE
Sbjct: 181 LEKDGFTVKGPFVLARYNPP-FTLPPLRTNEVMVPVE 216
>UniRef100_Q8KDD5 Hypothetical protein CT1119 [Chlorobium tepidum]
Length = 215
Score = 110 bits (276), Expect = 2e-23
Identities = 71/198 (35%), Positives = 105/198 (52%), Gaps = 9/198 (4%)
Query: 4 VFGRFSAETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFG 63
V G+ A P YE+LK + +R+Y P ++AE D ++ GF L YI FG
Sbjct: 18 VLGKREAAEPPYELLKHDGAFEVRRYGPMVIAETILDEKSYSAASGKGFNRLAGYI--FG 75
Query: 64 KPQNTKTEKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVP 123
K N ISMT PV+ + SSEKI+MT PV+ + +M F LP + ++ P
Sbjct: 76 K--NRSKTSISMTAPVLQER---SSEKISMTAPVLQQPQKGGWSMAFVLPEGFT-LQSAP 129
Query: 124 KPIDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNP 183
+P+D V +RE VVTF G+ S +++ +L+ L+K G++ + + L Y+P
Sbjct: 130 EPLDPEVKLRELPPSTIAVVTFSGLHSAANLEKYSRQLQAWLKKQGYRALSEPKLASYDP 189
Query: 184 PAITIPMFRTNEVLIPVE 201
P TIP R NEV I +E
Sbjct: 190 P-WTIPFLRRNEVQIRIE 206
>UniRef100_UPI000029E73E UPI000029E73E UniRef100 entry
Length = 206
Score = 105 bits (263), Expect = 5e-22
Identities = 72/196 (36%), Positives = 106/196 (53%), Gaps = 17/196 (8%)
Query: 9 SAETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDG---GFKVLVDYIGIFGKP 65
+ E P Y+++++ QN IR+Y P ++AE+ F KD GF++L DYI FG
Sbjct: 22 NVEKPDYKVVQSEQNIEIRQYEPMIIAEV----EVFGKRKDAINDGFRLLADYI--FGN- 74
Query: 66 QNTKTEKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKP 125
NT + ISMT PV KEN +KIAMT PV + F +PS Y K++ +P P
Sbjct: 75 -NTLEKSISMTAPVQQKEN----QKIAMTAPVQQQLTENSWKISFVMPSKY-KMDSLPVP 128
Query: 126 IDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPA 185
+ RV ++E KK+ V+ F G S E + E +L +E+ ++IG YN P
Sbjct: 129 NNNRVRLKEIFTKKFVVIEFSGTNSTENLNEHENELMNYIERKQIQIIGSPKYAFYNAP- 187
Query: 186 ITIPMFRTNEVLIPVE 201
T+P R NEV+I ++
Sbjct: 188 WTLPFLRRNEVMIEIK 203
>UniRef100_UPI0000344277 UPI0000344277 UniRef100 entry
Length = 203
Score = 104 bits (260), Expect = 1e-21
Identities = 65/190 (34%), Positives = 102/190 (53%), Gaps = 18/190 (9%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNTKT 70
E PKY+I+K+ +NY IRKY L E+ Y ++D GF+ L +YI +NT
Sbjct: 26 EEPKYQIIKSNKNYEIRKYKDRLAVEVEY------SNEDSGFRYLFNYI----SGENTSL 75
Query: 71 EKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPIDERV 130
EKI+MT PV + S KI MT PV KN + MQF LPS + ++ PKP +++V
Sbjct: 76 EKINMTVPV------TQSVKINMTAPVTQTTKNNKMLMQFFLPSKF-SLDTAPKPTNKKV 128
Query: 131 VIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAITIPM 190
+ Y V+ + G +++ ++ ++L+ L KD + +N P T+P+
Sbjct: 129 RLVILKSCYYAVIKYSGRVTNKNFQKHYKELKKYLIKDNINFTEPAIRATFNGP-FTLPV 187
Query: 191 FRTNEVLIPV 200
FR NEV++ +
Sbjct: 188 FRRNEVMMKI 197
>UniRef100_UPI00002EBB25 UPI00002EBB25 UniRef100 entry
Length = 194
Score = 103 bits (257), Expect = 3e-21
Identities = 68/206 (33%), Positives = 104/206 (50%), Gaps = 18/206 (8%)
Query: 1 MGLVFGRFSAETPKYEILKTTQNYVIRKYAPSLVAEITYDPST--FKGDKDGGFKVLVDY 58
MG + G+ + E PK+E++ T Y +R+YAP +VAE ++ S F GD+ G F L +
Sbjct: 1 MGSIIGKITEELPKHEVVGKTAAYELRRYAPCVVAETSFASSKGMFGGDQGGSFMRLAKF 60
Query: 59 IGIFGKPQNTKTEKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLP-SMYL 117
IG+ KPQN + E I+MT PV+ + + S + M F LP S +
Sbjct: 61 IGVMAKPQNEQAEPIAMTAPVLMQRDDS------------RRGADGAYKMAFFLPASRFA 108
Query: 118 KVEEVPKPIDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDG--FKVIGD 175
K + PKP + V + + + TF G ++ EK +LR LE+DG K +
Sbjct: 109 KAADAPKPTNPDVTLVDLPARVVAAKTFSGNMRAGLIAEKERELREELERDGVRLKPGAE 168
Query: 176 FLLGRYNPPAITIPMFRTNEVLIPVE 201
+ +NPP T +TNEV++ VE
Sbjct: 169 VMAAGFNPP-WTPSFLKTNEVMLEVE 193
>UniRef100_Q9SR77 T22K18.4 protein [Arabidopsis thaliana]
Length = 309
Score = 102 bits (255), Expect = 5e-21
Identities = 75/199 (37%), Positives = 104/199 (51%), Gaps = 16/199 (8%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGG---FKVLVDYIGIFGKPQN 67
ET + +L T Y IR+ P VAE T P D G F VL +Y+ FGK N
Sbjct: 115 ETMNFRVLFRTDKYEIRQVEPYFVAE-TIMPGETGFDSYGASKSFNVLAEYL--FGK--N 169
Query: 68 TKTEKISMTTPVITKENKSSSEKIAMTVPVVTN--EKNKMVTMQFTLPSMYLKVEEVPKP 125
T EK+ MTTPV+T++ +S EK+ MT PV+T+ + M F +PS Y +P P
Sbjct: 170 TIKEKMEMTTPVVTRKVQSVGEKMEMTTPVITSKAKDQNQWRMSFVMPSKY--GSNLPLP 227
Query: 126 IDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGD---FLLGRYN 182
D V I++ K VV F G +DE ++ + +LR L+ D + D F + +YN
Sbjct: 228 KDPSVKIQQVPRKIVAVVAFSGYVTDEEIERRERELRRALQNDKKFRVRDGVSFEVAQYN 287
Query: 183 PPAITIPMFRTNEVLIPVE 201
PP T+P R NEV + VE
Sbjct: 288 PP-FTLPFMRRNEVSLEVE 305
>UniRef100_Q8L605 Hypothetical protein At3g10130 [Arabidopsis thaliana]
Length = 309
Score = 102 bits (255), Expect = 5e-21
Identities = 75/199 (37%), Positives = 104/199 (51%), Gaps = 16/199 (8%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGG---FKVLVDYIGIFGKPQN 67
ET + +L T Y IR+ P VAE T P D G F VL +Y+ FGK N
Sbjct: 115 ETMNFRVLFRTDKYEIRQVEPYFVAE-TIMPGETGFDSYGASKSFNVLAEYL--FGK--N 169
Query: 68 TKTEKISMTTPVITKENKSSSEKIAMTVPVVTN--EKNKMVTMQFTLPSMYLKVEEVPKP 125
T EK+ MTTPV+T++ +S EK+ MT PV+T+ + M F +PS Y +P P
Sbjct: 170 TIKEKMEMTTPVVTRKVQSVGEKMEMTTPVITSKAKDQNQWRMSFVMPSKY--GSNLPLP 227
Query: 126 IDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGD---FLLGRYN 182
D V I++ K VV F G +DE ++ + +LR L+ D + D F + +YN
Sbjct: 228 KDPSVKIQQVPRKIVAVVAFSGYVTDEEIERRERELRRALQNDKKFRVRDGVSFEVAQYN 287
Query: 183 PPAITIPMFRTNEVLIPVE 201
PP T+P R NEV + VE
Sbjct: 288 PP-FTLPFMRRNEVSLEVE 305
>UniRef100_Q6EPY8 SOUL heme-binding protein-like [Oryza sativa]
Length = 287
Score = 100 bits (250), Expect = 2e-20
Identities = 72/200 (36%), Positives = 107/200 (53%), Gaps = 18/200 (9%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPST---FKGDKDGGFKVLVDYIGIFGKPQN 67
ET + +LK Y IR+ VAE T + F G F VL Y+ FGK N
Sbjct: 91 ETVPFRVLKREAEYEIREVESYYVAETTMPGRSGFDFNGSSQS-FNVLASYL--FGK--N 145
Query: 68 TKTEKISMTTPVITKENKSSSEKIAMTVPVVTNE---KNKMVTMQFTLPSMYLKVEEVPK 124
T +E++ MTTPV T++ + EK+ MT PV+T + +NK M F +PS Y ++P
Sbjct: 146 TTSEQMEMTTPVFTRKGEPDGEKMDMTTPVITKKSANENKW-KMSFVMPSKY--GPDLPL 202
Query: 125 PIDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDG-FKVIGDFL--LGRY 181
P D V I+E K V F G+ +D+ + ++ +LR L+KD F+V D + + +Y
Sbjct: 203 PKDPSVTIKEVPAKIVAVAAFSGLVTDDDISQRESRLRETLQKDSQFRVKDDSVVEIAQY 262
Query: 182 NPPAITIPMFRTNEVLIPVE 201
NPP T+P R NE+ + V+
Sbjct: 263 NPP-FTLPFTRRNEIALEVK 281
>UniRef100_UPI0000298E21 UPI0000298E21 UniRef100 entry
Length = 196
Score = 100 bits (248), Expect = 3e-20
Identities = 71/206 (34%), Positives = 97/206 (46%), Gaps = 18/206 (8%)
Query: 1 MGLVFGRFSAETPKYEILKTTQNYVIRKYAPSLVAEITYDPST--FKGDKDGGFKVLVDY 58
MG FGR + E P Y ++T Y IR+Y P V E +Y S GD+ F L Y
Sbjct: 1 MGSAFGRITEEQPSYATIRTADEYEIRRYEPCCVIETSYASSAGMVSGDQGTSFMALARY 60
Query: 59 IGIFGKPQNTKTEKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLP-SMYL 117
IG+ +P N + EKI MT PV + ++S+ A+ M+F LP SM+
Sbjct: 61 IGVLSEPANERREKIDMTAPVFMTTDGAASDTDAI-----------RYVMRFVLPKSMFA 109
Query: 118 K-VEEVPKPIDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDG--FKVIG 174
+ P ERV +R+ + V F G ++ V+ E LR L +DG F
Sbjct: 110 DGAKSAPASTQERVRVRDLDARVVAVRRFSGRMREDAVRAHAEILREALRRDGVAFAENA 169
Query: 175 DFLLGRYNPPAITIPMFRTNEVLIPV 200
YNPP T +FRTNEV+I V
Sbjct: 170 ASEYAGYNPP-WTPGLFRTNEVMIEV 194
>UniRef100_UPI00002E5468 UPI00002E5468 UniRef100 entry
Length = 202
Score = 97.8 bits (242), Expect = 1e-19
Identities = 67/192 (34%), Positives = 103/192 (52%), Gaps = 11/192 (5%)
Query: 9 SAETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNT 68
+ E P Y+I+++ QN IR+Y P ++AE+ + + GF+++ DY IFG +
Sbjct: 22 NVEKPDYKIIESQQNIEIREYDPMIIAEVEVGGKR-EDAINNGFRIIADY--IFGNNEVK 78
Query: 69 KTEKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPIDE 128
KT ISMT PV K+N KIAMT PV K K + F +PS Y ++ +P P +
Sbjct: 79 KT--ISMTAPVQQKKN----HKIAMTAPVQQQLKGKSWRISFVMPSKY-SMDSLPVPNNN 131
Query: 129 RVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAITI 188
RV ++E KK+ V+ F G S E + + +L ++ + + IG YN P T+
Sbjct: 132 RVHLKEILAKKFVVIQFSGTNSHENLTKHENQLMNYVKVNKIEFIGSPKYAYYNAP-WTL 190
Query: 189 PMFRTNEVLIPV 200
P R NEV+I +
Sbjct: 191 PFLRRNEVMIEI 202
>UniRef100_O26218 Hypothetical protein MTH115 [Methanobacterium thermoautotrophicum]
Length = 189
Score = 95.9 bits (237), Expect = 6e-19
Identities = 60/195 (30%), Positives = 110/195 (55%), Gaps = 12/195 (6%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNTKT 70
E+P+Y + + IR+Y ++A++ + S F+ GF +L +YI FG N +
Sbjct: 3 ESPEYTVELKDGKFEIRRYPGYILAQVDVEAS-FRDAMVIGFSILANYI--FGG--NRRK 57
Query: 71 EKISMTTPVITKENKSSSEKIAMTVPVVT----NEKNKMVTMQFTLPSMYLKVEEVPKPI 126
E++ MT+PV T N SSE+I M VPV + + + FT+PS Y +E +P+P+
Sbjct: 58 EELPMTSPV-TGVNLGSSERIPMKVPVTEEVPDDADSGKYRISFTMPSSYT-LETLPEPL 115
Query: 127 DERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAI 186
D+R+ REE +++ F G + ++ +++ +L+ LE++ + +F++ +YN PA+
Sbjct: 116 DDRIRFREEKDQRFAAYRFSGRVNSDMAAQRIAELKEWLERNSIEPRSNFIIAQYNHPAV 175
Query: 187 TIPMFRTNEVLIPVE 201
R NEVL+ ++
Sbjct: 176 P-GFLRKNEVLVKID 189
>UniRef100_UPI00002C69CE UPI00002C69CE UniRef100 entry
Length = 211
Score = 93.6 bits (231), Expect = 3e-18
Identities = 61/188 (32%), Positives = 97/188 (51%), Gaps = 11/188 (5%)
Query: 9 SAETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNT 68
+ E P Y+IL+ T+N IR+Y P ++AE+ S K GF++L D+I FGK +
Sbjct: 29 NVEIPDYKILEKTENIEIRRYPPLIIAEVRTMGSR-KDSIGDGFRILADFI--FGKYKGE 85
Query: 69 KTEKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPIDE 128
KISMTTPV+ +E + MT PV K + F +PS + + +PKP ++
Sbjct: 86 N--KISMTTPVMQQEGTKNE----MTAPVQQENTGKEWLVSFIMPSKF-SMNTLPKPTND 138
Query: 129 RVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAITI 188
++ I + K Y + F G + + + +L+ L + + + G+ YNPP T+
Sbjct: 139 KIKIVQMPSKNYAAIVFSGRGTKDNLTANTLELKEYLSRSNYSIRGNPKYAFYNPP-WTL 197
Query: 189 PMFRTNEV 196
P R NEV
Sbjct: 198 PFMRRNEV 205
>UniRef100_Q8VZ90 Hypothetical protein At5g20140 [Arabidopsis thaliana]
Length = 378
Score = 93.6 bits (231), Expect = 3e-18
Identities = 67/191 (35%), Positives = 95/191 (49%), Gaps = 23/191 (12%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNTKT 70
ETPKY+ILK T NY +R Y P +V E D K GF + YI FGK N+
Sbjct: 206 ETPKYQILKRTANYEVRNYEPFIVVETIGD----KLSGSSGFNNVAGYI--FGK--NSTM 257
Query: 71 EKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPIDERV 130
EKI MTTPV T+ + + + V++Q +PS + +P P +E+V
Sbjct: 258 EKIPMTTPVFTQTTDT--------------QLSSDVSVQIVIPSGK-DLSSLPMPNEEKV 302
Query: 131 VIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAITIPM 190
+++ G V F G +++VV+ K +LR L KDG + +L RYN P T
Sbjct: 303 NLKKLEGGFAAAVKFSGKPTEDVVQAKENELRSSLSKDGLRAKKGCMLARYNDPGRTWNF 362
Query: 191 FRTNEVLIPVE 201
NEV+I +E
Sbjct: 363 IMRNEVIIWLE 373
>UniRef100_UPI0000278AFB UPI0000278AFB UniRef100 entry
Length = 149
Score = 91.3 bits (225), Expect = 1e-17
Identities = 58/163 (35%), Positives = 88/163 (53%), Gaps = 17/163 (10%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNTKT 70
E PKY+ILK+ Y +RKY L E+ Y ++D GF+ L +YI +NT++
Sbjct: 4 EEPKYQILKSNGKYEVRKYNDRLAVEVEY------SNEDSGFRYLFNYI----SGENTQS 53
Query: 71 EKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPIDERV 130
EK++MT PV + S KI MT PV ++++ + MQF LPS + E PKP + RV
Sbjct: 54 EKVNMTVPV------TQSVKIDMTAPVTQSKEDGKMVMQFFLPSKF-TFETAPKPTNNRV 106
Query: 131 VIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVI 173
+ G Y V+ + G +D+ ++ E+L+ L KD I
Sbjct: 107 SLVVLEGGVYAVIKYSGRLTDKNFQKHYEELKDYLIKDKINFI 149
>UniRef100_UPI000032CEC3 UPI000032CEC3 UniRef100 entry
Length = 189
Score = 90.5 bits (223), Expect = 2e-17
Identities = 60/185 (32%), Positives = 95/185 (50%), Gaps = 12/185 (6%)
Query: 2 GLVFGRFSA--ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYI 59
GL++G + E PKY+++ + + +R Y P++VAE+ D + G F++L DYI
Sbjct: 15 GLLWGPIMSQVERPKYKVIMSKGSIEVRSYLPTIVAEVEVDGEREQAINQG-FRLLADYI 73
Query: 60 GIFGKPQNTKTEKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKV 119
FG N K +KI+MT PV + SS+KIAMT PV N +QF +PS Y +
Sbjct: 74 --FGN--NIKKDKIAMTAPV----TQQSSQKIAMTAPVTQQVVNDKWRVQFIMPSEY-TL 124
Query: 120 EEVPKPIDERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLG 179
E +P P+ V+I++ K Y + F G ++ + + +L L + +
Sbjct: 125 ESLPSPVQPEVLIKKIPAKVYAAIKFNGNGDNQNITKHQSQLNKFLTDKKLLSKSEPMYA 184
Query: 180 RYNPP 184
YNPP
Sbjct: 185 FYNPP 189
>UniRef100_UPI000033FBAD UPI000033FBAD UniRef100 entry
Length = 211
Score = 90.1 bits (222), Expect = 3e-17
Identities = 65/191 (34%), Positives = 99/191 (51%), Gaps = 8/191 (4%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNTKT 70
E PKY +LK +N+ IR YA LVAE+ + S K + F++L Y IFG N +
Sbjct: 26 EEPKYSVLKEYENFEIRNYASYLVAEVDIEGSYNKSGNE-AFRILAGY--IFG--DNQSS 80
Query: 71 EKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPIDERV 130
K++MT PV E SSEK+ MT PV +N+ T +F + S Y + E +P P + ++
Sbjct: 81 TKMNMTAPV-ESEAIQSSEKMNMTAPVFSNKNFNGYTYRFVMESKYTQ-ETLPVPNNSKI 138
Query: 131 VIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAITIPM 190
I E + V++F G S + ++ + L L+ G V + + RYN P T
Sbjct: 139 RITEIKDRVMAVISFSGRWSQKNFEKHEQILVNDLKNAGIGVASEAIYARYNAP-FTPWF 197
Query: 191 FRTNEVLIPVE 201
R NE++ +E
Sbjct: 198 LRRNEIMFEIE 208
>UniRef100_UPI0000278014 UPI0000278014 UniRef100 entry
Length = 169
Score = 89.7 bits (221), Expect = 4e-17
Identities = 56/161 (34%), Positives = 84/161 (51%), Gaps = 17/161 (10%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNTKT 70
E P Y I+K+ +NY IRKY L E+ + +D GF+ L +YI +NT +
Sbjct: 26 EEPSYSIVKSNENYEIRKYKDRLAVEVEFK------SEDSGFQYLFNYI----SGENTTS 75
Query: 71 EKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPIDERV 130
EK++MT PV + S KI MT PV + K+ + MQF LPS + ++ P P ++RV
Sbjct: 76 EKVNMTVPV------TQSAKIDMTAPVTQSNKDGKMLMQFFLPSKF-TIDTAPTPTNKRV 128
Query: 131 VIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFK 171
+ G Y + + G SD K+ +EKL+ L D +
Sbjct: 129 KLVIIEGGYYAIKRYSGRTSDTNYKKHLEKLKESLTSDNIE 169
>UniRef100_UPI00002A2065 UPI00002A2065 UniRef100 entry
Length = 182
Score = 88.2 bits (217), Expect = 1e-16
Identities = 64/191 (33%), Positives = 91/191 (47%), Gaps = 31/191 (16%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDKDGGFKVLVDYIGIFGKPQNTKT 70
E YEI+K +NY IRKY+ LV E ++ +G+ GF+ L +YI
Sbjct: 22 EEANYEIVKENKNYEIRKYSDRLVIET----NSIQGN---GFRKLFNYIS---------- 64
Query: 71 EKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPIDERV 130
N +E+I MTVPV KN +TMQF LPS Y + PKP++ V
Sbjct: 65 ------------GNNKKNEEIKMTVPVTQEVKNGNMTMQFYLPSKY-NEDNAPKPVNSEV 111
Query: 131 VIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAITIPM 190
I G Y V+ + G SD+ + E L L++D V+ + YN P T+PM
Sbjct: 112 KILTIEGGYYAVIKYSGRTSDKNFLKNKEILDKKLKQDNITVLSPPIRASYNSP-FTLPM 170
Query: 191 FRTNEVLIPVE 201
+ NEV+ V+
Sbjct: 171 LKRNEVMYRVD 181
>UniRef100_Q69VD1 SOUL heme-binding protein-like [Oryza sativa]
Length = 381
Score = 88.2 bits (217), Expect = 1e-16
Identities = 73/191 (38%), Positives = 93/191 (48%), Gaps = 31/191 (16%)
Query: 11 ETPKYEILKTTQNYVIRKYAPSLVAEITYDPSTFKGDK---DGGFKVLVDYIGIFGKPQN 67
ETPKY ILK T NY IR Y P L+ E KGDK GF + Y IFGK N
Sbjct: 211 ETPKYLILKRTANYEIRSYPPFLIVEA-------KGDKLTGSSGFNNVTGY--IFGK--N 259
Query: 68 TKTEKISMTTPVITKENKSSSEKIAMTVPVVTNEKNKMVTMQFTLPSMYLKVEEVPKPID 127
+E I+MTTPV T+ +++K V++Q LP M ++ +P P
Sbjct: 260 ASSETIAMTTPVFTQ---------------ASDDKLSDVSIQIVLP-MNKDLDSLPAPNT 303
Query: 128 ERVVIREEGGKKYGVVTFGGVASDEVVKEKVEKLRLCLEKDGFKVIGDFLLGRYNPPAIT 187
E V +R+ G V F G +E+V +K ++LR L KD K LL RYN P T
Sbjct: 304 EAVNLRKVEGGIAAVKKFSGRPKEEIVIQKEKELRSQLLKDVLKPQHGCLLARYNDPR-T 362
Query: 188 IPMFRTNEVLI 198
NEVLI
Sbjct: 363 QSFIMRNEVLI 373
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.137 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 352,843,274
Number of Sequences: 2790947
Number of extensions: 15254256
Number of successful extensions: 43134
Number of sequences better than 10.0: 215
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 115
Number of HSP's that attempted gapping in prelim test: 42677
Number of HSP's gapped (non-prelim): 226
length of query: 201
length of database: 848,049,833
effective HSP length: 121
effective length of query: 80
effective length of database: 510,345,246
effective search space: 40827619680
effective search space used: 40827619680
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC144474.2


