

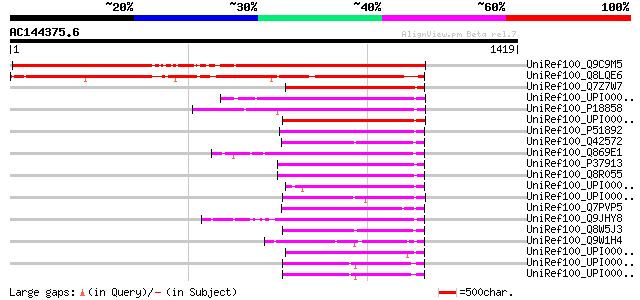
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144375.6 - phase: 0
(1419 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C9M5 DNA ligase I, putative [Arabidopsis thaliana] 1217 0.0
UniRef100_Q8LQE6 Putative DNA ligase [Oryza sativa] 1025 0.0
UniRef100_Q7Z7W7 DNA ligase I [Coprinus cinereus] 302 6e-80
UniRef100_UPI00004375A9 UPI00004375A9 UniRef100 entry 294 2e-77
UniRef100_P18858 DNA ligase I [Homo sapiens] 292 6e-77
UniRef100_UPI00004375AA UPI00004375AA UniRef100 entry 291 8e-77
UniRef100_P51892 DNA ligase I [Xenopus laevis] 290 3e-76
UniRef100_Q42572 DNA ligase [Arabidopsis thaliana] 289 4e-76
UniRef100_Q869E1 Similar to Xenopus laevis (African clawed frog)... 287 1e-75
UniRef100_P37913 DNA ligase I [Mus musculus] 284 1e-74
UniRef100_Q8R055 Lig1 protein [Mus musculus] 283 3e-74
UniRef100_UPI000042C223 UPI000042C223 UniRef100 entry 281 8e-74
UniRef100_UPI00004375AB UPI00004375AB UniRef100 entry 280 2e-73
UniRef100_Q7PVP5 ENSANGP00000010547 [Anopheles gambiae str. PEST] 279 4e-73
UniRef100_Q9JHY8 DNA ligase I [Rattus norvegicus] 278 1e-72
UniRef100_Q8W5J3 Putative DNA ligase [Oryza sativa] 276 3e-72
UniRef100_Q9W1H4 CG5602-PA [Drosophila melanogaster] 268 7e-70
UniRef100_UPI00003C2646 UPI00003C2646 UniRef100 entry 267 2e-69
UniRef100_UPI00004314CE UPI00004314CE UniRef100 entry 265 8e-69
UniRef100_UPI00004314CD UPI00004314CD UniRef100 entry 265 8e-69
>UniRef100_Q9C9M5 DNA ligase I, putative [Arabidopsis thaliana]
Length = 1417
Score = 1217 bits (3150), Expect = 0.0
Identities = 668/1174 (56%), Positives = 826/1174 (69%), Gaps = 57/1174 (4%)
Query: 7 SQTLESLNSTQLYLNALQSLNLPPPTLPLPPLPSSTIPHSKLIPNTRFLIDSFR--HTTP 64
S+TL +LN+T+LY +A+ S++ P+ P P +IP+SK IPNT F++D FR H +
Sbjct: 20 SETL-NLNTTKLYSSAISSISPQFPS-PKPTSSCPSIPNSKRIPNTNFIVDLFRLPHQS- 76
Query: 65 SSFTYFLSHFHSDHYSPLSSSWSHGIIFCSPITSHLLINILHIPSPFVHPLSLNQSVVID 124
SS +FLSHFHSDHYS LSSSWS GII+CS T+ L+ IL +PS FV L +NQ V ID
Sbjct: 77 SSVAFFLSHFHSDHYSGLSSSWSKGIIYCSHKTARLVAEILQVPSQFVFALPMNQMVKID 136
Query: 125 GSVVTLIDANHCPGAVQFLFKVNETES--PRYVHTGDFRFNREMLLDLNLGEFIGADAVF 182
GS V LI+ANHCPGAVQFLFKV S +YVHTGDFRF EM D L F+G D VF
Sbjct: 137 GSEVVLIEANHCPGAVQFLFKVKLESSGFEKYVHTGDFRFCDEMRFDPFLNGFVGCDGVF 196
Query: 183 LDTTYCHPKFVFPTQNESVDYIVDVVKECDGENVLFLVATYVVGKEKILLEIARRCGKKV 242
LDTTYC+PKFVFP+Q ESV Y+V V+ + E VLFLVATYVVGKEKIL+EIARRC +K+
Sbjct: 197 LDTTYCNPKFVFPSQEESVGYVVSVIDKISEEKVLFLVATYVVGKEKILVEIARRCKRKI 256
Query: 243 CVDGKKMEVLRALGYGESGEFTEDRLESNVHVVGWNVLGETWPYFRPNFVRMKEIMVERG 302
VD +KM +L LG GE G FTED ES+VHVVGWNVLGETWPYFRPNFV+M EIMVE+G
Sbjct: 257 VVDARKMSMLSVLGCGEEGMFTEDENESDVHVVGWNVLGETWPYFRPNFVKMNEIMVEKG 316
Query: 303 YSKVVGFVPTGWTYEVKRDKFKVREKDSCKIHLVPYSEHSNYEELREYVRFLKPKKVVPT 362
Y KVVGFVPTGWTYEVKR+KF VR KDS +IHLVPYSEHSNY+ELRE+++FLKPK+V+PT
Sbjct: 317 YDKVVGFVPTGWTYEVKRNKFAVRFKDSMEIHLVPYSEHSNYDELREFIKFLKPKRVIPT 376
Query: 363 VGLDVEKSDSKHVDKMRKYFAGLVDETANKHEFLKGFKQCDSGRSGFEVGKDVGNDTEPG 422
VG+D+EK D K V+KM+K+F+GLVDE ANK +FL GF R ++ + D
Sbjct: 377 VGVDIEKFDCKEVNKMQKHFSGLVDEMANKKDFLLGFY-----RQSYQKNEKSDVDV-VS 430
Query: 423 HSVEKEVKPSDVGGDKSIDQDVAMSLSSCMGETCIEDPTLLNDEEKEKVVQELSCCLPTW 482
HS E + +K+ +D ++ S G + D T +D +++ +L LP W
Sbjct: 431 HSAEVYEEE-----EKNACEDGGENVPSSRG-PILHDTTPSSD---SRLLIKLRDSLPAW 481
Query: 483 VTRSQMLDLISISGSNVVEAVSNFFERETEFHEQVNSSQTPVPTHRSCSSNDTSPLSKSN 542
VT QMLDLI N V+ VSNF+E E E ++Q + P P S N+ + L +
Sbjct: 482 VTEEQMLDLIKKHAGNPVDIVSNFYEYEAELYKQ---ASLPTP-----SLNNQAVLFDDD 533
Query: 543 LKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSNQISPSKRKKGSESK 602
+ N K + + K DL R NL K ISP KR K S SK
Sbjct: 534 VTDLQPNPV----KGICPDVQAIQKGFDLPRKM-----NLTK---GTISPGKRGKSSGSK 581
Query: 603 SNKKVKVKAKSESSGSKQATITKFFGKAMPVMPGDTQSDQFGSKPGESPEVEELVPTDAG 662
SNKK K KS+ G Q T+ KFF K V+ G + S GS+ E +++V DA
Sbjct: 582 SNKKAKKDPKSKPVGPGQPTLFKFFNK---VLDGGSNSVSVGSETEECNTDKKMVHIDAS 638
Query: 663 NMYKQEIDQFMQIINGDESLKKQAITIIEEAKGDINKALDIYYSNSCNL-----GEREIS 717
YK+ DQF+ I+NG ESL+ A +II+EAKGDI++AL+IYYS + GER +S
Sbjct: 639 EAYKEVTDQFIDIVNGSESLRDYAASIIDEAKGDISRALNIYYSKPREIPGDHAGERGLS 698
Query: 718 VQ----GEC-KVDRPLEKKYVSKELNVIPDISMHRVLRDNVDATHVSLPSDKYNPKEHAC 772
+ +C + E K S+ +I + ++VD +VSLP +KY PKEHAC
Sbjct: 699 SKTIQYPKCSEACSSQEDKKASENSGHAVNICVQTSAEESVDKNYVSLPPEKYQPKEHAC 758
Query: 773 WRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVYLCTNKIAA 832
WR+GQPAPY+HL RTF+ +E EKGKIKA S+LCNMFRSL LSPEDVLPAVYLCTNKIAA
Sbjct: 759 WREGQPAPYIHLVRTFASVESEKGKIKAMSMLCNMFRSLFALSPEDVLPAVYLCTNKIAA 818
Query: 833 DHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRLLAPPTPLL 892
DHEN+ELNIGGSL+++ALEEACG +R +++MYN LGDLGDVAQ CRQTQ+LL PP PLL
Sbjct: 819 DHENIELNIGGSLISSALEEACGISRSTVRDMYNSLGDLGDVAQLCRQTQKLLVPPPPLL 878
Query: 893 IKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVL 952
++D++S LRKISVQTG GST KK +I+ LMRSCREKE+KFLVRTL RNLRIGAMLRTVL
Sbjct: 879 VRDVFSTLRKISVQTGTGSTRLKKNLIVKLMRSCREKEIKFLVRTLARNLRIGAMLRTVL 938
Query: 953 PALAHAVVMNSRPTVYEEGTAEN-LKAALQVLSVAVVEAYNILPNLDIIVPTLMNKGIEF 1011
PAL A+VMNS + + +E+ + L+ +S AVVEAYNILP+LD++VP+LM+K IEF
Sbjct: 939 PALGRAIVMNSFWNDHNKELSESCFREKLEGVSAAVVEAYNILPSLDVVVPSLMDKDIEF 998
Query: 1012 SVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVDGSVLVF 1071
S S+LSMVPGIPIKPMLAKI G+ + L Q KAFTCEYKYDGQRAQIHKL+DG+V +F
Sbjct: 999 STSTLSMVPGIPIKPMLAKIAKGVQEFFNLSQEKAFTCEYKYDGQRAQIHKLLDGTVCIF 1058
Query: 1072 SRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCRIMSFQELSSRGRGGK 1131
SRNGDE+TSRFPDLVD+IK+ P A TF++DAEVV DR NG ++MSFQELS+R RG K
Sbjct: 1059 SRNGDETTSRFPDLVDVIKQFSCPAAETFMLDAEVVATDRINGNKLMSFQELSTRERGSK 1118
Query: 1132 DTLVTKESIKVGICIFVFDIMFANGEH-LADPLK 1164
D L+T ESIKV +C+FVFDIMF NGE LA PL+
Sbjct: 1119 DALITTESIKVEVCVFVFDIMFVNGEQLLALPLR 1152
>UniRef100_Q8LQE6 Putative DNA ligase [Oryza sativa]
Length = 1419
Score = 1025 bits (2649), Expect = 0.0
Identities = 576/1202 (47%), Positives = 766/1202 (62%), Gaps = 151/1202 (12%)
Query: 2 APKSKSQTLESLNSTQLYLNALQSLNLP---PPTL----PLPPLPSSTIPHSKLIPNTRF 54
+P SKS S+ ++ L+L++L SL P PP+ PL P+P+S +P + LIP +RF
Sbjct: 7 SPDSKSL---SVATSTLFLSSLASLPSPHSEPPSASAASPLSPVPAS-VPPTALIPGSRF 62
Query: 55 LIDSFRHTTPSSFTYFLSHFHSDHYSPLSSSWSHGIIFCSPITSHLLINILHIPSPFVHP 114
L+D+FRH + +YFLSHFHSDHY+ L SW G++FCSP+T+ LL+++L +P V
Sbjct: 63 LVDAFRHAGDFTASYFLSHFHSDHYTGLGPSWRRGLVFCSPLTARLLVSVLSVPPQLVVV 122
Query: 115 LSLNQSVVIDGSVVTLIDANHCPGAVQFLFKVNETESPRYVHTGDFRFNREMLLDLNLGE 174
L V +DG V +DANHCPGAVQFLF+ + + RYVHTGDFRF++ M+ + NL E
Sbjct: 123 LDAGVRVTVDGWCVVAVDANHCPGAVQFLFRSSGPNAERYVHTGDFRFSQSMITEPNLLE 182
Query: 175 FIGADAVFLDTTYCHPKFVFPTQNESVDYIVDVVKE------CDGENVLFLVATYVVGKE 228
FIGADAVFLDTTYC+PKF FP Q ES++Y+V+ +K GE VL L+ATYVVGKE
Sbjct: 183 FIGADAVFLDTTYCNPKFTFPPQKESLEYVVNSIKRVKEESRASGERVLCLIATYVVGKE 242
Query: 229 KILLEIARRCGKKVCVDGKKMEVLRALGYG-ESGEFTEDRLESNVHVVGWNVLGETWPYF 287
+ILLE+ARRCG K+ VD +KME+L LG G E G FTED ++VHV GWN+LGETWPYF
Sbjct: 243 RILLEVARRCGCKIHVDSRKMEILTLLGIGGEDGVFTEDAAATDVHVTGWNILGETWPYF 302
Query: 288 RPNFVRMKEIMVERGYSKVVGFVPTGWTYEVKRDKFKVREKDSCKIHLVPYSEHSNYEEL 347
RPNFV+MKEIMVERGY+K VGFVPTGW YE K++ F VR KDS +IHLVPYSEHS+Y EL
Sbjct: 303 RPNFVKMKEIMVERGYNKAVGFVPTGWMYETKKEGFAVRTKDSLEIHLVPYSEHSSYNEL 362
Query: 348 REYVRFLKPKKVVPTVGLDVEKSDSKHVDKMRKYFAGLVDETANKHEFLKGFKQCDSGRS 407
R+YV+FL PK+V+PTVGLD K DSK ++K+FAGLVDETANK EFL F
Sbjct: 363 RDYVKFLHPKRVIPTVGLDGGKLDSKEAFALQKHFAGLVDETANKQEFLMAF-------- 414
Query: 408 GFEVGKDVGNDTEPGHSVEKEVKPSDVGGDKSIDQDVAMSLSSCMGETC-IEDPTLL--- 463
+ + + +G +D LS GE +E+ TLL
Sbjct: 415 ------------------HRSSRNATLG-----PEDAVTGLSQQEGEVQELEEATLLPAS 451
Query: 464 -----NDEEKEKVV----QELSCCLPTWVTRSQMLDLISISGSNVVEAVSNFFERETEFH 514
+D +EK+ +ELS LP+WV++ +LDL+ SG +VV+A ++FFE+E +F
Sbjct: 452 LAFERSDSFQEKITVEMKKELSDFLPSWVSQDLILDLLIKSGGDVVQAATDFFEKERDFF 511
Query: 515 EQVN--SSQTPVPTHRSCSSNDTSPLSKSNLKSFSSNDASPFSKSNLNNTNSTTKKLDLF 572
E+ N +S+TP S D S S+ + S + FS+ ++++
Sbjct: 512 EEANVYNSETPK------SEIDLSSDHGSSADASSQQEVPLFSQKPMDHS---------- 555
Query: 573 RSQESKLTNLR-KALSNQISPSKRKKGSES-----KSNKKVKVKAKSESSGSKQATITKF 626
SKL NL + + +S +RK+GS S K + K +ESSG KQ+TIT +
Sbjct: 556 ----SKLLNLNAMRMKSNLSKRERKRGSNSADKPKKKGRSTAFKPLTESSGRKQSTITNY 611
Query: 627 FGKAMPVMPGDTQSDQFGSKPGESPEVEELVPTDAGNMYKQEIDQFMQIINGDESLKKQA 686
F + M SD+ ++ + T+ KQ + Q +QI++G S ++ A
Sbjct: 612 FARTMLAASKSDTSDKVTVDANQNNVRNDDQFTEVVESEKQSVSQLLQIVDGGMS-RESA 670
Query: 687 ITIIEEAKGDINKALDIYYS--NSCNLGEREISVQGECKVDRPLEK-------KYVSKEL 737
I+++E+AKGD+N A+DI+YS ++ N+ E ++++ + + +K + S+
Sbjct: 671 ISLLEKAKGDVNVAVDIFYSKTDNSNVLENDMNIVTQNTENEMTDKSSSTGLLRNSSEAT 730
Query: 738 NVIPDISMHRVLRDNVDATHVSLPSDKYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGK 797
+P++ + + D+ +SLP +KY P EHACW GQPAPYLHLARTF L+E EKGK
Sbjct: 731 PKMPNLCVQSYVA-QADSVCISLPIEKYLPIEHACWTAGQPAPYLHLARTFDLVEREKGK 789
Query: 798 IKATSILCNMFRSLLVLSPEDVLPAVYLCTNKIAADHENVELNIGGSLVTTALEEACGTN 857
IK T++LCNMFRSLL LSP+DVLPAVYLCTNKI+ DHEN+ EA G++
Sbjct: 790 IKTTAMLCNMFRSLLALSPDDVLPAVYLCTNKISPDHENI--------------EALGSS 835
Query: 858 RLKIKEMYNKLGDLGDVAQECRQTQRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKG 917
R KI EMY GDLGDVAQECRQ Q LLAPP PL I+D++S LRK+S G+GST R+K
Sbjct: 836 RSKIHEMYKTFGDLGDVAQECRQNQMLLAPPRPLSIRDVFSTLRKLSGIAGSGSTGRRKI 895
Query: 918 IILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVLPALAHAVVMNSRPTVYEEGTAENLK 977
++LHL+RSCRE EMKFLVRTLVRNLRIG M++T+LPALAHAVV++ + + + E +K
Sbjct: 896 LVLHLIRSCREMEMKFLVRTLVRNLRIGVMMKTILPALAHAVVIDGKYSNSPVLSLEGIK 955
Query: 978 AALQVLSVAVVEAYNILPNLDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQ 1037
LQ LS V EAYN++PNLD+++P+L+ + FS SSL+M+ G PI PMLA+ITNG+ Q
Sbjct: 956 PQLQELSTEVAEAYNVIPNLDLLIPSLLREDTAFSASSLAMITGTPIPPMLARITNGLTQ 1015
Query: 1038 ALKLFQNKAFTCEYKYDGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVA 1097
+LKLF +AFTCEYKYDGQRAQIH+ DGSV +FSR ESTSRFPDLV MIKE C
Sbjct: 1016 SLKLFNGRAFTCEYKYDGQRAQIHRSNDGSVQIFSRQMKESTSRFPDLVGMIKELCSIEV 1075
Query: 1098 STFIIDAEVVGIDRKNGCRIMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGE 1157
S+FI+DAE V IC+FVFDIMF NG+
Sbjct: 1076 SSFILDAE------------------------------------VDICVFVFDIMFCNGQ 1099
Query: 1158 HL 1159
L
Sbjct: 1100 SL 1101
>UniRef100_Q7Z7W7 DNA ligase I [Coprinus cinereus]
Length = 825
Score = 302 bits (773), Expect = 6e-80
Identities = 166/391 (42%), Positives = 244/391 (61%), Gaps = 9/391 (2%)
Query: 773 WRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPED----VLPAVYLCTN 828
W+ GQP PY LA+TFSL+E +++ +IL ++ S +D +L AVYLC N
Sbjct: 193 WKVGQPVPYAALAKTFSLIEATTKRLEKNAILTAFLLLVIQRSAKDDHKSLLQAVYLCIN 252
Query: 829 KIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRLLAPP 888
+++ D+ +EL IG SL+ A+ E+ G + IK K GDLG VA + +Q+ L P
Sbjct: 253 RLSPDYVGIELGIGESLLIKAIAESTGRSLAVIKADLKKEGDLGLVAMNSKNSQKTLFKP 312
Query: 889 TPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRIGAML 948
PL + +++ L+ I++ TGN S +K GII L+ +C++ E K++VR+L LRIG
Sbjct: 313 KPLTLPFVFTQLKDIALTTGNASQAKKVGIITKLLAACQDVEAKYIVRSLEGKLRIGNAE 372
Query: 949 RTVLPALAHAVVMNSRPTVYEEGTAENLKAALQVLSVAVVEAYNILPNLDIIVPTLMNKG 1008
R+VL +LAHA V+ R ++ + + L A L+ + + Y+ LP+ D +VP L+ G
Sbjct: 373 RSVLVSLAHAAVLAERERADKKWSQDKLTARLEEGAEIMKAVYSELPSYDSVVPALLEGG 432
Query: 1009 IEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVDGSV 1068
I+ + PGIP+KPMLAK T I + L F+NK FTCEYKYDG+RAQIH+L DG+V
Sbjct: 433 IKGLRERCKLTPGIPLKPMLAKPTKAIREVLDRFENKRFTCEYKYDGERAQIHRLDDGTV 492
Query: 1069 LVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCRIMSFQELSSRGR 1128
VFSRN ++ + ++PDLVD + + K +F++D+E V IDR G ++M FQELS R R
Sbjct: 493 GVFSRNSEDMSKKYPDLVDQLSKCIKKNTKSFVLDSEAVAIDRTTG-KLMPFQELSRRKR 551
Query: 1129 GGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
KD V E I+V +CIF FD+++ NGE L
Sbjct: 552 --KDVKV--EDIQVRVCIFAFDLLYLNGEPL 578
>UniRef100_UPI00004375A9 UPI00004375A9 UniRef100 entry
Length = 808
Score = 294 bits (752), Expect = 2e-77
Identities = 192/591 (32%), Positives = 309/591 (51%), Gaps = 35/591 (5%)
Query: 590 ISPS---KRKKGSESKSNKKVKVKAKSE---SSGSKQATITKFFGKAMPVMPGDTQSDQF 643
+SPS KRK E + K V SE KQ T K + D+
Sbjct: 5 VSPSGIPKRKTVEEKEDKAKTSVVVDSEMKDEDDKKQKINTNNEAK---------EEDKN 55
Query: 644 GSKPGES--PEVEELVPTDAGNMYKQEIDQFMQIINGDESLKKQAITIIEEAKGDINKAL 701
SK E E+E GN +++ + + D+S KKQ E+ K D +
Sbjct: 56 ESKQEEKIDTEMETKEKDKKGNKQQEKKETKKETKEKDKSDKKQE----EKNKSDKKQEE 111
Query: 702 DIYYSNSCNLGEREISVQGECKVDRPLEKKYVSKELNVIPDISMHRVLRDNVDATHVSLP 761
N + + + + +K V KE + + + + +
Sbjct: 112 KDKTEEKVNGESEKEKIPDKKGKESSSPEKVVKKEKDTDGGSAKKKPISSFFVQSQYDPS 171
Query: 762 SDKYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLP 821
Y+P +HACW+ GQ PYL +ARTF +E++ G++K L N RS+++LSP+D+L
Sbjct: 172 RAHYHPIDHACWKKGQKVPYLAVARTFEKIEEDSGRLKNIETLSNFLRSVILLSPDDLLC 231
Query: 822 AVYLCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQT 881
VYLC N++ ++ +EL IG +++ A+ +A G KIK + GDLG VA+ R
Sbjct: 232 CVYLCLNQLGPAYQGLELGIGETVLMKAVAQATGRQLDKIKAEAQEKGDLGLVAESSRSN 291
Query: 882 QRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRN 941
QR++ P L +++ L++I+ +GN + +K II L +CR E +++VR+L
Sbjct: 292 QRMMFAPANLTAGGVFNKLKEIAHMSGNSAMNKKIDIIKGLFVACRFSEARYIVRSLAGK 351
Query: 942 LRIGAMLRTVLPALAHAVVMNS-----RPTVYEEG---TAENLKAALQVLSVAVVEAYNI 993
LRIG ++VL AL+ AV + P V + G + EN + L+ ++ + + Y
Sbjct: 352 LRIGLAEQSVLAALSQAVCLTPPGQEFPPAVLDAGKGMSTENRRVFLEEKALILKQTYCE 411
Query: 994 LPNLDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKY 1053
+PN D I+P L+ +GI+ + + PG+P++PMLA T G+ + +K F +FTCEYKY
Sbjct: 412 MPNYDAIIPVLLKEGIDELPNHCKLTPGVPLRPMLAHPTKGVGEVMKRFDEASFTCEYKY 471
Query: 1054 DGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKN 1113
DG+RAQIH L +G V +FSRN +++TS++PD++ I + K + ++D+E V DR+
Sbjct: 472 DGERAQIHILENGEVRIFSRNQEDNTSKYPDIISRIPQVKKESVRSCVLDSEAVAWDREK 531
Query: 1114 GCRIMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHLA-DPL 1163
+I FQ L++R R KD V IKV +C++ FD+++ NGE L +PL
Sbjct: 532 K-QIQPFQVLTTRKR--KD--VDASEIKVQVCVYAFDLLYLNGESLVREPL 577
>UniRef100_P18858 DNA ligase I [Homo sapiens]
Length = 919
Score = 292 bits (747), Expect = 6e-77
Identities = 201/675 (29%), Positives = 327/675 (47%), Gaps = 32/675 (4%)
Query: 513 FHEQVNSSQTPVPTHRSCSSNDTSPLSKSNLKSFS---SNDASPFSKSNLNNTNSTTKKL 569
FH + S SS +T P K+ LK ++ S SP + +
Sbjct: 9 FHPKKEGKAKKPEKEASNSSRETEPPPKAALKEWNGVVSESDSPVKRPGRKAARVLGSEG 68
Query: 570 DLFRSQESKLTNLRKALS-NQISPSKRKKGSESKSNKKVKVKAKSESSG-SKQATITKFF 627
+ S + AL +Q+SP + E+ ++ S SG K+ T K
Sbjct: 69 EEEDEALSPAKGQKPALDCSQVSPPRPATSPENNASLSDTSPMDSSPSGIPKRRTARKQL 128
Query: 628 GKA--MPVMPGDTQSDQFGSKPGESPEVEELVPTDAGNMYKQEIDQFMQIINGDESLKKQ 685
K V+ ++ + +K + E EE T ++ + E+ + +GD+
Sbjct: 129 PKRTIQEVLEEQSEDEDREAKRKKEEEEEE---TPKESLTEAEVATEKEGEDGDQPTTPP 185
Query: 686 AITIIEEAKGDINKALDIYYSNSCNLGEREISVQGECKVDRPLEKKYVSKELNVIPDISM 745
+A+ + + L E E + + + L + ++ V ++
Sbjct: 186 KPLKTSKAETPTESVSEPEVATKQELQEEEEQTKPPRRAPKTLSSFFTPRKPAVKKEVKE 245
Query: 746 HR--------VLRDNVDATHVSLPSDKYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGK 797
+D + + + Y+P E ACW+ GQ PYL +ARTF +E+ +
Sbjct: 246 EEPGAPGKEGAAEGPLDPSGYNPAKNNYHPVEDACWKPGQKVPYLAVARTFEKIEEVSAR 305
Query: 798 IKATSILCNMFRSLLVLSPEDVLPAVYLCTNKIAADHENVELNIGGSLVTTALEEACGTN 857
++ L N+ RS++ LSP D+LP +YL N + + +EL +G ++ A+ +A G
Sbjct: 306 LRMVETLSNLLRSVVALSPPDLLPVLYLSLNHLGPPQQGLELGVGDGVLLKAVAQATGRQ 365
Query: 858 RLKIKEMYNKLGDLGDVAQECRQTQRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKG 917
++ + GD+G VA+ R TQRL+ PP PL ++S R I+ TG+ ST +K
Sbjct: 366 LESVRAEAAEKGDVGLVAENSRSTQRLMLPPPPLTASGVFSKFRDIARLTGSASTAKKID 425
Query: 918 IILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVLPALAHAVVMNS-----RPTVYEEG- 971
II L +CR E +F+ R+L LR+G ++VL AL+ AV + P + + G
Sbjct: 426 IIKGLFVACRHSEARFIARSLSGRLRLGLAEQSVLAALSQAVSLTPPGQEFPPAMVDAGK 485
Query: 972 --TAENLKAALQVLSVAVVEAYNILPNLDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLA 1029
TAE K L+ + + + + +P+LD I+P L+ G+E + PGIP+KPMLA
Sbjct: 486 GKTAEARKTWLEEQGMILKQTFCEVPDLDRIIPVLLEHGLERLPEHCKLSPGIPLKPMLA 545
Query: 1030 KITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMI 1089
T GI + LK F+ AFTCEYKYDGQRAQIH L G V +FSRN +++T ++PD++ I
Sbjct: 546 HPTRGISEVLKRFEEAAFTCEYKYDGQRAQIHALEGGEVKIFSRNQEDNTGKYPDIISRI 605
Query: 1090 KESCKPVASTFIIDAEVVGIDRKNGCRIMSFQELSSRGRGGKDTLVTKESIKVGICIFVF 1149
+ P ++FI+D E V DR+ +I FQ L++R R D I+V +C++ F
Sbjct: 606 PKIKLPSVTSFILDTEAVAWDREKK-QIQPFQVLTTRKRKEVDA----SEIQVQVCLYAF 660
Query: 1150 DIMFANGEHLA-DPL 1163
D+++ NGE L +PL
Sbjct: 661 DLIYLNGESLVREPL 675
>UniRef100_UPI00004375AA UPI00004375AA UniRef100 entry
Length = 652
Score = 291 bits (746), Expect = 8e-77
Identities = 156/408 (38%), Positives = 250/408 (61%), Gaps = 14/408 (3%)
Query: 765 YNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVY 824
Y+P +HACW+ GQ PYL +ARTF +E++ G++K L N RS+++LSP+D+L VY
Sbjct: 10 YHPIDHACWKKGQKVPYLAVARTFEKIEEDSGRLKNIETLSNFLRSVILLSPDDLLCCVY 69
Query: 825 LCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRL 884
LC N++ ++ +EL IG +++ A+ +A G KIK + GDLG VA+ R QR+
Sbjct: 70 LCLNQLGPAYQGLELGIGETVLMKAVAQATGRQLDKIKAEAQEKGDLGLVAESSRSNQRM 129
Query: 885 LAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRI 944
+ P L +++ L++I+ +GN + +K II L +CR E +++VR+L LRI
Sbjct: 130 MFAPANLTAGGVFNKLKEIAHMSGNSAMNKKIDIIKGLFVACRFSEARYIVRSLAGKLRI 189
Query: 945 GAMLRTVLPALAHAVVMNS-----RPTVYEEG---TAENLKAALQVLSVAVVEAYNILPN 996
G ++VL AL+ AV + P V + G + EN + L+ ++ + + Y +PN
Sbjct: 190 GLAEQSVLAALSQAVCLTPPGQEFPPAVLDAGKGMSTENRRVFLEEKALILKQTYCEMPN 249
Query: 997 LDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQ 1056
D I+P L+ +GI+ + + PG+P++PMLA T G+ + +K F +FTCEYKYDG+
Sbjct: 250 YDAIIPVLLKEGIDELPNHCKLTPGVPLRPMLAHPTKGVGEVMKRFDEASFTCEYKYDGE 309
Query: 1057 RAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCR 1116
RAQIH L +G V +FSRN +++TS++PD++ I + K + ++D+E V DR+ +
Sbjct: 310 RAQIHILENGEVRIFSRNQEDNTSKYPDIISRIPQVKKESVRSCVLDSEAVAWDREKK-Q 368
Query: 1117 IMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHLA-DPL 1163
I FQ L++R R KD V IKV +C++ FD+++ NGE L +PL
Sbjct: 369 IQPFQVLTTRKR--KD--VDASEIKVQVCVYAFDLLYLNGESLVREPL 412
>UniRef100_P51892 DNA ligase I [Xenopus laevis]
Length = 1070
Score = 290 bits (741), Expect = 3e-76
Identities = 160/414 (38%), Positives = 245/414 (58%), Gaps = 13/414 (3%)
Query: 754 DATHVSLPSDKYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLV 813
+ T + KY+P + ACW +GQ PYL +ARTF +E+E ++K L N RS++
Sbjct: 415 EQTEYNPSKSKYHPIDDACWCNGQKVPYLAVARTFERIEEESARLKNVETLSNFLRSVIA 474
Query: 814 LSPEDVLPAVYLCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGD 873
L+PED+LP +YLC N++ +E +EL IG +++ A+ +A G KIK + GDLG
Sbjct: 475 LTPEDLLPCIYLCLNRLGPAYEGLELGIGETILMKAVAQATGRQLEKIKAEAQEKGDLGL 534
Query: 874 VAQECRQTQRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKF 933
VA+ R QR + P L+ ++ L+ I+ TGN S +K II L +CR E ++
Sbjct: 535 VAESSRSNQRTMFTPPKLMASGVFGKLKDIARMTGNASMNKKIDIIKGLFVACRHSEARY 594
Query: 934 LVRTLVRNLRIGAMLRTVLPALAHAVVM-----NSRPTVYEEG---TAENLKAALQVLSV 985
+ R+L LRIG ++VL ++A AV + ++ PTV + G +A+ K+ ++ ++
Sbjct: 595 IARSLGGKLRIGLAEQSVLSSIAQAVCLTPPGRDAPPTVMDAGKGMSADARKSWIEEKAM 654
Query: 986 AVVEAYNILPNLDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNK 1045
+ + + LPN D I+P L+ GI+ + PGIP+KPMLA T GI + LK F
Sbjct: 655 ILKQTFCELPNYDAIIPILLEHGIDDLPKHCRLTPGIPLKPMLAHPTKGIGEVLKRFDEA 714
Query: 1046 AFTCEYKYDGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAE 1105
AFTCEYKYDG+RAQIH L +G V V+SRN + +T+++PD++ I + K + I+D E
Sbjct: 715 AFTCEYKYDGERAQIHILENGEVHVYSRNQENNTTKYPDIISRIPKIKKESVKSCILDTE 774
Query: 1106 VVGIDRKNGCRIMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
V D + +I FQ L++R R KD V IKV +C++ FD+++ NGE L
Sbjct: 775 AVAGDAEKK-QIQPFQVLTTRKR--KD--VDASEIKVQVCVYAFDMLYLNGESL 823
>UniRef100_Q42572 DNA ligase [Arabidopsis thaliana]
Length = 790
Score = 289 bits (740), Expect = 4e-76
Identities = 156/399 (39%), Positives = 235/399 (58%), Gaps = 10/399 (2%)
Query: 761 PSDKYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVL 820
P+D ++P++ +CW G+ P+L +A F L+ +E G+I T ILCNM R+++ +PED++
Sbjct: 158 PND-FDPEKMSCWEKGERVPFLFVALAFDLISNESGRIVITDILCNMLRTVIATTPEDLV 216
Query: 821 PAVYLCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQ 880
VYL N+IA HE VEL IG S + A+ EA G +K+ +LGDLG VA+ R
Sbjct: 217 ATVYLSANEIAPAHEGVELGIGESTIIKAISEAFGRTEDHVKKQNTELGDLGLVAKGSRS 276
Query: 881 TQRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVR 940
TQ ++ P PL + ++ R+I+ ++G S +KK + L+ + + E +L R L
Sbjct: 277 TQTMMFKPEPLTVVKVFDTFRQIAKESGKDSNEKKKNRMKALLVATTDCEPLYLTRLLQA 336
Query: 941 NLRIGAMLRTVLPALAHAVVMNSRPTVYEEGTAENLKAALQVLSVAVVEAYNILPNLDII 1000
LR+G +TVL AL A V N N K+ L+ + V + + +LP DII
Sbjct: 337 KLRLGFSGQTVLAALGQAAVYNEE----HSKPPPNTKSPLEEAAKIVKQVFTVLPVYDII 392
Query: 1001 VPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQI 1060
VP L++ G+ + + G+PI PMLAK T G+ + L FQ+ FTCEYKYDG+RAQI
Sbjct: 393 VPALLSGGVWNLPKTCNFTLGVPIGPMLAKPTKGVAEILNKFQDIVFTCEYKYDGERAQI 452
Query: 1061 HKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCRIMSF 1120
H + DG+ ++SRN + +T ++PD+ + KP +FI+D EVV DR+ +I+ F
Sbjct: 453 HFMEDGTFEIYSRNAERNTGKYPDVALALSRLKKPSVKSFILDCEVVAFDREKK-KILPF 511
Query: 1121 QELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
Q LS+R R V IKVG+CIF FD+++ NG+ L
Sbjct: 512 QILSTRARKN----VNVNDIKVGVCIFAFDMLYLNGQQL 546
>UniRef100_Q869E1 Similar to Xenopus laevis (African clawed frog). DNA ligase I
[Dictyostelium discoideum]
Length = 1192
Score = 287 bits (735), Expect = 1e-75
Identities = 199/614 (32%), Positives = 313/614 (50%), Gaps = 31/614 (5%)
Query: 564 STTKKLDLFRSQESKLTNLRKALSNQISPSKRKKGSESKSNKKVKVKAKSESSGSKQATI 623
++ KK + S K N S S SK SK+NKK KV K ++ K + +
Sbjct: 350 TSPKKETIDISDLFKRANAEAKSSVPTSTSKN-----SKTNKKQKVDHKPTATTKKPSPV 404
Query: 624 ------TKFFGKAMPVMPGDTQSDQFGSKPGESPEVEELVPTDAGNMYKQEIDQFMQIIN 677
T T S + S P + E E + K E+ + +
Sbjct: 405 LEAKQSTTTTTTTTTTSTATTISSKSISSPSKKEEKEVITSKKQVEATKVEVKKEKE--K 462
Query: 678 GDESLKKQAITIIEEAKGDINKALDIYYSNSCNLGEREISVQGECKVDRPLEKKYVSKEL 737
E K+ EE + D K DI E E +G + + EKK +
Sbjct: 463 EKEKEKEDDEEEEEEEEDDDEKLEDIDEEEYEE--EEEEDEEGISENEEEEEKKSTQIKS 520
Query: 738 NVIPDISMHRVLRDNVDATHVSLPS-DKYNPKEHACWRDGQPAPYLHLARTFSLLEDEKG 796
I + + + + N L KY P E A W+ G+ PY+ LA+TF ++E
Sbjct: 521 KFIKKVPISKK-KGNAKTIQADLKVIGKYRPIEDAQWKKGEAVPYMVLAKTFEMMESTSS 579
Query: 797 KIKATSILCNMFRSLLVLSPEDVLPAVYLCTNKIAADHENVELNIGGSLVTTALEEACGT 856
++ L N+FRS+++LSP+D++ +YL NKI +++ EL IG ++ +L E+ G
Sbjct: 580 RLIIIEHLANLFRSIMLLSPKDLVMTIYLSINKIGPSYQSKELGIGEHVLIKSLAESTGR 639
Query: 857 NRLKIKEMYNKLGDLGDVAQECRQTQRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKK 916
+ IK+ ++GDLG +AQ R TQ L+ PTPL I+ ++ ++I+ +G G +KK
Sbjct: 640 SVDVIKQELTEVGDLGIIAQNSRSTQTLMGKPTPLTIQSVFKTFQQIADLSGTGGQQKKK 699
Query: 917 GIILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVLPALAHAVVM-----NSRPTVYE-- 969
+I L+ SC++ E +++R+L LRIG R+VL ALA +V++ S +++
Sbjct: 700 DLIKKLLVSCKDCETLYIIRSLQGKLRIGLAERSVLMALAKSVLVTPPIDGSGQQIFDIR 759
Query: 970 -EGTAENLKAALQVLSVAVVEAYNILPNLDIIVPTLMN-KGIEFSVSSLSMVPGIPIKPM 1027
+ E + Q + V AY+ LPN D+ VP L+ GI+ +S+ S+ GIP+KPM
Sbjct: 760 KQMKQEEFEERYQNVVSKVTRAYSQLPNYDLFVPHLIAVNGIDNILSTCSLKVGIPVKPM 819
Query: 1028 LAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVD 1087
LA+ T GI Q L F + FTCE+KYDG+RAQIH+L DG+ +++RN ++ T ++PD+V
Sbjct: 820 LAQPTTGISQMLDRFSDMEFTCEFKYDGERAQIHRLPDGTTHIYTRNLEDYTQKYPDIVA 879
Query: 1088 MIKESCKPVASTFIIDAEVVGIDRKNGCRIMSFQELSSRGRGGKDTLVTKESIKVGICIF 1147
+ + P +FI+D E V D +I+SFQ LS+R R V IKV +C+F
Sbjct: 880 NVTKFVGPNVKSFILDCEAVAFDAATK-KILSFQVLSTRARKS----VQLSQIKVPVCVF 934
Query: 1148 VFDIMFANGEHLAD 1161
FD+++ NG+ L D
Sbjct: 935 AFDLLYLNGQSLID 948
>UniRef100_P37913 DNA ligase I [Mus musculus]
Length = 916
Score = 284 bits (727), Expect = 1e-74
Identities = 154/418 (36%), Positives = 241/418 (56%), Gaps = 13/418 (3%)
Query: 750 RDNVDATHVSLPSDKYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFR 809
+ +D + + + Y+P E ACW+ GQ P+L +ARTF +E+ ++K L N+ R
Sbjct: 256 KGTLDPANYNPSKNNYHPIEDACWKHGQKVPFLAVARTFEKIEEVSARLKMVETLSNLLR 315
Query: 810 SLLVLSPEDVLPAVYLCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLG 869
S++ LSP D+LP +YL N++ + +EL +G ++ A+ +A G I+ + G
Sbjct: 316 SVVALSPPDLLPVLYLSLNRLGPPQQGLELGVGDGVLLKAVAQATGRQLESIRAEVAEKG 375
Query: 870 DLGDVAQECRQTQRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREK 929
D+G VA+ R TQRL+ PP PL I +++ I+ TG+ S +K II L +CR
Sbjct: 376 DVGLVAENSRSTQRLMLPPPPLTISGVFTKFCDIARLTGSASMAKKMDIIKGLFVACRHS 435
Query: 930 EMKFLVRTLVRNLRIGAMLRTVLPALAHAVVM----NSRPTVYEEG----TAENLKAALQ 981
E +++ R+L LR+G ++VL ALA AV + PTV + TAE K L+
Sbjct: 436 EARYIARSLSGRLRLGLAEQSVLAALAQAVSLTPPGQEFPTVVVDAGKGKTAEARKMWLE 495
Query: 982 VLSVAVVEAYNILPNLDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKL 1041
+ + + + +P+LD I+P L+ G+E + PG+P+KPMLA T G+ + LK
Sbjct: 496 EQGMILKQTFCEVPDLDRIIPVLLEHGLERLPEHCKLSPGVPLKPMLAHPTRGVSEVLKR 555
Query: 1042 FQNKAFTCEYKYDGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFI 1101
F+ FTCEYKYDGQRAQIH L G V +FSRN +++T ++PD++ I + P ++FI
Sbjct: 556 FEEVDFTCEYKYDGQRAQIHVLEGGEVKIFSRNQEDNTGKYPDIISRIPKIKHPSVTSFI 615
Query: 1102 IDAEVVGIDRKNGCRIMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
+D E V DR+ +I FQ L++R R D I+V +C++ FD+++ NGE L
Sbjct: 616 LDTEAVAWDREKK-QIQPFQVLTTRKRKEVDA----SEIQVQVCLYAFDLIYLNGESL 668
>UniRef100_Q8R055 Lig1 protein [Mus musculus]
Length = 916
Score = 283 bits (723), Expect = 3e-74
Identities = 153/418 (36%), Positives = 240/418 (56%), Gaps = 13/418 (3%)
Query: 750 RDNVDATHVSLPSDKYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFR 809
+ +D + + + Y+P E ACW+ GQ P+L +ARTF +E+ ++K L N+ R
Sbjct: 256 KGTLDPANYNPSKNNYHPIEDACWKHGQKVPFLAVARTFEKIEEVSARLKMVETLSNLLR 315
Query: 810 SLLVLSPEDVLPAVYLCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLG 869
S++ LSP D+LP +YL N++ + +EL +G ++ A+ +A G I+ + G
Sbjct: 316 SVVALSPPDLLPVLYLSLNRLGPPQQGLELGVGDGVLLKAVAQATGRQLESIRAEVAEKG 375
Query: 870 DLGDVAQECRQTQRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREK 929
D+G VA+ R TQRL+ PP PL I +++ I+ TG+ S +K II L +CR
Sbjct: 376 DVGLVAENSRSTQRLMLPPPPLTISGVFTKFCDIARLTGSASMAKKMDIIKGLFVACRHS 435
Query: 930 EMKFLVRTLVRNLRIGAMLRTVLPALAHAVVM----NSRPTVYEEG----TAENLKAALQ 981
E +++ R+L LR+G ++VL ALA AV + PT + TAE K L+
Sbjct: 436 EARYIARSLSGRLRLGLAEQSVLAALAQAVSLTPPGQEFPTAVVDAGKGKTAEARKMWLE 495
Query: 982 VLSVAVVEAYNILPNLDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKL 1041
+ + + + +P+LD I+P L+ G+E + PG+P+KPMLA T G+ + LK
Sbjct: 496 EQGMILKQTFCEVPDLDRIIPVLLEHGLERLPEHCKLSPGVPLKPMLAHPTRGVSEVLKR 555
Query: 1042 FQNKAFTCEYKYDGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFI 1101
F+ FTCEYKYDGQRAQIH L G V +FSRN +++T ++PD++ I + P ++FI
Sbjct: 556 FEEVDFTCEYKYDGQRAQIHVLEGGEVKIFSRNQEDNTGKYPDIISRIPKIKHPSVTSFI 615
Query: 1102 IDAEVVGIDRKNGCRIMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
+D E V DR+ +I FQ L++R R D I+V +C++ FD+++ NGE L
Sbjct: 616 LDTEAVAWDREKK-QIQPFQVLTTRKRKEVDA----SEIQVQVCLYAFDLIYLNGESL 668
>UniRef100_UPI000042C223 UPI000042C223 UniRef100 entry
Length = 803
Score = 281 bits (720), Expect = 8e-74
Identities = 163/396 (41%), Positives = 230/396 (57%), Gaps = 19/396 (4%)
Query: 773 WRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPED---------VLPAV 823
W+DG+ PY L TF +E K IL + + LLV++ D +L V
Sbjct: 172 WKDGEAVPYPALVSTFEKIE---ATTKRLEILEFLTQFLLVVAKRDTATDAKGSMLLKVV 228
Query: 824 YLCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQR 883
YLC N++ D+ +EL IG SL+ A+ E+ G KIKE K GDLG VA R TQ
Sbjct: 229 YLCINRLCPDYMGIELGIGESLLIKAIAESTGRAMPKIKEDLKKEGDLGKVAMNSRNTQS 288
Query: 884 LLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLR 943
+ P PL + ++ L I+ +GN S +K GII L+ +C+ E KF++R+L LR
Sbjct: 289 TMFKPKPLTVPYVFQCLTDIAKASGNASQAKKVGIIKKLLAACQGTEAKFIIRSLEGKLR 348
Query: 944 IGAMLRTVLPALAHAVVMNSRPTVYEEGTAENLKAALQVLSVAVVEAYNILPNLDIIVPT 1003
IG +TV+ ALAHA+V+ + ++ E L L+ + V Y+ LP+ D+++P
Sbjct: 349 IGLADKTVVVALAHAIVLKTIGD--KKPPPEKLAIMLEQGAETVKAVYSELPSYDLVIPA 406
Query: 1004 LMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKL 1063
L+ G+E + PG+P+KPMLAK T I + L F+ K FTCEYKYDG+RAQ+H L
Sbjct: 407 LLQGGVEGLRERCKLTPGVPLKPMLAKPTKAITEVLDRFEGKEFTCEYKYDGERAQVHLL 466
Query: 1064 VDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCRIMSFQEL 1123
DG++ VFSRN + ++++PDLVD I S KP A TF+IDAE V D + +I+ FQ+L
Sbjct: 467 EDGTISVFSRNSENMSAKYPDLVDQIPRSMKPTAKTFVIDAEAVAYDLETK-KILPFQDL 525
Query: 1124 SSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
S R R KD V E I V + IF FD+++ NGE L
Sbjct: 526 SRRKR--KD--VKAEDITVRVHIFAFDLLYLNGESL 557
>UniRef100_UPI00004375AB UPI00004375AB UniRef100 entry
Length = 628
Score = 280 bits (717), Expect = 2e-73
Identities = 153/409 (37%), Positives = 246/409 (59%), Gaps = 19/409 (4%)
Query: 765 YNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVY 824
Y+P +HACW+ GQ PYL +ARTF +E++ G++K L N RS+++LSP+D+L VY
Sbjct: 10 YHPIDHACWKKGQKVPYLAVARTFEKIEEDSGRLKNIETLSNFLRSVILLSPDDLLCCVY 69
Query: 825 LCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRL 884
LC N++ ++ +EL IG +++ A+ +A G KIK + GDLG VA+ R QR+
Sbjct: 70 LCLNQLGPAYQGLELGIGETVLMKAVAQATGRQLDKIKAEAQEKGDLGLVAESSRSNQRM 129
Query: 885 LAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRI 944
+ P L +++ L++I+ +GN + +K II L +CR E +++VR+L LRI
Sbjct: 130 MFAPANLTAGGVFNKLKEIAHMSGNSAMNKKIDIIKGLFVACRFSEARYIVRSLAGKLRI 189
Query: 945 GAMLRTVLPALAHAVVMNSRPTVYEEGTAENLKAALQVLSVAVVEAYNI---------LP 995
G ++VL AL+ AV + T +G+ +++ + A N+ +P
Sbjct: 190 GLAEQSVLAALSQAVCL----TPPGQGSLILTNWTGVAVAITSMHANNVFCAFSHISEMP 245
Query: 996 NLDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDG 1055
N D I+P L+ +GI+ + + PG+P++PMLA T G+ + +K F +FTCEYKYDG
Sbjct: 246 NYDAIIPVLLKEGIDELPNHCKLTPGVPLRPMLAHPTKGVGEVMKRFDEASFTCEYKYDG 305
Query: 1056 QRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGC 1115
+RAQIH L +G V +FSRN +++TS++PD++ I + K + ++D+E V DR+
Sbjct: 306 ERAQIHILENGEVRIFSRNQEDNTSKYPDIISRIPQVKKESVRSCVLDSEAVAWDREKK- 364
Query: 1116 RIMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHLA-DPL 1163
+I FQ L++R R KD V IKV +C++ FD+++ NGE L +PL
Sbjct: 365 QIQPFQVLTTRKR--KD--VDASEIKVQVCVYAFDLLYLNGESLVREPL 409
>UniRef100_Q7PVP5 ENSANGP00000010547 [Anopheles gambiae str. PEST]
Length = 661
Score = 279 bits (714), Expect = 4e-73
Identities = 159/409 (38%), Positives = 247/409 (59%), Gaps = 14/409 (3%)
Query: 761 PSDK-YNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDV 819
PS K Y+P + A W+ G PYL LARTF ++E+ G+++ IL N FRS+++LSP D+
Sbjct: 12 PSKKNYHPLKDAFWKQGDRVPYLALARTFQMIEETSGRLRMIEILSNYFRSVILLSPRDL 71
Query: 820 LPAVYLCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECR 879
L +VYLC N++A +E VEL I + A+ ++ G + +IK GDLG VA++ +
Sbjct: 72 LASVYLCLNQLAPAYEGVELGIAEFSLMKAIAQSTGRSLAQIKADAQTTGDLGLVAEQSK 131
Query: 880 QTQRLLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLV 939
+Q L+ P PL ++ ++ LR+I+ TG+ S +K I + +CR E ++++R+L
Sbjct: 132 SSQHLMFRPAPLSVEVVFGKLREIASMTGSASMAKKMDKIQSMFVACRHSESRYIIRSLA 191
Query: 940 RNLRIGAMLRTVLPALAHAVVM---NSRPTVYEEGTAEN---LKAALQVLSVAVVEAYNI 993
LRIG +++L ALA A + ++ P V E+ +KA + +++A+ Y
Sbjct: 192 GKLRIGVAEQSLLQALAQACALTPPHADPPVVNALAGESEARVKARVDEVALALKTVYCQ 251
Query: 994 LPNLDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKY 1053
PN + IVP L+ G++ M PG P+KPMLA T G+ + L+ F FTCE+KY
Sbjct: 252 CPNYNRIVPVLLEHGVQRLSEHCPMEPGTPLKPMLAHPTKGVQEVLQRFDGIDFTCEWKY 311
Query: 1054 DGQRAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIK-ESCKPVASTFIIDAEVVGIDRK 1112
DG+RAQIH L DGSV +FSRN + +TS++PD++ ++ +PVAS I+D E V D
Sbjct: 312 DGERAQIHLLADGSVQIFSRNQENNTSKYPDVIARLEFTRTEPVASA-ILDCEAVAWDTD 370
Query: 1113 NGCRIMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHLAD 1161
+I+ FQ LS+R R KD + IKV +C+F+FD+++ NGE L +
Sbjct: 371 KR-QILPFQVLSTRKR--KD--ANEADIKVQVCVFMFDLLYLNGEPLVE 414
>UniRef100_Q9JHY8 DNA ligase I [Rattus norvegicus]
Length = 918
Score = 278 bits (710), Expect = 1e-72
Identities = 201/635 (31%), Positives = 317/635 (49%), Gaps = 58/635 (9%)
Query: 537 PLSKSNLKSFSSNDASPFSKSNLNNTNSTTKKLDLFRSQESKLTNLRKALSNQISPSKRK 596
P+S S S S D+ P + N + S +D+ S K RK L + +
Sbjct: 84 PVSDSKQSSPPSPDSCPENSPVFNCSPS----MDISPSGFPKRRTARKQLPKRTIQDTLE 139
Query: 597 KGSESKSNKKVKVKAKSESSGSKQATITKFFGKAMPVMPGDTQSDQFGS---KPGESPEV 653
+ +E K+ K VK K K E + ++T+ A V + Q + + +P ESPE
Sbjct: 140 EPNEDKA-KAVK-KRKKEDPQTPPESLTE----AEEVNQKEEQVEDQPTVPPEPTESPES 193
Query: 654 EELVPTDAGNMYKQEIDQFMQIINGDESLKKQAITIIEEAKGDINKALDIYYSNSCNLGE 713
L T+ M K + Q Q +E K A+G K L +++
Sbjct: 194 VTLTKTENIPMCKAGVKQKPQ----EEEQSKPP------ARGA--KPLSSFFT------P 235
Query: 714 REISVQGECKVDRPLEKKYVSKELNVIPDISMHRVLRDNVDATHVSLPSDKYNPKEHACW 773
R+ +V+ E K + D + D T+ + Y+P E ACW
Sbjct: 236 RKPAVKTEVKQEES--------------DTPRKEETKGAPDPTNYNPSKSNYHPIEDACW 281
Query: 774 RDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVYLCTNKIAAD 833
+ GQ P+L +ARTF +E+ ++K L N+ RS++ LSP D+LP +YL N++
Sbjct: 282 KHGQKVPFLAVARTFEKIEEVSARLKMVETLSNLLRSVVALSPTDLLPVLYLSLNRLGPP 341
Query: 834 HENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRLLAPPTPLLI 893
+ +EL +G ++ A+ +A G I+ + GD+G VA+ R TQRL+ P PL +
Sbjct: 342 QQGLELGVGDGVLLKAVAQATGRQLESIRAEVAEKGDVGLVAENSRSTQRLMLPSPPLTV 401
Query: 894 KDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVLP 953
+++ I+ TG+ S +K II L +CR E +F+ R+L LR+G ++VL
Sbjct: 402 SGVFTKFCDIARLTGSASMAKKMDIIKGLFVACRYSEARFIARSLSGRLRLGLAEQSVLA 461
Query: 954 ALAHAVVM----NSRPTVYEEG----TAENLKAALQVLSVAVVEAYNILPNLDIIVPTLM 1005
ALA A + PTV + TAE K L+ + + + + +P+LD I+P L+
Sbjct: 462 ALAQAGSLTPPGQEFPTVVVDAGKGKTAEARKMWLEEQGMILKQTFCEVPDLDRIIPVLL 521
Query: 1006 NKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVD 1065
G+E + PG+P+KPMLA T G+ + LK F+ FTCEYKY GQRAQIH L
Sbjct: 522 EHGLESLPEHCKLSPGVPLKPMLAHPTRGVREVLKRFEEVDFTCEYKYYGQRAQIHVLEG 581
Query: 1066 GSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCRIMSFQELSS 1125
G V +FSRN ++++ ++PD++ I + P ++FI+D E V DR+ +I FQ L++
Sbjct: 582 GEVKIFSRNQEDNSGKYPDIISRIPKIKHPSVTSFILDTEAVAWDREKK-QIQPFQVLTT 640
Query: 1126 RGRGGKDTLVTKESIKVGICIFVFDIMFANGEHLA 1160
R R D I+V +C++ FD+++ NGE LA
Sbjct: 641 RKRKEVDA----SEIQVQVCLYAFDLIYLNGESLA 671
>UniRef100_Q8W5J3 Putative DNA ligase [Oryza sativa]
Length = 810
Score = 276 bits (706), Expect = 3e-72
Identities = 148/396 (37%), Positives = 228/396 (57%), Gaps = 10/396 (2%)
Query: 764 KYNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAV 823
++NP A W +P P+L LAR L+ +E G+I T IL N+FR+++ +PED+L V
Sbjct: 180 EFNPMAAAYWSPEEPVPFLFLARALDLISNESGRIVITEILSNVFRTVIATTPEDLLATV 239
Query: 824 YLCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQR 883
YL N+IA HE EL IG + + AL EA G +K+ +LGDLG VA+ R +Q+
Sbjct: 240 YLSANRIAPPHEGTELGIGDASIIRALAEAYGRREEHVKKNLKELGDLGLVAKASRLSQK 299
Query: 884 LLAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLR 943
++ P PL I + + R I+ ++G S +K+ I L+ + + E +++ R L +R
Sbjct: 300 MMYKPKPLTISHVLAKFRTIAKESGKDSQDKKRNHIKGLLVAATDCEPQYITRLLQSKMR 359
Query: 944 IGAMLRTVLPALAHAVVMNSRPTVYEEGTAENLKAALQVLSVAVVEAYNILPNLDIIVPT 1003
IG +TV AL A V + + +++ + + + + Y++LP D IVP
Sbjct: 360 IGLAEKTVQMALGQAAVYSEK-----HSPPSKIQSPFEEAAKIIKQVYSVLPIYDKIVPA 414
Query: 1004 LMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKL 1063
++ G+ S G+P+ PMLAK T + + + FQ +TCEYKYDG+RAQIH L
Sbjct: 415 ILEVGVWKLPEICSFSIGVPVGPMLAKATKSVSEIIDKFQGLEYTCEYKYDGERAQIHCL 474
Query: 1064 VDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCRIMSFQEL 1123
DGSV ++SRN + +T ++PD+VD + KP +F++D E+V DR+ +I+ FQ L
Sbjct: 475 EDGSVEIYSRNAERNTGKYPDVVDAVSRFRKPTVKSFVLDCEIVAYDREKK-KILPFQIL 533
Query: 1124 SSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
S+R R G VT IKV +C F FDI++ NG+ L
Sbjct: 534 STRARKG----VTISDIKVSVCTFGFDILYINGKPL 565
>UniRef100_Q9W1H4 CG5602-PA [Drosophila melanogaster]
Length = 747
Score = 268 bits (686), Expect = 7e-70
Identities = 151/458 (32%), Positives = 258/458 (55%), Gaps = 33/458 (7%)
Query: 713 EREISVQGECKVDRPLEKKYVSKELNVIPDISMHRVLRDNVDATHVSLPSDKYNPKEHAC 772
E+ V+ K + ++ K K++ I ++ D + +D Y+P ++A
Sbjct: 62 EKTSPVKNVKKEPKEVDDKTTDKKVTTI---GLNSTAATKEDVENYDPSADSYHPLKNAY 118
Query: 773 WRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVYLCTNKIAA 832
W+D + PYL LARTF ++E+ KG++K L N F S++++SPED++P+VYL N++A
Sbjct: 119 WKDKKVTPYLALARTFQVIEETKGRLKMIDTLSNFFCSVMLVSPEDLVPSVYLSINQLAP 178
Query: 833 DHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRLLAPPTPLL 892
+E +EL + + + A+ +A G N IK GDLG VA++ R +QR++ P PL
Sbjct: 179 AYEGLELGVAETTLMKAICKATGRNLAHIKSQTQLTGDLGIVAEQSRVSQRMMFQPAPLN 238
Query: 893 IKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVL 952
++D++ LR+I+ +G K ++ ++ +CR E +F +R+L+ LRIG +++L
Sbjct: 239 VRDVFRKLREIAKLSGQS----KMDLVYNMFVACRSSEARFFIRSLIGKLRIGIAEQSLL 294
Query: 953 PALAHAVVMNSR---------PTVYEEGTAENLKAALQVLSVAVVEAYNILPNLDIIVPT 1003
ALA +V + P VY++ + ++ + AY PN DII+P
Sbjct: 295 TALAIGLVKKNHIDDCKASKVPDVYKDEIVDT--------TLLLKTAYCQCPNYDIIIPA 346
Query: 1004 LMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKL 1063
++ I+ M PG+P++PMLA+ T G+ + + F TCE+KYDG+RAQIH+
Sbjct: 347 ILKYDIKELQERCPMHPGMPLRPMLAQPTKGVHEVFERFGGMQITCEWKYDGERAQIHRN 406
Query: 1064 VDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVG--IDRKNGCRIMSFQ 1121
G + +FSRN + +T+++PDL+ K ++IID+E+V ++RK +I+ FQ
Sbjct: 407 EKGEISIFSRNSENNTAKYPDLIARSTALLKGDVKSYIIDSEIVAWDVERK---QILPFQ 463
Query: 1122 ELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
LS+R R D E IKV +C+++FD+++ NG L
Sbjct: 464 VLSTRKRKNVDI----EEIKVQVCVYIFDLLYINGTAL 497
>UniRef100_UPI00003C2646 UPI00003C2646 UniRef100 entry
Length = 892
Score = 267 bits (682), Expect = 2e-69
Identities = 148/395 (37%), Positives = 229/395 (57%), Gaps = 12/395 (3%)
Query: 773 WRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVYLCTNKIAA 832
W++G+ PY LA TF+ ++ +++ T IL ++ SP+++L VYLC N++
Sbjct: 255 WKEGEAVPYAALATTFADIQATTKRLEITEILTQFLVRVIKRSPDNLLQVVYLCINRLCP 314
Query: 833 DHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRLLAPPTPLL 892
D+E +EL IG SL+ A+ ++ G +IK+ GDLG VA ++ Q + + L
Sbjct: 315 DYEGLELGIGESLLIKAIAQSTGREVARIKKDLEAQGDLGLVALHSKKNQPTMFKVSSLK 374
Query: 893 IKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRIGAMLRTVL 952
+ ++ L++I++ +GN S RK G+I L+ SC+ E KFL+R+L LRIG R+VL
Sbjct: 375 VPQVFKQLKEIALVSGNKSQDRKIGMIKKLLASCQGDEPKFLIRSLEGKLRIGLAERSVL 434
Query: 953 PALAHAVVMNSRPTVYEEGTAENLKAALQVLSVAVVEAYNILPNLDIIVPTLMNKGIEFS 1012
+LA AVV+ + + E+L L+ + V Y+ LP+ D++VP L+ G+E
Sbjct: 435 VSLARAVVIAKLGKTISKLSQESLAKQLEDATELVKAVYSELPSYDLVVPALLKGGVEHL 494
Query: 1013 VSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQRAQIHKLVDGSVLVFS 1072
S + PG+P+KPMLAK T I + L F+ K FTCEYKYDG+RAQ+H L + + VFS
Sbjct: 495 RSECKLTPGVPLKPMLAKPTKAISEVLDRFEGKPFTCEYKYDGERAQVHLLPNRQLAVFS 554
Query: 1073 RNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRK--------NGCRIMSFQELS 1124
RN + + ++PDLV+ I KP +F++DAE + +++ FQELS
Sbjct: 555 RNSENMSVKYPDLVEQIPRCIKPTVKSFVLDAEAAAWKKAQLNAEGILEPAKLLPFQELS 614
Query: 1125 SRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
R R KD V + IKV + +F FD++F NGE L
Sbjct: 615 RRKR--KD--VKAQDIKVKVKLFAFDLLFLNGESL 645
>UniRef100_UPI00004314CE UPI00004314CE UniRef100 entry
Length = 655
Score = 265 bits (677), Expect = 8e-69
Identities = 145/403 (35%), Positives = 229/403 (55%), Gaps = 23/403 (5%)
Query: 765 YNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVY 824
YNP ++ACW+ + PY+ L RT L+ED ++K IL N FRS++VLS +D+LP++Y
Sbjct: 28 YNPIDNACWKHNEKVPYIALTRTLELIEDTSARLKIIEILSNYFRSVIVLSSDDLLPSIY 87
Query: 825 LCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRL 884
LC N++A +E +EL + + + A+ + G +IK ++GDLG VA+ R QR
Sbjct: 88 LCLNQLAPAYEGIELGVAETNLMKAIAQCTGRTLAQIKTDVQEVGDLGIVAEGSRSNQRT 147
Query: 885 LAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRI 944
+ P PL + +Y+ L++I+ TG+ S +K I L +CR E ++L+R+L LRI
Sbjct: 148 MFQPAPLTVSSVYTRLKEIAQMTGSASVTKKLDKIQSLFVACRFTEARYLIRSLAGKLRI 207
Query: 945 GAMLRTVLPALAHAVVMN-SRPT-------VYEEGTAENLKAALQVLSVAVVEAYNILPN 996
G ++ ALA A M +PT V ++ + + K ++ + Y PN
Sbjct: 208 GLAEQS---ALALACTMTPPKPTFPPEILDVSKKMSNDTFKQKYDETALILKTTYCECPN 264
Query: 997 LDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQ 1056
I+P L+ +GI+ + + PGIP+KPMLA T G+ + L F FTCE+KYDG+
Sbjct: 265 YGKIIPVLLKEGIKELPNKCKITPGIPMKPMLAHPTKGVQEVLTRFDGLKFTCEWKYDGE 324
Query: 1057 RAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCR 1116
RAQIH DG + ++SRN + +TS++PD++ K + + I+D E V D +
Sbjct: 325 RAQIHLAEDGKISIYSRNQENNTSKYPDIIGRFKNTQGENVKSCILDCEAVAWDNDKK-Q 383
Query: 1117 IMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
I+ FQ LS T++ IKV +C+F+FD+++ NGE L
Sbjct: 384 ILPFQILS-----------TRKHIKVQVCVFMFDLLYLNGEPL 415
>UniRef100_UPI00004314CD UPI00004314CD UniRef100 entry
Length = 648
Score = 265 bits (677), Expect = 8e-69
Identities = 145/403 (35%), Positives = 229/403 (55%), Gaps = 23/403 (5%)
Query: 765 YNPKEHACWRDGQPAPYLHLARTFSLLEDEKGKIKATSILCNMFRSLLVLSPEDVLPAVY 824
YNP ++ACW+ + PY+ L RT L+ED ++K IL N FRS++VLS +D+LP++Y
Sbjct: 14 YNPIDNACWKHNEKVPYIALTRTLELIEDTSARLKIIEILSNYFRSVIVLSSDDLLPSIY 73
Query: 825 LCTNKIAADHENVELNIGGSLVTTALEEACGTNRLKIKEMYNKLGDLGDVAQECRQTQRL 884
LC N++A +E +EL + + + A+ + G +IK ++GDLG VA+ R QR
Sbjct: 74 LCLNQLAPAYEGIELGVAETNLMKAIAQCTGRTLAQIKTDVQEVGDLGIVAEGSRSNQRT 133
Query: 885 LAPPTPLLIKDIYSALRKISVQTGNGSTLRKKGIILHLMRSCREKEMKFLVRTLVRNLRI 944
+ P PL + +Y+ L++I+ TG+ S +K I L +CR E ++L+R+L LRI
Sbjct: 134 MFQPAPLTVSSVYTRLKEIAQMTGSASVTKKLDKIQSLFVACRFTEARYLIRSLAGKLRI 193
Query: 945 GAMLRTVLPALAHAVVMN-SRPT-------VYEEGTAENLKAALQVLSVAVVEAYNILPN 996
G ++ ALA A M +PT V ++ + + K ++ + Y PN
Sbjct: 194 GLAEQS---ALALACTMTPPKPTFPPEILDVSKKMSNDTFKQKYDETALILKTTYCECPN 250
Query: 997 LDIIVPTLMNKGIEFSVSSLSMVPGIPIKPMLAKITNGIPQALKLFQNKAFTCEYKYDGQ 1056
I+P L+ +GI+ + + PGIP+KPMLA T G+ + L F FTCE+KYDG+
Sbjct: 251 YGKIIPVLLKEGIKELPNKCKITPGIPMKPMLAHPTKGVQEVLTRFDGLKFTCEWKYDGE 310
Query: 1057 RAQIHKLVDGSVLVFSRNGDESTSRFPDLVDMIKESCKPVASTFIIDAEVVGIDRKNGCR 1116
RAQIH DG + ++SRN + +TS++PD++ K + + I+D E V D +
Sbjct: 311 RAQIHLAEDGKISIYSRNQENNTSKYPDIIGRFKNTQGENVKSCILDCEAVAWDNDKK-Q 369
Query: 1117 IMSFQELSSRGRGGKDTLVTKESIKVGICIFVFDIMFANGEHL 1159
I+ FQ LS T++ IKV +C+F+FD+++ NGE L
Sbjct: 370 ILPFQILS-----------TRKHIKVQVCVFMFDLLYLNGEPL 401
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.327 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,299,582,546
Number of Sequences: 2790947
Number of extensions: 99021080
Number of successful extensions: 375850
Number of sequences better than 10.0: 885
Number of HSP's better than 10.0 without gapping: 215
Number of HSP's successfully gapped in prelim test: 695
Number of HSP's that attempted gapping in prelim test: 371904
Number of HSP's gapped (non-prelim): 2693
length of query: 1419
length of database: 848,049,833
effective HSP length: 140
effective length of query: 1279
effective length of database: 457,317,253
effective search space: 584908766587
effective search space used: 584908766587
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 81 (35.8 bits)
Medicago: description of AC144375.6


