

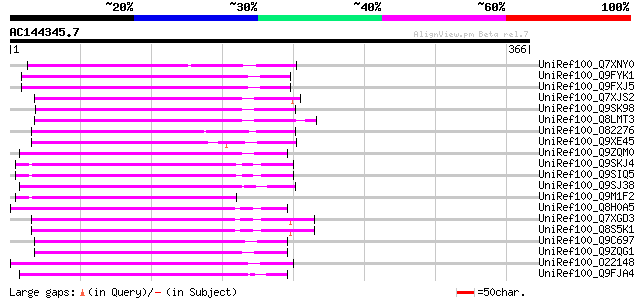
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144345.7 - phase: 0 /pseudo
(366 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa] 135 2e-30
UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana] 129 1e-28
UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana] 129 1e-28
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 125 2e-27
UniRef100_Q9SK98 Very similar to retrotransposon reverse transcr... 124 4e-27
UniRef100_Q8LMT3 Putative retroelement [Oryza sativa] 124 5e-27
UniRef100_O82276 Putative non-LTR retroelement reverse transcrip... 123 9e-27
UniRef100_Q9XE45 Putative non-LTR retrolelement reverse transcri... 122 2e-26
UniRef100_Q9ZQM0 Putative non-LTR retroelement reverse transcrip... 122 2e-26
UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcrip... 121 3e-26
UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcrip... 121 3e-26
UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcrip... 121 3e-26
UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis tha... 118 2e-25
UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa] 118 3e-25
UniRef100_Q7XGD3 Putative retroelement [Oryza sativa] 117 4e-25
UniRef100_Q8S5K1 Putative retroelement [Oryza sativa] 117 4e-25
UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [A... 117 7e-25
UniRef100_Q9ZQG1 Putative non-LTR retroelement reverse transcrip... 113 7e-24
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 113 7e-24
UniRef100_Q9FJA4 Non-LTR retroelement reverse transcriptase-like... 112 2e-23
>UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa]
Length = 1026
Score = 135 bits (340), Expect = 2e-30
Identities = 69/190 (36%), Positives = 103/190 (53%), Gaps = 9/190 (4%)
Query: 13 WYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNA 72
W ++++ +L W+ R S+GGRLILL LSS+P+Y +SF++ P G+ ++
Sbjct: 571 WKLAENKMEHKLGCWQGRLQSIGGRLILLNSTLSSVPMYMISFYRLPKGVQERIDYFRKR 630
Query: 73 FFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRV 132
F W + IRK + W VCS + +GGLGV L N A+LGKW WR+ E G W +
Sbjct: 631 FLWQEDQGIRKYHLVNWPLVCSPRDQGGLGVLDLEAMNKAMLGKWIWRLENEEG-WWQEI 689
Query: 133 LCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWV 192
+ A+Y + + G S +WQ +++V+D ++ VGNG TLFW
Sbjct: 690 IYAKYCSDKPLSGLRLKAGSSHFWQGVMEVKDD--------FFSFCTKIVGNGEKTLFWE 741
Query: 193 DPWLEEKPLS 202
D WL KPL+
Sbjct: 742 DSWLGGKPLA 751
>UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana]
Length = 1270
Score = 129 bits (325), Expect = 1e-28
Identities = 70/190 (36%), Positives = 104/190 (53%), Gaps = 8/190 (4%)
Query: 9 KLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLES 68
K+ + L DR+K +L W +R LS GG+ +LLK V ++PV+ +S FK P LES
Sbjct: 724 KVDMLHYLKDRLKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLES 783
Query: 69 LFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTL 128
+F+W C+ RKI W WE +C K GGLG R ++ FN ALL K WR+L L
Sbjct: 784 AMASFWWDSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCL 843
Query: 129 WYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLST 188
R+L +RY + L + S W++IL R+ + + + +RVG+G S
Sbjct: 844 LSRLLKSRYFDATDFLDAALSQRPSFGWRSILFGRELLSK--------GLQKRVGDGASL 895
Query: 189 LFWVDPWLEE 198
W+DPW+++
Sbjct: 896 FVWIDPWIDD 905
>UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana]
Length = 1254
Score = 129 bits (325), Expect = 1e-28
Identities = 70/190 (36%), Positives = 104/190 (53%), Gaps = 8/190 (4%)
Query: 9 KLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLES 68
K+ + L DR+K +L W +R LS GG+ +LLK V ++PV+ +S FK P LES
Sbjct: 727 KVDMLHYLKDRLKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLES 786
Query: 69 LFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTL 128
+F+W C+ RKI W WE +C K GGLG R ++ FN ALL K WR+L L
Sbjct: 787 AMASFWWDSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCL 846
Query: 129 WYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLST 188
R+L +RY + L + S W++IL R+ + + + +RVG+G S
Sbjct: 847 LSRLLKSRYFDATDFLDAALSQRPSFGWRSILFGRELLSK--------GLQKRVGDGASL 898
Query: 189 LFWVDPWLEE 198
W+DPW+++
Sbjct: 899 FVWIDPWIDD 908
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 125 bits (313), Expect = 2e-27
Identities = 65/192 (33%), Positives = 105/192 (53%), Gaps = 12/192 (6%)
Query: 18 DRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFWGG 77
++++ RL+ W ++ LS GG+ +LLK + ++PVY +S FK P + L ++ F+W
Sbjct: 748 EKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPKNLCQKLTTVMMDFWWNS 807
Query: 78 CEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCARY 137
+ RKI W+ W+ + K +GG G + L+ FN ALL K WR+LQE+G+L+ RV +RY
Sbjct: 808 MQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVFQSRY 867
Query: 138 GEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPWLE 197
L + S W++IL R+ ++ + +GNG T W D WL
Sbjct: 868 FSNSDFLSATRGSRPSYAWRSILFGRE--------LLMQGLRTVIGNGQKTFVWTDKWLH 919
Query: 198 E----KPLS*RR 205
+ +PL+ RR
Sbjct: 920 DGSNRRPLNRRR 931
>UniRef100_Q9SK98 Very similar to retrotransposon reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 124 bits (311), Expect = 4e-27
Identities = 70/183 (38%), Positives = 99/183 (53%), Gaps = 8/183 (4%)
Query: 19 RIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFWGGC 78
R+K +LS W S++LS GG+ I+LK V ++PV+ +S FK P + L S F+W
Sbjct: 619 RLKSKLSSWFSKSLSQGGKEIMLKAVAMAMPVFAMSCFKLPKTTCTNLASAMADFWWSAN 678
Query: 79 EDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCARYG 138
+ RKI W+ WE +C K GGLG R + FN ALL K WR+LQ L+ R++ +RY
Sbjct: 679 DHQRKIHWLSWEKLCLPKEHGGLGFRDIGLFNQALLAKQAWRLLQFPDCLFARLIKSRYF 738
Query: 139 EEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPWLEE 198
G L S W++IL RD + + +RVGNG S W+D WL++
Sbjct: 739 PVGEFLDSDVGSRPSFGWRSILHGRD--------LLCRGLVKRVGNGKSIRVWIDYWLDD 790
Query: 199 KPL 201
L
Sbjct: 791 NGL 793
>UniRef100_Q8LMT3 Putative retroelement [Oryza sativa]
Length = 764
Score = 124 bits (310), Expect = 5e-27
Identities = 71/199 (35%), Positives = 108/199 (53%), Gaps = 14/199 (7%)
Query: 18 DRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFWGG 77
++I++RL+ WK LS GG+ IL+ LSSIP+Y + + P G+ + ++S+ FFW G
Sbjct: 307 NKIEKRLATWKCGYLSYGGKAILINSCLSSIPLYMMGVYLLPEGVHNKMDSIRARFFWEG 366
Query: 78 CEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCARY 137
E RK IKWE +C K GGLG R+ N+ALL KW +R+ + +L +Y
Sbjct: 367 LEKKRKYHMIKWEALCRPKEFGGLGFIDTRKMNIALLCKWIYRLESGKEDPCCVLLRNKY 426
Query: 138 GEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPWLE 197
++GG S + S +W+ + +V+ WM S +VGNG +T FW D W+
Sbjct: 427 MKDGGGFFQSKAEESSQFWKGLHEVKK--------WMDLGSSYKVGNGKATNFWSDVWIG 478
Query: 198 EKPLS*RRINYTRWPSCLR 216
E PL T++P+ R
Sbjct: 479 ETPLK------TQYPNIYR 491
>UniRef100_O82276 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 123 bits (308), Expect = 9e-27
Identities = 65/186 (34%), Positives = 103/186 (54%), Gaps = 5/186 (2%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
+++R+ RL+ WK R+LS+ GR+ L K VLSSIPV+ +S P + TL+ F W
Sbjct: 631 VLERVSARLAGWKGRSLSLAGRITLTKAVLSSIPVHVMSAILLPVSTLDTLDRYSRTFLW 690
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
G + +K + W +C K EGG+G+R R+ N AL+ K WR+LQ++ +LW RV+
Sbjct: 691 GSTMEKKKQHLLSWRKICKPKAEGGIGLRSARDMNKALVAKVGWRLLQDKESLWARVVRK 750
Query: 136 RYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPW 195
+Y + GG S W T V G+ ++ ++ + G+G + FW+D W
Sbjct: 751 KY-KVGGVQDTSWLKPQPRWSSTWRSVAVGLREV----VVKGVGWVPGDGCTIRFWLDRW 805
Query: 196 LEEKPL 201
L ++PL
Sbjct: 806 LLQEPL 811
>UniRef100_Q9XE45 Putative non-LTR retrolelement reverse transcriptase [Arabidopsis
thaliana]
Length = 732
Score = 122 bits (305), Expect = 2e-26
Identities = 67/196 (34%), Positives = 102/196 (51%), Gaps = 23/196 (11%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
+++++ RL+ WK R LS+ GR+ L K VLSSIPV+ +S P + L+ + +F W
Sbjct: 293 ILEKLTTRLAGWKGRFLSLAGRVTLTKAVLSSIPVHTMSTIALPKSTLDGLDKVSRSFLW 352
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
G RK I W+ VC + EGGLG+R+ ++ N ALL K WR++Q+ +LW R++
Sbjct: 353 GSSVTQRKQHLISWKRVCKPRSEGGLGIRKAQDMNKALLSKVGWRLIQDYHSLWARIMRC 412
Query: 136 RYGEEGGRLCCSCRDG---------GSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGL 186
Y + RDG S W L +R+ V + +S +G+G
Sbjct: 413 NYRVQ------DVRDGAWTKVRSVCSSTWRSVALGMREVV--------IPGLSWVIGDGR 458
Query: 187 STLFWVDPWLEEKPLS 202
LFW+D WL PL+
Sbjct: 459 EILFWMDKWLTNIPLA 474
>UniRef100_Q9ZQM0 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1044
Score = 122 bits (305), Expect = 2e-26
Identities = 65/189 (34%), Positives = 96/189 (50%), Gaps = 8/189 (4%)
Query: 8 RKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLE 67
+K + +VDRI++R W SR LS G+ +LK VL+S+P Y +S FK + ++
Sbjct: 586 KKRDLFNQIVDRIRQRSLSWSSRFLSTAGKTTMLKSVLASMPTYTMSCFKLLVSLCKRIQ 645
Query: 68 SLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGT 127
S F+W D +K+ WI W + K EGGLG + + FN ALL K WRI+Q
Sbjct: 646 SALTHFWWDSSADKKKMCWIAWSKMAKNKKEGGLGFKDITNFNDALLAKLSWRIVQSPSC 705
Query: 128 LWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLS 187
+ R+L +Y L CS S W+ I +D + + + +G+GL
Sbjct: 706 VLVRILLGKYCRTSSFLDCSVTAASSHGWRGICTGKD--------LIKSQLGKVIGSGLD 757
Query: 188 TLFWVDPWL 196
TL W +PWL
Sbjct: 758 TLVWNEPWL 766
>UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1750
Score = 121 bits (304), Expect = 3e-26
Identities = 72/196 (36%), Positives = 108/196 (54%), Gaps = 9/196 (4%)
Query: 5 GDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIIS 64
G +K F Y ++DR+K+R S W +R LS G+ I+LK V ++PVY +S FK P GI+S
Sbjct: 1140 GRKKKEMFEY-IIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKGIVS 1198
Query: 65 TLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQE 124
+ESL F+W + R I W+ W+ + K EGGLG R L +FN ALL K WR++Q
Sbjct: 1199 EIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQY 1258
Query: 125 RGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGN 184
+L+ RV+ ARY ++ L R S W ++L DG+ + G +G+
Sbjct: 1259 PNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLL---DGIALLKKG-----TRHLIGD 1310
Query: 185 GLSTLFWVDPWLEEKP 200
G + +D ++ P
Sbjct: 1311 GQNIRIGLDNIVDSHP 1326
>UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1524
Score = 121 bits (304), Expect = 3e-26
Identities = 72/196 (36%), Positives = 108/196 (54%), Gaps = 9/196 (4%)
Query: 5 GDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIIS 64
G +K F Y ++DR+K+R S W +R LS G+ I+LK V ++PVY +S FK P GI+S
Sbjct: 914 GRKKKEMFEY-IIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKGIVS 972
Query: 65 TLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQE 124
+ESL F+W + R I W+ W+ + K EGGLG R L +FN ALL K WR++Q
Sbjct: 973 EIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQY 1032
Query: 125 RGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGN 184
+L+ RV+ ARY ++ L R S W ++L DG+ + G +G+
Sbjct: 1033 PNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLL---DGIALLKKG-----TRHLIGD 1084
Query: 185 GLSTLFWVDPWLEEKP 200
G + +D ++ P
Sbjct: 1085 GQNIRIGLDNIVDSHP 1100
>UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1229
Score = 121 bits (303), Expect = 3e-26
Identities = 65/195 (33%), Positives = 103/195 (52%), Gaps = 9/195 (4%)
Query: 8 RKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLE 67
RK + ++D+I+++ W SR LS G+ ++LK VL+S+P+Y +S FK PS + ++
Sbjct: 655 RKRDIFGAIIDKIRQKSHSWASRFLSQAGKQVMLKAVLASMPLYSMSCFKLPSALCRKIQ 714
Query: 68 SLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGT 127
SL F+W D+RK +W+ W + + K GGLG R + N +LL K WR+L +
Sbjct: 715 SLLTRFWWDTKPDVRKTSWVAWSKLTNPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPES 774
Query: 128 LWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLS 187
L R+L +Y + C S W++I+ R+ + + GW+ + NG
Sbjct: 775 LLSRILLGKYCHSSSFMECKLPSQPSHGWRSIIAGRE-ILKEGLGWL-------ITNGEK 826
Query: 188 TLFWVDPWLE-EKPL 201
W DPWL KPL
Sbjct: 827 VSIWNDPWLSISKPL 841
>UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis thaliana]
Length = 851
Score = 118 bits (296), Expect = 2e-25
Identities = 64/156 (41%), Positives = 92/156 (58%), Gaps = 1/156 (0%)
Query: 5 GDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIIS 64
G +K F Y ++DR+K R + W ++ LS G+ ILLK V ++PVY +S FK P GI+S
Sbjct: 379 GRKKKEMFNY-IIDRVKERTASWSAKFLSPAGKEILLKSVALAMPVYAMSCFKLPQGIVS 437
Query: 65 TLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQE 124
+ESL F+W + R I W+ W+ + K EGGLG R L +FN ALL K WRI+Q
Sbjct: 438 EIESLLMNFWWEKASNKRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRIIQY 497
Query: 125 RGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTIL 160
+L+ RV+ ARY ++ + R S W ++L
Sbjct: 498 PNSLFARVMKARYFKDNSIIDAKTRSQQSYGWSSLL 533
>UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa]
Length = 1557
Score = 118 bits (295), Expect = 3e-25
Identities = 69/196 (35%), Positives = 97/196 (49%), Gaps = 8/196 (4%)
Query: 1 LPIGGDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPS 60
LP G + L +RI +++ W LS GG+ +L+K V+ +IPVY + FK P
Sbjct: 1122 LPTPGGRMHKGRFQSLRERIWKKILQWGENYLSSGGKEVLIKAVIQAIPVYVMGIFKLPE 1181
Query: 61 GIISTLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWR 120
+ L SL F+WG + RK W W+++ K GGLG + +R FN ALL + WR
Sbjct: 1182 SVCEDLTSLTRNFWWGAEKGKRKTHWKAWKSLTKSKSLGGLGFKDIRLFNQALLARQAWR 1241
Query: 121 ILQERGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISR 180
++ +L RVL A+Y G + S S WQ I G+ + G I
Sbjct: 1242 LIDNPDSLCARVLKAKYYPNGSIVDTSFGGNASPGWQAI---EHGLELVKKG-----IIW 1293
Query: 181 RVGNGLSTLFWVDPWL 196
R+GNG S W DPWL
Sbjct: 1294 RIGNGRSVRVWQDPWL 1309
>UniRef100_Q7XGD3 Putative retroelement [Oryza sativa]
Length = 1694
Score = 117 bits (294), Expect = 4e-25
Identities = 70/204 (34%), Positives = 104/204 (50%), Gaps = 12/204 (5%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
L +R+ +R+ W + LS GG+ IL+K VL ++PVY + FK P + L L F+W
Sbjct: 1381 LQERLWKRIMLWGEKFLSTGGKEILIKAVLQAMPVYVMGLFKLPESVCEELTKLVRNFWW 1440
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
G + +R+ W W+ + ++K +GG+G R R FN ALL + WR+L +L R+L A
Sbjct: 1441 GAEKGMRRTHWKSWDCLIAQKLKGGMGFRDFRLFNQALLARQAWRLLDRPDSLCARILKA 1500
Query: 136 RYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPW 195
Y G + S S W+ I G+ + G I R+GNG S W DPW
Sbjct: 1501 IYYPNGSLIDTSFGGNASPGWRAI---EYGLELLKKG-----IIWRIGNGKSVRLWRDPW 1552
Query: 196 L----EEKPLS*RRINYTRWPSCL 215
+ +P+S +R RW S L
Sbjct: 1553 IPRSYSRRPISAKRNCRLRWVSDL 1576
>UniRef100_Q8S5K1 Putative retroelement [Oryza sativa]
Length = 1888
Score = 117 bits (294), Expect = 4e-25
Identities = 70/204 (34%), Positives = 104/204 (50%), Gaps = 12/204 (5%)
Query: 16 LVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFW 75
L +R+ +R+ W + LS GG+ IL+K VL ++PVY + FK P + L L F+W
Sbjct: 1338 LQERLWKRIMLWGEKFLSTGGKEILIKAVLQAMPVYVMGLFKLPESVCEELTKLVRNFWW 1397
Query: 76 GGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCA 135
G + +R+ W W+ + ++K +GG+G R R FN ALL + WR+L +L R+L A
Sbjct: 1398 GAEKGMRRTHWKSWDCLIAQKLKGGMGFRDFRLFNQALLARQAWRLLDRPDSLCARILKA 1457
Query: 136 RYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPW 195
Y G + S S W+ I G+ + G I R+GNG S W DPW
Sbjct: 1458 IYYPNGSLIDTSFGGNASPGWRAI---EYGLELLKKG-----IIWRIGNGKSVRLWRDPW 1509
Query: 196 L----EEKPLS*RRINYTRWPSCL 215
+ +P+S +R RW S L
Sbjct: 1510 IPRSYSRRPISAKRNCRLRWVSDL 1533
>UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
Length = 1142
Score = 117 bits (292), Expect = 7e-25
Identities = 59/179 (32%), Positives = 96/179 (52%), Gaps = 8/179 (4%)
Query: 18 DRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFWGG 77
DR++ R++ W ++ LS GG+ +++K V +++P Y +S F+ P I S L S F+W
Sbjct: 554 DRLQSRINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCFRLPKAITSKLTSAVAKFWWSS 613
Query: 78 CEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCARY 137
D R + W+ W+ +CS K +GGLG R + +FN ALL K WR++ +L+ +V RY
Sbjct: 614 NGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGRY 673
Query: 138 GEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPWL 196
+ L S W++++ R V + +RVG+G S W DPW+
Sbjct: 674 FRKSNPLDSIKSYSPSYGWRSMISARSLV--------YKGLIKRVGSGASISVWNDPWI 724
>UniRef100_Q9ZQG1 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1138
Score = 113 bits (283), Expect = 7e-24
Identities = 67/179 (37%), Positives = 91/179 (50%), Gaps = 8/179 (4%)
Query: 18 DRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLESLFNAFFWGG 77
D++K RLS W +R LS+GG+ ILLK V ++ VY +S FK L S + F+W
Sbjct: 546 DKLKSRLSGWFARTLSLGGKEILLKAVAMAMQVYAMSCFKLTKTTCKNLTSAMSDFWWNA 605
Query: 78 CEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGTLWYRVLCARY 137
E RK W+ WE +C K GGLG R + FN ALL K WRILQ L+ R +RY
Sbjct: 606 LEHKRKTHWVSWEKLCLAKESGGLGFRDIESFNQALLAKQSWRILQYPSFLFARFFKSRY 665
Query: 138 GEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLSTLFWVDPWL 196
++ L S +IL R+ + + + VGNG S W+DPW+
Sbjct: 666 FDDEEFLEADLGVRPSYACCSILFGRE--------LLAKGLRKEVGNGKSLNVWMDPWI 716
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 113 bits (283), Expect = 7e-24
Identities = 66/200 (33%), Positives = 101/200 (50%), Gaps = 8/200 (4%)
Query: 1 LPIGGDPRKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPS 60
LP K+A L DR+ +++ W+S LS GG+ ILLK V ++P Y +S FK P
Sbjct: 753 LPESFQGSKVATLSYLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPK 812
Query: 61 GIISTLESLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWR 120
I +ES+ F+W ++ R + W W + K GGLG + + FN+ALLGK WR
Sbjct: 813 TICQQIESVMAEFWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWR 872
Query: 121 ILQERGTLWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISR 180
++ E+ +L +V +RY + L S W++I + + + Q I
Sbjct: 873 MITEKDSLMAKVFKSRYFSKSDPLNAPLGSRPSFAWKSIYEAQVLIKQ--------GIRA 924
Query: 181 RVGNGLSTLFWVDPWLEEKP 200
+GNG + W DPW+ KP
Sbjct: 925 VIGNGETINVWTDPWIGAKP 944
>UniRef100_Q9FJA4 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 1197
Score = 112 bits (280), Expect = 2e-23
Identities = 63/189 (33%), Positives = 96/189 (50%), Gaps = 8/189 (4%)
Query: 8 RKLAFWYPLVDRIKRRLSDWKSRNLSMGGRLILLKPVLSSIPVYFLSFFKAPSGIISTLE 67
RK + +VD+IK++ W SR LS G+ ++L+ VL++IPVY +S F P + ++
Sbjct: 590 RKKDLFTSIVDKIKQKAISWSSRFLSGAGKQVMLQSVLTAIPVYAMSCFLLPLSLCDRIQ 649
Query: 68 SLFNAFFWGGCEDIRKITWIKWETVCSRKGEGGLGVRRLREFNLALLGKWWWRILQERGT 127
S F+W + RK+ W+ W+ + K GGLG + + +N+ALL K WR+L
Sbjct: 650 STLTRFWWDKTDQQRKMCWVGWDRLAVPKVRGGLGFKNFKAYNVALLAKQSWRVLPHPSC 709
Query: 128 LWYRVLCARYGEEGGRLCCSCRDGGSVWWQTILDVRDGVGQIDSGWMLDNISRRVGNGLS 187
L RVL +Y + L S W+ +L RD + + GW +GN S
Sbjct: 710 LLARVLLGKYCQNESFLKALSPTFASHGWRDLLAGRDLLRK-QLGW-------TIGNDKS 761
Query: 188 TLFWVDPWL 196
L W D WL
Sbjct: 762 VLVWQDAWL 770
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.350 0.159 0.603
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 605,160,999
Number of Sequences: 2790947
Number of extensions: 25153065
Number of successful extensions: 88955
Number of sequences better than 10.0: 276
Number of HSP's better than 10.0 without gapping: 204
Number of HSP's successfully gapped in prelim test: 72
Number of HSP's that attempted gapping in prelim test: 88464
Number of HSP's gapped (non-prelim): 360
length of query: 366
length of database: 848,049,833
effective HSP length: 129
effective length of query: 237
effective length of database: 488,017,670
effective search space: 115660187790
effective search space used: 115660187790
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144345.7


