

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144404.22 + phase: 0
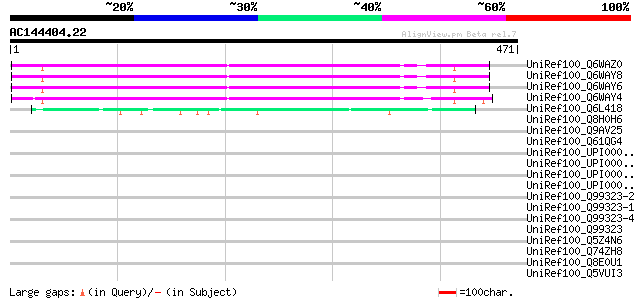
(471 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum] 277 5e-73
UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum] 274 3e-72
UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum] 274 5e-72
UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum] 269 1e-70
UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demi... 63 2e-08
UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum] 47 0.001
UniRef100_Q9AV25 Putative kinesin-related protein [Oryza sativa] 41 0.085
UniRef100_Q61QG4 Hypothetical protein CBG07027 [Caenorhabditis b... 40 0.14
UniRef100_UPI0000080FE8 UPI0000080FE8 UniRef100 entry 39 0.25
UniRef100_UPI0000079B2E UPI0000079B2E UniRef100 entry 39 0.25
UniRef100_UPI000042FC3C UPI000042FC3C UniRef100 entry 39 0.25
UniRef100_UPI000042C8BB UPI000042C8BB UniRef100 entry 39 0.25
UniRef100_Q99323-2 Splice isoform 2 of Q99323 [Drosophila melano... 39 0.25
UniRef100_Q99323-1 Splice isoform 1 of Q99323 [Drosophila melano... 39 0.25
UniRef100_Q99323-4 Splice isoform 4 of Q99323 [Drosophila melano... 39 0.25
UniRef100_Q99323 Myosin heavy chain, non-muscle [Drosophila mela... 39 0.25
UniRef100_Q5Z4N6 Epstein-Barr virus EBNA-1-like protein [Oryza s... 39 0.42
UniRef100_Q74ZH8 AGR229Wp [Ashbya gossypii] 39 0.42
UniRef100_Q8E0U1 Conserved domain protein [Streptococcus agalact... 38 0.55
UniRef100_Q5VUI3 Trichohyalin [Homo sapiens] 38 0.72
>UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum]
Length = 562
Score = 277 bits (708), Expect = 5e-73
Identities = 166/451 (36%), Positives = 237/451 (51%), Gaps = 18/451 (3%)
Query: 2 RSLRQYSFRKPDLLSLRKLGKKVLCTDD--FFEQHGNLLGVVKTDIDEGLLNAFVQFYDP 59
R YSF + L SL +L + + F +Q+G++L ++K +D L +QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 60 GYHCFTFQDYQILPTLEEYSCWINLPVLDKVPFSGLEETPKHSTIAATLHLETDEVKTHL 119
HCFTFQDYQ+ PTLEEYS +++PV +VPF + + +A LHL EV +
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKEVSDNW 144
Query: 120 ITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFVDIKAIQIFL 179
+ G +G FL + G AF++ LA+++YG+VLFPS+ NFVD A+ IF+
Sbjct: 145 KSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 180 SKNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSLLPRTPHFTTNPENLLWSKR 239
NPVPTLL DTY +IH R G G I CC LL RW SLLP + F W++R
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 240 LMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARRQFGYPMEKKPL 299
+MSLT ++ W D +I SCG F NVPL+G G I+YNP L+ RQ G+ M+ +PL
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 300 SIYLENVYYFNTEDSTGMREQVVRAWHTIRRRDKGQLGKKTGAVHESYTQWVIDRAVQIR 359
+ YF + EQ+ RAW +I +D LGKK YT WV +R +
Sbjct: 324 EAEIAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETLL 383
Query: 360 MPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQE-----YDT 414
+PY + + PPL + + AE +++ M R+ R+KEQ+ Y
Sbjct: 384 LPYD---RMEPLQEQPPLIL-------AESVPAEHYKQALMENRRLREKEQDTQMELYKA 433
Query: 415 LKNLLDQQIQANRQEKEENIRLRAALQKKED 445
L+ Q +E+ RLR+ + E+
Sbjct: 434 KAGRLNLAHQLRGVREEDASRLRSKKRSYEE 464
>UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum]
Length = 562
Score = 274 bits (701), Expect = 3e-72
Identities = 165/451 (36%), Positives = 235/451 (51%), Gaps = 18/451 (3%)
Query: 2 RSLRQYSFRKPDLLSLRKLGKKVLCTDD--FFEQHGNLLGVVKTDIDEGLLNAFVQFYDP 59
R YSF + L SL +L + + F +Q+G++L ++K +D L +QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 60 GYHCFTFQDYQILPTLEEYSCWINLPVLDKVPFSGLEETPKHSTIAATLHLETDEVKTHL 119
HCFTFQDYQ+ PTLEEYS +++PV +VPF + + +A LHL EV
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKEVSDSW 144
Query: 120 ITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFVDIKAIQIFL 179
+ G +G FL + G AF++ LA+++YG+VLFPS+ NFVD A+ IF+
Sbjct: 145 KSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 180 SKNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSLLPRTPHFTTNPENLLWSKR 239
NPVPTLL DTY +IH R G G I CC LL RW SLLP + F W++R
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 240 LMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARRQFGYPMEKKPL 299
+MSLT ++ W D +I SCG F NVPL+G G I+YNP L+ RQ G+ M+ +PL
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLIGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 300 SIYLENVYYFNTEDSTGMREQVVRAWHTIRRRDKGQLGKKTGAVHESYTQWVIDRAVQIR 359
+ F EQ+ RAW +I +D LGKK YT WV +R +
Sbjct: 324 EAEVAESVCFEKRSDPARLEQIGRAWKSIGMKDGSVLGKKFAVAMPDYTDWVKERVETLL 383
Query: 360 MPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQE-----YDT 414
+PY + + PPL + + AE +++ M R+ R+KEQ+ Y
Sbjct: 384 LPYD---RMEPLQEQPPLIL-------AESVPAEHYKQALMENRRLREKEQDTQMELYKA 433
Query: 415 LKNLLDQQIQANRQEKEENIRLRAALQKKED 445
+ L+ Q +E+ RLR+ + E+
Sbjct: 434 KADSLNLAHQLRGVREEDASRLRSKKRSYEE 464
>UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum]
Length = 562
Score = 274 bits (700), Expect = 5e-72
Identities = 166/451 (36%), Positives = 233/451 (50%), Gaps = 18/451 (3%)
Query: 2 RSLRQYSFRKPDLLSLRKLGKKVLCTDD--FFEQHGNLLGVVKTDIDEGLLNAFVQFYDP 59
R YSF + L SL +L + + F +QHG++L ++K +D L +QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQHGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 60 GYHCFTFQDYQILPTLEEYSCWINLPVLDKVPFSGLEETPKHSTIAATLHLETDEVKTHL 119
HCFTFQDYQ+ PTLEEYS +++PV +VPF + + +A LHL EV
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFGVVARALHLGIKEVSDSW 144
Query: 120 ITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFVDIKAIQIFL 179
G +G FL + G AF + LA+++YG+VLFPS+ NFVD A+ IF+
Sbjct: 145 KYSGDVVGLPLKFLLRVAREEAEKGSWEAFRAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 180 SKNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSLLPRTPHFTTNPENLLWSKR 239
NPVPTLL DTY +IH R G G I CC LL RW SLLP + F W++R
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 240 LMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARRQFGYPMEKKPL 299
+MSLT ++ W D +I SCG F NVPL+G G I+YNP L+ RQ G+ M+ +PL
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 300 SIYLENVYYFNTEDSTGMREQVVRAWHTIRRRDKGQLGKKTGAVHESYTQWVIDRAVQIR 359
+ YF + EQ+ RAW +I +D LGKK YT WV +R +
Sbjct: 324 EAEVAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETLL 383
Query: 360 MPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQE-----YDT 414
+PY + + PP + + AE +++ M R+ R KEQ+ Y
Sbjct: 384 LPYD---RMEPLQEQPPSIL-------AESVPAEHYKQALMENRRLRGKEQDTQLELYKA 433
Query: 415 LKNLLDQQIQANRQEKEENIRLRAALQKKED 445
+ L+ Q +E+ RLR+ + E+
Sbjct: 434 KADKLNLAHQLRGVREEDASRLRSKKRSYEE 464
>UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum]
Length = 546
Score = 269 bits (687), Expect = 1e-70
Identities = 166/458 (36%), Positives = 238/458 (51%), Gaps = 23/458 (5%)
Query: 2 RSLRQYSFRKPDLLSLRKLGKKVLCTDD---FFEQHGNLLGVVKTDIDEGLLNAFVQFYD 58
R YSF + L SL +LG ++ +D F Q G++L ++KT +D L +QFYD
Sbjct: 9 RKTCSYSFYREPLTSLIELGS-LMPSDQLKGFVGQFGDILTLLKTVVDPVPLQTLLQFYD 67
Query: 59 PGYHCFTFQDYQILPTLEEYSCWINLPVLDKVPFSGLEETPKHSTIAATLHLETDEVKTH 118
P CFTFQDYQ+ PTLEEYS +++P+ +VPF + + IA L + EV +
Sbjct: 68 PELRCFTFQDYQLAPTLEEYSILMSIPIQHQVPFLDVPKEVDFRVIARALRMSVKEVCDN 127
Query: 119 LITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFVDIKAIQIF 178
G+ +G FL + G F++ LA ++YG+VLFPS+ NF+D AI IF
Sbjct: 128 WKPSGEVVGMPLKFLLRVAREEAEKGNWEVFHAQLAAMIYGIVLFPSMPNFIDHAAISIF 187
Query: 179 LSKNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSLLPRTPHFTTNPENLLWSK 238
+ NPVPTLL DTY +IH R G G I CC LL RW SLLP + F L W++
Sbjct: 188 IGGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFMSLLPASGPFMDTQSTLKWTQ 246
Query: 239 RLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARRQFGYPMEKKP 298
R+MSLT ++ W D ++ SCG+F NVPL+G +G I+YNP L+ RQ G+ M ++P
Sbjct: 247 RVMSLTSYDIRWQSYRMDVKDVVMSCGEFRNVPLVGTKGCINYNPVLSLRQLGFIMSRRP 306
Query: 299 LSIYLENVYYFNTEDSTGMREQVVRAWHTIRRRDKGQLGKKTGAVHESYTQWVIDRAVQI 358
L + YF EQ+ RAW +I +D LGKK YT WV R +
Sbjct: 307 LEAEIVESVYFEKRTDPVRLEQIGRAWKSIGVKDGSVLGKKFAIAMPDYTDWVKKRVETL 366
Query: 359 RMPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQE-----YD 413
+PY + + PPL + AE +++ M R+ R+KEQ+ Y
Sbjct: 367 LLPYD---RMEPLQEQPPLILADSVP-------AEHYKQALMENRRLREKEQDTRMELYK 416
Query: 414 TLKNLLDQQIQANRQEKEENIRLRA---ALQKKEDFLD 448
+ L+ Q + E+ R R+ + ++ E LD
Sbjct: 417 AKADKLNLAHQLREMQGEDASRARSKKRSYKEMESMLD 454
>UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demissum]
Length = 607
Score = 63.2 bits (152), Expect = 2e-08
Identities = 105/474 (22%), Positives = 181/474 (38%), Gaps = 78/474 (16%)
Query: 21 GKKVLCTDDFFEQHGNLLGVVKTDIDEGLLNAFVQFYDPGYHCFTFQDYQILPTLEEYSC 80
GK+V+ F++ G L ++ + L+ A V F+DP + F F D+++ PTLEE
Sbjct: 40 GKEVV-----FKRLGFLRSLLYVEPRRDLIEALVHFWDPIKNVFHFSDFELTPTLEEIGA 94
Query: 81 WINLPVLDKVPFSGLEETPKH---STIAATLHLETDEVKTHLIT-----------RGKFL 126
+I K G PKH LH+ +E+ L G+
Sbjct: 95 FIG---RGKNLHEGEPMIPKHINGRRFLELLHINENEIGGCLDNGWVPLEFLYKRYGRKD 151
Query: 127 GFSTDFLYERTTFFDKMGVAYAFNSILALLV--YGLVLFPSLDNFVDIK--AIQIFLSKN 182
GF LY + + + + +S A+ V G+++F ++I A+ K
Sbjct: 152 GFE---LYGKKLHNNGCRLTWETHSYDAITVAFLGIMVFLKRGGKININLAAVITAFEKK 208
Query: 183 P----VPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSLLPRTPHFT---------- 228
P VP +L D Y ++ + G C LL W+ + P+
Sbjct: 209 PNITLVPMILADIYRAL-TICKNGGDYFEGCNMLLQLWMIEHIRHHPYVVDFKVECNDYI 267
Query: 229 ----------TNPENL-LWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEG 277
+ P+ + W K L +LT ++VW + +I + + L+G+ G
Sbjct: 268 GGHEERIKDHSFPKGIEAWKKYLNNLTADKIVWNYHWFPSAEVIYMSTFRSFIVLMGLRG 327
Query: 278 GISYNPTLARRQFGYPMEKKPLSIYLENVYYFNTEDSTGMREQVVRAW-HTIRRRDKGQL 336
Y P RQ G P E +Y F+ E + ++ + W + ++
Sbjct: 328 VQPYMPLRVMRQLGRRQFLPPNEDIREFMYEFHPEIPL-RKSEIFKIWGGCMLSSPHDRV 386
Query: 337 GKKT-GAVHESYTQWV-----------------IDRAVQIRMPYKISRFVSAITPAPPLP 378
+T G V ++Y +WV +DR +I + K +R + L
Sbjct: 387 EDRTKGEVDQAYLEWVHDQPSPKVLPEGSVKGPVDREAEIEVRIKQARLEVERSYRSTLD 446
Query: 379 MTFDTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNLLDQQIQANRQEKEE 432
+ K +E EL + ++E + RK TL+ L I A Q++EE
Sbjct: 447 CLSNDLKNAKE---ELAQRDAIFEVRVRKHRSTILTLQEDLGIVISAMEQQEEE 497
>UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum]
Length = 464
Score = 47.0 bits (110), Expect = 0.001
Identities = 67/310 (21%), Positives = 116/310 (36%), Gaps = 70/310 (22%)
Query: 35 GNLLGVVKTDIDEGLLNAFVQFYDPGYHCFTFQDYQILPTLEEYSCWINLPVLDKVPFSG 94
G L ++K + L+ A V F+DP ++ F F D+++ PTLEE +SG
Sbjct: 39 GALTDIMKIKPRDDLIEALVTFWDPVHNVFCFSDFELTPTLEEID-----------GYSG 87
Query: 95 LEETPKHSTI----AATLHLETD------EVKTHLITRGKFLGFSTDFLYER-------- 136
++ + A ++H D +++ + +G S FLY R
Sbjct: 88 FGRDLRNQELIFPRALSVHRFFDLLNISKQIRKTNVVKG---CCSFYFLYSRFGQPNGFE 144
Query: 137 --------TTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFVD---IKAIQIFLSKNP-- 183
D + F I+A L G+++FP+ + +D + +Q+ +K
Sbjct: 145 MHEKGLNNKQNKDTWHIHRCFAFIMAFL--GIMVFPNRERTIDTRIARVVQVLTTKEHHT 202
Query: 184 -VPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSLLPRTPHFTT------------- 229
P +L D Y ++ ++G C LL W+ L P F +
Sbjct: 203 LAPIILSDIYRAL-TLCKSGAKFFEGCNILLQMWLIEHLRHHPKFMSYGPSKDNFIDSYE 261
Query: 230 -------NPENL-LWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISY 281
+PE + W L SL ++ W +I +++ LLG+ Y
Sbjct: 262 ERVKEYNSPEGVETWISHLRSLNACQIEWTLGWLPLREVIHMSALKSHLLLLGLRSVQPY 321
Query: 282 NPTLARRQFG 291
P RQ G
Sbjct: 322 MPHRVLRQLG 331
>UniRef100_Q9AV25 Putative kinesin-related protein [Oryza sativa]
Length = 1578
Score = 40.8 bits (94), Expect = 0.085
Identities = 27/82 (32%), Positives = 45/82 (53%), Gaps = 2/82 (2%)
Query: 382 DTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNL-LDQQIQANRQEKEEN-IRLRAA 439
D + +E I EL E+ MW+RK RK +Q +TLK + D+ Q + Q+ E+ R
Sbjct: 286 DVLENAEETIDELRGEAKMWQRKTRKLKQGLETLKKVSTDKSKQRSEQDLEKMWQRKTRK 345
Query: 440 LQKKEDFLDKVCPGRKKRRMDL 461
L++ + L K C + K++ +L
Sbjct: 346 LKQGLETLKKECADKSKQQSEL 367
>UniRef100_Q61QG4 Hypothetical protein CBG07027 [Caenorhabditis briggsae]
Length = 2177
Score = 40.0 bits (92), Expect = 0.14
Identities = 37/179 (20%), Positives = 78/179 (42%), Gaps = 10/179 (5%)
Query: 281 YNPTLARRQ---FGYPME-KKPLSIYLENVYYFNTEDSTGMREQVVRAWHTIRRRDKGQ- 335
YNP L ++Q FG+PM P+ + + Y R+Q HT ++ + Q
Sbjct: 48 YNPMLLQQQYLNFGFPMGYNNPMFDFQQQQQYQMQLQHQMQRQQPQHLQHTPHQQQQQQH 107
Query: 336 -LGKKTGAVHESYTQWVIDRAVQIRMPYKISRFVSAIT----PAPPLPMTFDTEKECQEK 390
++ H Q + R+ Q ++ +S+ ++ + T++ Q
Sbjct: 108 MFNQQQMLQHMMQQQQQVQRSQQQKLQQPVSQHIAPTSHQFQQVQQAQHQQPTQQPSQAA 167
Query: 391 IAELTRESYMWERKYRKKEQEYDTLKNLLDQQIQANRQEKEENIRLRAALQKKEDFLDK 449
+ E + ++E + R+ EQ+ + L +Q + R++++E + RA KK+ L++
Sbjct: 168 MFESSTHQKLYEAQRRQIEQQQKEQEELRRKQEEIRRKQEQEEAKKRAEEAKKQRILEE 226
>UniRef100_UPI0000080FE8 UPI0000080FE8 UniRef100 entry
Length = 2056
Score = 39.3 bits (90), Expect = 0.25
Identities = 24/92 (26%), Positives = 43/92 (46%)
Query: 359 RMPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNL 418
R+ K+ + L D K+ +EK+ L + + +ERKY++ E TL
Sbjct: 919 RLYTKVKPLLEVTKQEEKLVQKEDELKQVREKLDTLAKNTQEYERKYQQALVEKTTLAEQ 978
Query: 419 LDQQIQANRQEKEENIRLRAALQKKEDFLDKV 450
L +I+ + +E RL A Q+ ED + ++
Sbjct: 979 LQAEIELCAEAEESRSRLMARKQELEDMMQEL 1010
>UniRef100_UPI0000079B2E UPI0000079B2E UniRef100 entry
Length = 2011
Score = 39.3 bits (90), Expect = 0.25
Identities = 24/92 (26%), Positives = 43/92 (46%)
Query: 359 RMPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNL 418
R+ K+ + L D K+ +EK+ L + + +ERKY++ E TL
Sbjct: 874 RLYTKVKPLLEVTKQEEKLVQKEDELKQVREKLDTLAKNTQEYERKYQQALVEKTTLAEQ 933
Query: 419 LDQQIQANRQEKEENIRLRAALQKKEDFLDKV 450
L +I+ + +E RL A Q+ ED + ++
Sbjct: 934 LQAEIELCAEAEESRSRLMARKQELEDMMQEL 965
>UniRef100_UPI000042FC3C UPI000042FC3C UniRef100 entry
Length = 572
Score = 39.3 bits (90), Expect = 0.25
Identities = 25/78 (32%), Positives = 48/78 (61%), Gaps = 2/78 (2%)
Query: 384 EKECQEKIAELTRESYMWERKYRKKEQEYDTLKNLLDQQIQANRQEKEENIRLRAALQKK 443
EKE Q++ +L +E E+K RK+E+E + + L+++ + +Q++EE R + L+K+
Sbjct: 83 EKEKQKQEEKLRKEQEKLEKK-RKQEEERELKRKRLEEEKELKKQKQEEERRAK-ELKKE 140
Query: 444 EDFLDKVCPGRKKRRMDL 461
E+ L K ++K R+ L
Sbjct: 141 EERLQKEKEKQEKERLRL 158
>UniRef100_UPI000042C8BB UPI000042C8BB UniRef100 entry
Length = 572
Score = 39.3 bits (90), Expect = 0.25
Identities = 25/78 (32%), Positives = 48/78 (61%), Gaps = 2/78 (2%)
Query: 384 EKECQEKIAELTRESYMWERKYRKKEQEYDTLKNLLDQQIQANRQEKEENIRLRAALQKK 443
EKE Q++ +L +E E+K RK+E+E + + L+++ + +Q++EE R + L+K+
Sbjct: 83 EKEKQKQEEKLRKEQEKLEKK-RKQEEERELKRKRLEEEKELKKQKQEEERRAK-ELKKE 140
Query: 444 EDFLDKVCPGRKKRRMDL 461
E+ L K ++K R+ L
Sbjct: 141 EERLQKEKEKQEKERLRL 158
>UniRef100_Q99323-2 Splice isoform 2 of Q99323 [Drosophila melanogaster]
Length = 1972
Score = 39.3 bits (90), Expect = 0.25
Identities = 24/92 (26%), Positives = 43/92 (46%)
Query: 359 RMPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNL 418
R+ K+ + L D K+ +EK+ L + + +ERKY++ E TL
Sbjct: 835 RLYTKVKPLLEVTKQEEKLVQKEDELKQVREKLDTLAKNTQEYERKYQQALVEKTTLAEQ 894
Query: 419 LDQQIQANRQEKEENIRLRAALQKKEDFLDKV 450
L +I+ + +E RL A Q+ ED + ++
Sbjct: 895 LQAEIELCAEAEESRSRLMARKQELEDMMQEL 926
>UniRef100_Q99323-1 Splice isoform 1 of Q99323 [Drosophila melanogaster]
Length = 2017
Score = 39.3 bits (90), Expect = 0.25
Identities = 24/92 (26%), Positives = 43/92 (46%)
Query: 359 RMPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNL 418
R+ K+ + L D K+ +EK+ L + + +ERKY++ E TL
Sbjct: 880 RLYTKVKPLLEVTKQEEKLVQKEDELKQVREKLDTLAKNTQEYERKYQQALVEKTTLAEQ 939
Query: 419 LDQQIQANRQEKEENIRLRAALQKKEDFLDKV 450
L +I+ + +E RL A Q+ ED + ++
Sbjct: 940 LQAEIELCAEAEESRSRLMARKQELEDMMQEL 971
>UniRef100_Q99323-4 Splice isoform 4 of Q99323 [Drosophila melanogaster]
Length = 2012
Score = 39.3 bits (90), Expect = 0.25
Identities = 24/92 (26%), Positives = 43/92 (46%)
Query: 359 RMPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNL 418
R+ K+ + L D K+ +EK+ L + + +ERKY++ E TL
Sbjct: 875 RLYTKVKPLLEVTKQEEKLVQKEDELKQVREKLDTLAKNTQEYERKYQQALVEKTTLAEQ 934
Query: 419 LDQQIQANRQEKEENIRLRAALQKKEDFLDKV 450
L +I+ + +E RL A Q+ ED + ++
Sbjct: 935 LQAEIELCAEAEESRSRLMARKQELEDMMQEL 966
>UniRef100_Q99323 Myosin heavy chain, non-muscle [Drosophila melanogaster]
Length = 2057
Score = 39.3 bits (90), Expect = 0.25
Identities = 24/92 (26%), Positives = 43/92 (46%)
Query: 359 RMPYKISRFVSAITPAPPLPMTFDTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNL 418
R+ K+ + L D K+ +EK+ L + + +ERKY++ E TL
Sbjct: 920 RLYTKVKPLLEVTKQEEKLVQKEDELKQVREKLDTLAKNTQEYERKYQQALVEKTTLAEQ 979
Query: 419 LDQQIQANRQEKEENIRLRAALQKKEDFLDKV 450
L +I+ + +E RL A Q+ ED + ++
Sbjct: 980 LQAEIELCAEAEESRSRLMARKQELEDMMQEL 1011
>UniRef100_Q5Z4N6 Epstein-Barr virus EBNA-1-like protein [Oryza sativa]
Length = 93
Score = 38.5 bits (88), Expect = 0.42
Identities = 19/51 (37%), Positives = 26/51 (50%)
Query: 35 GNLLGVVKTDIDEGLLNAFVQFYDPGYHCFTFQDYQILPTLEEYSCWINLP 85
GNLL + K ID L NA + YD FT ++ TL++ C + LP
Sbjct: 23 GNLLKINKIYIDRNLCNAITRSYDKEKKAFTINGTFVMMTLDDVDCLLGLP 73
>UniRef100_Q74ZH8 AGR229Wp [Ashbya gossypii]
Length = 705
Score = 38.5 bits (88), Expect = 0.42
Identities = 20/49 (40%), Positives = 29/49 (58%)
Query: 383 TEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNLLDQQIQANRQEKE 431
+ E K+AELT E+ + ER+Y+K ++YDT L Q+Q QE E
Sbjct: 363 SSSELDAKVAELTEENALIEREYQKLLKDYDTETKQLGGQMQQLSQEAE 411
>UniRef100_Q8E0U1 Conserved domain protein [Streptococcus agalactiae]
Length = 354
Score = 38.1 bits (87), Expect = 0.55
Identities = 23/67 (34%), Positives = 39/67 (57%), Gaps = 4/67 (5%)
Query: 385 KECQEKIAELTRESYMWERKYRKKEQEYDTLKNLLDQQI----QANRQEKEENIRLRAAL 440
KE ++ IA+ E + E+K +++ +T K L ++Q +A R++KEE RL A
Sbjct: 251 KEQRKLIAKHEEEKRLAEKKVEEEKAAAETQKKLEEEQARLAAEAQRKQKEEQARLAAET 310
Query: 441 QKKEDFL 447
QKK++ L
Sbjct: 311 QKKQETL 317
>UniRef100_Q5VUI3 Trichohyalin [Homo sapiens]
Length = 1943
Score = 37.7 bits (86), Expect = 0.72
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 7/71 (9%)
Query: 384 EKECQEKIAELTRES------YMWERKYRKKEQEYDTLKNLLDQQIQANR-QEKEENIRL 436
E+E Q++ +L RE WER+YRKK++ + LL ++ + R QE+E R
Sbjct: 993 EEELQQEEEQLLREEREKRRRQEWERQYRKKDELQQEEEQLLREEREKRRLQERERQYRE 1052
Query: 437 RAALQKKEDFL 447
LQ++E+ L
Sbjct: 1053 EEELQQEEEQL 1063
Score = 35.4 bits (80), Expect = 3.6
Identities = 25/78 (32%), Positives = 43/78 (55%), Gaps = 2/78 (2%)
Query: 382 DTEKECQEKIAELTRESYMWERKYRKKEQEYDTLKNLL-DQQIQANRQEKEENIRLRAAL 440
+ E+E Q + E R ER+YR++EQ + LL +++ + RQE+E R L
Sbjct: 908 EEEEELQREEREKRRRQEQ-ERQYREEEQLQQEEEQLLREEREKRRRQERERQYRKDKKL 966
Query: 441 QKKEDFLDKVCPGRKKRR 458
Q+KE+ L P +++R+
Sbjct: 967 QQKEEQLLGEEPEKRRRQ 984
Score = 34.3 bits (77), Expect = 7.9
Identities = 28/95 (29%), Positives = 49/95 (51%), Gaps = 9/95 (9%)
Query: 384 EKECQEKIAEL------TRESYMWERKYRKKEQEYDTLKNLL-DQQIQANRQEKEENIRL 436
E+E Q++ +L TR ER+YRK+E+ + LL ++ + RQE+E R
Sbjct: 1053 EEELQQEEEQLLGEERETRRRQELERQYRKEEELQQEEEQLLREEPEKRRRQERERQCRE 1112
Query: 437 RAALQKKEDFLDKVCPGRKKRRMDLFDGPHSDFED 471
LQ++E+ L + R+KRR + + + E+
Sbjct: 1113 EEELQQEEEQL--LREEREKRRRQELERQYREEEE 1145
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 839,376,550
Number of Sequences: 2790947
Number of extensions: 37573063
Number of successful extensions: 106286
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 75
Number of HSP's that attempted gapping in prelim test: 105947
Number of HSP's gapped (non-prelim): 358
length of query: 471
length of database: 848,049,833
effective HSP length: 131
effective length of query: 340
effective length of database: 482,435,776
effective search space: 164028163840
effective search space used: 164028163840
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC144404.22


