

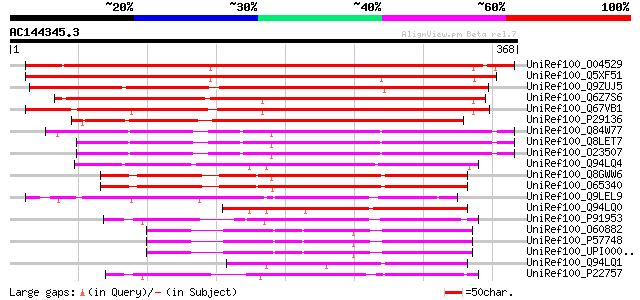
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144345.3 + phase: 0
(368 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O04529 F20P5.11 protein [Arabidopsis thaliana] 383 e-105
UniRef100_Q5XF51 At1g24140 [Arabidopsis thaliana] 377 e-103
UniRef100_Q9ZUJ5 T2K10.2 protein [Arabidopsis thaliana] 355 1e-96
UniRef100_Q6Z7S6 Putative zinc metalloproteinase [Oryza sativa] 348 1e-94
UniRef100_Q67VB1 Putative metalloproteinase [Oryza sativa] 334 3e-90
UniRef100_P29136 Metalloendoproteinase 1 precursor [Glycine max] 306 5e-82
UniRef100_Q84W77 Hypothetical protein At4g16640 [Arabidopsis tha... 273 6e-72
UniRef100_Q8LET7 Proteinase like protein [Arabidopsis thaliana] 273 7e-72
UniRef100_O23507 Proteinase like protein [Arabidopsis thaliana] 273 7e-72
UniRef100_Q94LQ4 Putative metalloproteinase [Oryza sativa] 258 1e-67
UniRef100_Q8GWW6 Putative metalloproteinase [Arabidopsis thaliana] 251 2e-65
UniRef100_O65340 Metalloproteinase [Arabidopsis thaliana] 251 2e-65
UniRef100_Q9LEL9 Matrix metalloproteinase [Cucumis sativus] 229 7e-59
UniRef100_Q94LQ0 Putative metalloproteinase [Oryza sativa] 160 4e-38
UniRef100_P91953 Hatching enzyme precursor [Hemicentrotus pulche... 154 4e-36
UniRef100_O60882 Matrix metalloproteinase-20 precursor [Homo sap... 149 9e-35
UniRef100_P57748 Matrix metalloproteinase-20 precursor [Mus musc... 149 2e-34
UniRef100_UPI000036F042 UPI000036F042 UniRef100 entry 147 3e-34
UniRef100_Q94LQ1 Putative metalloproteinase [Oryza sativa] 147 5e-34
UniRef100_P22757 Hatching enzyme precursor [Paracentrotus lividus] 146 8e-34
>UniRef100_O04529 F20P5.11 protein [Arabidopsis thaliana]
Length = 378
Score = 383 bits (983), Expect = e-105
Identities = 197/376 (52%), Positives = 251/376 (66%), Gaps = 24/376 (6%)
Query: 12 IFYITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLK 71
+F + + ++ SA FFP+ +++P + N WD + NFTGC G+ DGL ++K
Sbjct: 5 VFGFLSLFLIVSPASAWFFPNSTAVPP-SLRNTTRVFWDAFSNFTGCHHGQNVDGLYRIK 63
Query: 72 NYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVA 131
YF FGYIP NFTDDFD+ L++AV YQ NFNLN+TGELD T+ +IV PRCG
Sbjct: 64 KYFQRFGYIPETFSGNFTDDFDDILKAAVELYQTNFNLNVTGELDALTIQHIVIPRCGNP 123
Query: 132 DIINGTTSMNSGK-----FNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDD 186
D++NGT+ M+ G+ N S T+ H V Y+ FPG+PRWP + LTYAFDP L +
Sbjct: 124 DVVNGTSLMHGGRRKTFEVNFSRTHLHAVKRYTLFPGEPRWPRNRRDLTYAFDPKNPLTE 183
Query: 187 ATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSP 246
K VF+ AF +WS VT + FT + S+S+SDI IGFY+GDHGDGE FDGVLGTLAHAFSP
Sbjct: 184 EVKSVFSRAFGRWSDVTALNFTLSESFSTSDITIGFYTGDHGDGEPFDGVLGTLAHAFSP 243
Query: 247 TDGRLHLDKAEDWVVNGDVTE-SSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTIS 305
G+ HLD E+WVV+GD+ S++ AVDLESV VHEIGHLLGLGHSS+EE+IMYPTI+
Sbjct: 244 PSGKFHLDADENWVVSGDLDSFLSVTAAVDLESVAVHEIGHLLGLGHSSVEESIMYPTIT 303
Query: 306 SRTKKVELESDDIEGIQMLYGSNPNFTGTT---TATSRDRDSSFGGRHG----------- 351
+ +KV+L +DD+EGIQ LYG+NPNF GTT + T RD+ GG
Sbjct: 304 TGKRKVDLTNDDVEGIQYLYGANPNFNGTTSPPSTTKHQRDT--GGFSAAWRIDGSSRST 361
Query: 352 -VSLFLSLVFLGFGFL 366
VSL LS V L FL
Sbjct: 362 IVSLLLSTVGLVLWFL 377
>UniRef100_Q5XF51 At1g24140 [Arabidopsis thaliana]
Length = 384
Score = 377 bits (967), Expect = e-103
Identities = 184/353 (52%), Positives = 241/353 (68%), Gaps = 11/353 (3%)
Query: 12 IFYITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLK 71
+F + L+ + VSA F+ + S++P NA W+ + NFTGC GK YDGL LK
Sbjct: 6 VFMVFLLFFAPSPVSAGFYTNSSAIPPQLLRNATGNPWNSFLNFTGCHAGKKYDGLYMLK 65
Query: 72 NYFNHFGYIPNGPPS-NFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGV 130
YF HFGYI S NFTDDFD+ L++AV YQ+NF LN+TG LD+ T+ ++V PRCG
Sbjct: 66 QYFQHFGYITETNLSGNFTDDFDDILKNAVEMYQRNFQLNVTGVLDELTLKHVVIPRCGN 125
Query: 131 ADIINGTTSMNSGK------FNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENL 184
D++NGT++M+SG+ F FH V HYSFFPG+PRWP + LTYAFDP L
Sbjct: 126 PDVVNGTSTMHSGRKTFEVSFAGRGQRFHAVKHYSFFPGEPRWPRNRRDLTYAFDPRNAL 185
Query: 185 DDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAF 244
+ K VF+ AF +W +VT +TFT +S+SDI IGFYSG+HGDGE FDG + TLAHAF
Sbjct: 186 TEEVKSVFSRAFTRWEEVTPLTFTRVERFSTSDISIGFYSGEHGDGEPFDGPMRTLAHAF 245
Query: 245 SPTDGRLHLDKAEDWVVNGDVTES--SLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYP 302
SP G HLD E+W+V+G+ + S+S AVDLESV VHEIGHLLGLGHSS+E +IMYP
Sbjct: 246 SPPTGHFHLDGEENWIVSGEGGDGFISVSEAVDLESVAVHEIGHLLGLGHSSVEGSIMYP 305
Query: 303 TISSRTKKVELESDDIEGIQMLYGSNPNFTGTTT--ATSRDRDSSFGGRHGVS 353
TI + +KV+L +DD+EG+Q LYG+NPNF G+ + +++ RD+ G G S
Sbjct: 306 TIRTGRRKVDLTTDDVEGVQYLYGANPNFNGSRSPPPSTQQRDTGDSGAPGRS 358
>UniRef100_Q9ZUJ5 T2K10.2 protein [Arabidopsis thaliana]
Length = 360
Score = 355 bits (910), Expect = 1e-96
Identities = 177/336 (52%), Positives = 225/336 (66%), Gaps = 11/336 (3%)
Query: 15 ITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLKNYF 74
I + T+ +SA+F+ + SS+P NA AW+ + GC G+ +GLSKLK YF
Sbjct: 8 ILIFFFTVNPISAKFYTNVSSIPPLQFLNATQNAWETFSKLAGCHIGENINGLSKLKQYF 67
Query: 75 NHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVADII 134
FGYI N TDDFD+ L+SA+ TYQKNFNL +TG+LD +T+ IVKPRCG D+I
Sbjct: 68 RRFGYITT--TGNCTDDFDDVLQSAINTYQKNFNLKVTGKLDSSTLRQIVKPRCGNPDLI 125
Query: 135 NGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDDATKQVFAN 194
+G + MN GK T YSFFPG+PRWP+ + LTYAF P NL D K+VF+
Sbjct: 126 DGVSEMNGGKI------LRTTEKYSFFPGKPRWPKRKRDLTYAFAPQNNLTDEVKRVFSR 179
Query: 195 AFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLD 254
AF +W++VT + FT + S +DI IGF+SG+HGDGE FDG +GTLAHA SP G LHLD
Sbjct: 180 AFTRWAEVTPLNFTRSESILRADIVIGFFSGEHGDGEPFDGAMGTLAHASSPPTGMLHLD 239
Query: 255 KAEDWVV-NGDVTESSL--SNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKV 311
EDW++ NG+++ L + VDLESV VHEIGHLLGLGHSS+E+AIM+P IS +KV
Sbjct: 240 GDEDWLISNGEISRRILPVTTVVDLESVAVHEIGHLLGLGHSSVEDAIMFPAISGGDRKV 299
Query: 312 ELESDDIEGIQMLYGSNPNFTGTTTATSRDRDSSFG 347
EL DDIEGIQ LYG NPN G + SR+ S+ G
Sbjct: 300 ELAKDDIEGIQHLYGGNPNGDGGGSKPSRESQSTGG 335
>UniRef100_Q6Z7S6 Putative zinc metalloproteinase [Oryza sativa]
Length = 372
Score = 348 bits (894), Expect = 1e-94
Identities = 178/322 (55%), Positives = 230/322 (71%), Gaps = 14/322 (4%)
Query: 33 PSSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLKNYFNHFGYIPNGPPSN-FTDD 91
P +PA ++N P W ++N +GC G+ +GL +LK+Y +HFGY+P P S+ ++D
Sbjct: 30 PMGLPA--TANPFPNPWSAFQNLSGCHAGEEREGLGRLKDYLSHFGYLPPPPSSSPYSDA 87
Query: 92 FDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTN 151
FD++LE+A+ YQ+NF LN TGELD T++ +V PRCGVAD+INGT++M+ NSS+
Sbjct: 88 FDDSLEAAIAAYQRNFGLNATGELDTDTVDQMVAPRCGVADVINGTSTMDR---NSSAAA 144
Query: 152 FHTVAHYSFFPGQPRWPEGTQTLTYAFDPSE--NLDDAT-KQVFANAFNQWSKVTTITFT 208
YS+FPG P WP + L YA + ++D AT VFA AF++W+ T + FT
Sbjct: 145 LRGRHLYSYFPGGPMWPPFRRNLRYAITATSATSIDRATLSAVFARAFSRWAAATRLQFT 204
Query: 209 EATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDV-TE 267
E +S S++DI IGFYSGDHGDGEAFDG LGTLAHAFSPTDGR HLD AE WV +GDV T
Sbjct: 205 EVSSASNADITIGFYSGDHGDGEAFDGPLGTLAHAFSPTDGRFHLDAAEAWVASGDVSTS 264
Query: 268 SSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGS 327
SS AVDLESV VHEIGHLLGLGHSS+ ++IMYPTI + T+KV+LESDD+ GIQ LYG+
Sbjct: 265 SSFGTAVDLESVAVHEIGHLLGLGHSSVPDSIMYPTIRTGTRKVDLESDDVLGIQSLYGT 324
Query: 328 NPNFTGTT----TATSRDRDSS 345
NPNF G T + +SR+ D S
Sbjct: 325 NPNFKGVTPTSPSTSSREMDGS 346
>UniRef100_Q67VB1 Putative metalloproteinase [Oryza sativa]
Length = 371
Score = 334 bits (856), Expect = 3e-90
Identities = 174/346 (50%), Positives = 227/346 (65%), Gaps = 18/346 (5%)
Query: 12 IFYITLISITLTSVSARFFPDPSSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLK 71
+ + +++ S+ F P+ +P S P W +KN +GC G GL KLK
Sbjct: 10 VVLLAVVAAIAVSLVQPAFALPAGLPDIKSLTNP---WSAFKNLSGCHFGDERQGLGKLK 66
Query: 72 NYFNHFGYIPNGPPSN----FTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPR 127
+Y HFGY+ S+ F D FD +E A++ YQ NF L++TG+LD AT++ ++ PR
Sbjct: 67 DYLWHFGYLSYPSSSSLSPSFNDLFDADMELAIKMYQGNFGLDVTGDLDAATVSQMMAPR 126
Query: 128 CGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSE--NLD 185
CGVAD++NGT++M G YS+FPG PRWP TL YA + ++D
Sbjct: 127 CGVADVVNGTSTMGGGG------GVRGRGLYSYFPGSPRWPRSRTTLRYAITATSQTSID 180
Query: 186 DAT-KQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAF 244
AT +VFA+AF +WS TT+ FTEA S + +DI IGFY GDHGDGEAFDG LGTLAHAF
Sbjct: 181 RATLSKVFASAFARWSAATTLNFTEAASAADADITIGFYGGDHGDGEAFDGPLGTLAHAF 240
Query: 245 SPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTI 304
SPT+GRLHLD +E WV GDVT +S + AVDLESV VHEIGH+LGLGHSS ++IM+PT+
Sbjct: 241 SPTNGRLHLDASEAWVAGGDVTRASSNAAVDLESVAVHEIGHILGLGHSSAADSIMFPTL 300
Query: 305 SSRTKKVELESDDIEGIQMLYGSNPNFTGTT--TATSRDRDSSFGG 348
+SRTKKV L +DD+ GIQ LYG+NPNF G T +SR+ DS+ G
Sbjct: 301 TSRTKKVNLATDDVAGIQGLYGNNPNFKGVTPPATSSREMDSAGAG 346
>UniRef100_P29136 Metalloendoproteinase 1 precursor [Glycine max]
Length = 305
Score = 306 bits (785), Expect = 5e-82
Identities = 157/287 (54%), Positives = 193/287 (66%), Gaps = 15/287 (5%)
Query: 46 PGAWDG---YKNFTGCSRGKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRT 102
P AWDG YK FT G+ Y GLS +KNYF+H GYIPN P +F D+FD+ L SA++T
Sbjct: 31 PYAWDGEATYK-FTTYHPGQNYKGLSNVKNYFHHLGYIPNAP--HFDDNFDDTLVSAIKT 87
Query: 103 YQKNFNLNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFP 162
YQKN+NLN+TG+ D T+ I+ PRCGV DII T + +T+F ++ Y+FF
Sbjct: 88 YQKNYNLNVTGKFDINTLKQIMTPRCGVPDIIINT---------NKTTSFGMISDYTFFK 138
Query: 163 GQPRWPEGTQTLTYAFDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGF 222
PRW GT LTYAF P LDD K A AF++W+ V I F E TSY +++IKI F
Sbjct: 139 DMPRWQAGTTQLTYAFSPEPRLDDTFKSAIARAFSKWTPVVNIAFQETTSYETANIKILF 198
Query: 223 YSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVH 282
S +HGD FDG G L HAF+PTDGR H D E WV +GDVT+S +++A DLESV VH
Sbjct: 199 ASKNHGDPYPFDGPGGILGHAFAPTDGRCHFDADEYWVASGDVTKSPVTSAFDLESVAVH 258
Query: 283 EIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNP 329
EIGHLLGLGHSS AIMYP+I RT+KV L DDI+GI+ LYG NP
Sbjct: 259 EIGHLLGLGHSSDLRAIMYPSIPPRTRKVNLAQDDIDGIRKLYGINP 305
>UniRef100_Q84W77 Hypothetical protein At4g16640 [Arabidopsis thaliana]
Length = 364
Score = 273 bits (698), Expect = 6e-72
Identities = 154/346 (44%), Positives = 201/346 (57%), Gaps = 23/346 (6%)
Query: 27 ARFFPDP--SSMPAWNSSNAPPGAWDGYKNFTGCSRGKTYDGLSKLKNYFNHFGYIPNGP 84
AR P+ S+ A + W + G G+S+LK Y + FGY+ +G
Sbjct: 28 ARITPEAEQSTAKATQIIHVSNSTWHDFSRLVDVQIGSHVSGVSELKRYLHRFGYVNDGS 87
Query: 85 PSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKPRCGVADIINGTTSMNSGK 144
F+D FD LESA+ YQ+N L ITG LD +T+ + PRCGV+D
Sbjct: 88 EI-FSDVFDGPLESAISLYQENLGLPITGRLDTSTVTLMSLPRCGVSDT----------H 136
Query: 145 FNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSENLDDAT----KQVFANAFNQWS 200
++ HT AHY++F G+P+W TLTYA + LD T K VF AF+QWS
Sbjct: 137 MTINNDFLHTTAHYTYFNGKPKW--NRDTLTYAISKTHKLDYLTSEDVKTVFRRAFSQWS 194
Query: 201 KVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWV 260
V ++F E ++++D+KIGFY+GDHGDG FDGVLGTLAHAF+P +GRLHLD AE W+
Sbjct: 195 SVIPVSFEEVDDFTTADLKIGFYAGDHGDGLPFDGVLGTLAHAFAPENGRLHLDAAETWI 254
Query: 261 VNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEG 320
V+ D+ SS AVDLESV HEIGHLLGLGHSS E A+MYP++ RTKKV+L DD+ G
Sbjct: 255 VDDDLKGSS-EVAVDLESVATHEIGHLLGLGHSSQESAVMYPSLRPRTKKVDLTVDDVAG 313
Query: 321 IQMLYGSNPNFTGTTTATSRDRDSSFGGRHGVSLFLSLVFLGFGFL 366
+ LYG NP + S D + H FLS F+G+ L
Sbjct: 314 VLKLYGPNPKLRLDSLTQSEDSIKNGTVSH---RFLSGNFIGYVLL 356
>UniRef100_Q8LET7 Proteinase like protein [Arabidopsis thaliana]
Length = 364
Score = 273 bits (697), Expect = 7e-72
Identities = 149/322 (46%), Positives = 192/322 (59%), Gaps = 21/322 (6%)
Query: 49 WDGYKNFTGCSRGKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFN 108
W + G G+S+LK Y + FGY+ +G F+D FD LESA+ YQ+N
Sbjct: 52 WHDFSRLVDVQIGSHVSGVSELKRYLHRFGYVKDGSEI-FSDVFDGPLESAISLYQENLG 110
Query: 109 LNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWP 168
L ITG LD +T+ + PRCGV D ++ HT AHY++F G+P+W
Sbjct: 111 LPITGRLDTSTVTLMSLPRCGVXDT----------HMTINNDFLHTTAHYTYFNGKPKW- 159
Query: 169 EGTQTLTYAFDPSENLDDAT----KQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYS 224
TLTYA + LD T K VF AF+QWS V ++F E ++++D+KIGFY+
Sbjct: 160 -NRDTLTYAISKTHKLDYLTSEDVKTVFRRAFSQWSSVIPVSFEEVDDFTTADLKIGFYA 218
Query: 225 GDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEI 284
GDHGDG FDGVLGTLAHAF+P +GRLHLD AE W+V+ D+ SS AVDLESV HEI
Sbjct: 219 GDHGDGLPFDGVLGTLAHAFAPENGRLHLDAAETWIVDDDLKGSS-EVAVDLESVATHEI 277
Query: 285 GHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDS 344
GHLLGLGHSS E A+MYP++ RTKKV+L DD+ G+ LYG NP + S D
Sbjct: 278 GHLLGLGHSSQESAVMYPSLRPRTKKVDLTVDDVAGVLKLYGPNPKLRLDSLTQSEDSIK 337
Query: 345 SFGGRHGVSLFLSLVFLGFGFL 366
+ H FLS F+G+ L
Sbjct: 338 NGTVSH---RFLSGNFIGYVLL 356
>UniRef100_O23507 Proteinase like protein [Arabidopsis thaliana]
Length = 364
Score = 273 bits (697), Expect = 7e-72
Identities = 149/322 (46%), Positives = 193/322 (59%), Gaps = 21/322 (6%)
Query: 49 WDGYKNFTGCSRGKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFN 108
W + G G+S+LK Y + FGY+ +G F+D FD LESA+ YQ+N
Sbjct: 52 WHDFSRLVDVQIGSHVSGVSELKRYLHRFGYVNDGSEI-FSDVFDGPLESAISLYQENLG 110
Query: 109 LNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWP 168
L ITG LD +T+ + PRCGV+D ++ HT AHY++F G+P+W
Sbjct: 111 LPITGRLDTSTVTLMSLPRCGVSDT----------HMTINNDFLHTTAHYTYFNGKPKW- 159
Query: 169 EGTQTLTYAFDPSENLDDAT----KQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYS 224
TLTYA + LD T K VF AF+QWS V ++F E ++++D+KIGFY+
Sbjct: 160 -NRDTLTYAISKTHKLDYLTSEDVKTVFRRAFSQWSSVIPVSFEEVDDFTTADLKIGFYA 218
Query: 225 GDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEI 284
GDHGDG FDGVLGTLAHAF+P +GRLHLD AE W+V+ D+ SS AVDLESV HEI
Sbjct: 219 GDHGDGLPFDGVLGTLAHAFAPENGRLHLDAAETWIVDDDLKGSS-EVAVDLESVATHEI 277
Query: 285 GHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSRDRDS 344
GHLLGLGHSS E A+MYP++ RTKKV+L DD+ G+ LYG NP + S D
Sbjct: 278 GHLLGLGHSSQESAVMYPSLRPRTKKVDLTVDDVAGVLKLYGPNPKLRLDSLTQSEDSIK 337
Query: 345 SFGGRHGVSLFLSLVFLGFGFL 366
+ H FLS F+G+ L
Sbjct: 338 NGTVSH---RFLSGNFIGYVLL 356
>UniRef100_Q94LQ4 Putative metalloproteinase [Oryza sativa]
Length = 355
Score = 258 bits (660), Expect = 1e-67
Identities = 137/306 (44%), Positives = 183/306 (59%), Gaps = 19/306 (6%)
Query: 48 AWDGYKNFTGCSRGKTYDGLSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNF 107
AW +K RG GL++LK Y FGY+ P + TD FDE LE AVR YQ F
Sbjct: 35 AWHSFKQLLDAGRGSHVTGLAELKRYLARFGYMAK-PGRDTTDAFDEHLEVAVRRYQTRF 93
Query: 108 NLNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRW 167
+L +TG LD+AT++ I+ PRCGV D S V+ ++FF G+PRW
Sbjct: 94 SLPVTGRLDNATLDQIMSPRCGVGD---DDVERPVSVALSPGAQGGVVSRFTFFKGEPRW 150
Query: 168 PEGTQ--TLTYAFDPSENLD----DATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIG 221
L+YA P+ + A + VF AF +W++ + F E Y ++DIK+G
Sbjct: 151 TRSDPPIVLSYAVSPTATVGYLPPAAVRAVFQRAFARWARTIPVGFVETDDYEAADIKVG 210
Query: 222 FYSGDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVV 281
FY+G+HGDG FDG LG L HAFSP +GRLHLD +E W V+ DV + ++A+DLESV
Sbjct: 211 FYAGNHGDGVPFDGPLGILGHAFSPKNGRLHLDASEHWAVDFDV--DATASAIDLESVAT 268
Query: 282 HEIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQMLYGSNPNFT-------GT 334
HEIGH+LGLGHS+ A+MYP+I R KKV L DD+EG+Q LYGSNP F+ GT
Sbjct: 269 HEIGHVLGLGHSASPRAVMYPSIKPREKKVRLTVDDVEGVQALYGSNPQFSLSSLSEQGT 328
Query: 335 TTATSR 340
++++ R
Sbjct: 329 SSSSPR 334
>UniRef100_Q8GWW6 Putative metalloproteinase [Arabidopsis thaliana]
Length = 342
Score = 251 bits (642), Expect = 2e-65
Identities = 133/272 (48%), Positives = 172/272 (62%), Gaps = 25/272 (9%)
Query: 67 LSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKP 126
+ ++K + +GY+P S+ D + E A+ YQKN L ITG+ D T++ I+ P
Sbjct: 50 IPEIKRHLQQYGYLPQNKESD-----DVSFEQALVRYQKNLGLPITGKPDSDTLSQILLP 104
Query: 127 RCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQT-LTYAFDPSENLD 185
RCG D + T+ FHT Y +FPG+PRW LTYAF ENL
Sbjct: 105 RCGFPDDVEPKTAP-----------FHTGKKYVYFPGRPRWTRDVPLKLTYAFS-QENLT 152
Query: 186 DAT-----KQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTL 240
++VF AF +W+ V ++F E Y +DIKIGF++GDHGDGE FDGVLG L
Sbjct: 153 PYLAPTDIRRVFRRAFGKWASVIPVSFIETEDYVIADIKIGFFNGDHGDGEPFDGVLGVL 212
Query: 241 AHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIM 300
AH FSP +GRLHLDKAE W V+ D +SS+ AVDLESV VHEIGH+LGLGHSS+++A M
Sbjct: 213 AHTFSPENGRLHLDKAETWAVDFDEEKSSV--AVDLESVAVHEIGHVLGLGHSSVKDAAM 270
Query: 301 YPTISSRTKKVELESDDIEGIQMLYGSNPNFT 332
YPT+ R+KKV L DD+ G+Q LYG+NPNFT
Sbjct: 271 YPTLKPRSKKVNLNMDDVVGVQSLYGTNPNFT 302
>UniRef100_O65340 Metalloproteinase [Arabidopsis thaliana]
Length = 341
Score = 251 bits (641), Expect = 2e-65
Identities = 133/271 (49%), Positives = 171/271 (63%), Gaps = 24/271 (8%)
Query: 67 LSKLKNYFNHFGYIPNGPPSNFTDDFDEALESAVRTYQKNFNLNITGELDDATMNYIVKP 126
+ ++K + +GY+P S+ D + E A+ YQKN L ITG+ D T++ I+ P
Sbjct: 50 IPEIKRHLQQYGYLPQNKESD-----DVSFEQALVRYQKNLGLPITGKPDSDTLSQILLP 104
Query: 127 RCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQT-LTYAFDPSENLD 185
RCG D + T+ FHT Y +FPG+PRW LTYAF ENL
Sbjct: 105 RCGFPDDVEPKTAP-----------FHTGKKYVYFPGRPRWTRDVPLKLTYAFS-QENLT 152
Query: 186 DATK----QVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLA 241
+VF AF +W+ V ++F E Y +DIKIGF++GDHGDGE FDGVLG LA
Sbjct: 153 PYLAPTDIRVFRRAFGEWASVIPVSFIETEDYVIADIKIGFFNGDHGDGEPFDGVLGVLA 212
Query: 242 HAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMY 301
H FSP +GRLHLDKAE W V+ D +SS+ AVDLESV VHEIGH+LGLGHSS+++A MY
Sbjct: 213 HTFSPENGRLHLDKAETWAVDFDEEKSSV--AVDLESVAVHEIGHVLGLGHSSVKDAAMY 270
Query: 302 PTISSRTKKVELESDDIEGIQMLYGSNPNFT 332
PT+ R+KKV L DD+ G+Q LYG+NPNFT
Sbjct: 271 PTLKPRSKKVNLNMDDVVGVQSLYGTNPNFT 301
>UniRef100_Q9LEL9 Matrix metalloproteinase [Cucumis sativus]
Length = 320
Score = 229 bits (585), Expect = 7e-59
Identities = 139/329 (42%), Positives = 182/329 (55%), Gaps = 35/329 (10%)
Query: 12 IFYITLISITLTSVSARFFPDPS---------SMPAWNSSNAPPGAWDGYKNFTGCSRGK 62
IF TL+ ++L FP+P+ S NSSN+ KN GC G
Sbjct: 10 IFPFTLLFLSL-------FPNPNTSSPIILKHSSQNMNSSNSLMFL----KNLQGCHLGD 58
Query: 63 TYDGLSKLKNYFNHFGYIPNGPPSN----FTDDFDEALESAVRTYQKNFNLNITGELDDA 118
T G+ ++K Y FGYI + F D FD LESA++TYQ N NL +G LD
Sbjct: 59 TKQGIHQIKKYLQRFGYITTNIQKHSNPIFDDTFDHILESALKTYQTNHNLAPSGILDSN 118
Query: 119 TMNYIVKPRCGVADIING--TTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTY 176
T+ I PRCGV D+I T N N+ T+FH V+H++FF G +WP L+Y
Sbjct: 119 TIAQIAMPRCGVQDVIKNKKTKKRNQNFTNNGHTHFHKVSHFTFFEGNLKWPSSKLHLSY 178
Query: 177 AFDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGV 236
F P+ +D A K V + AF++WS T F+ Y +DIKI F G+HGD FDGV
Sbjct: 179 GFLPNYPID-AIKPV-SRAFSKWSLNTHFKFSHVADYRKADIKISFERGEHGDNAPFDGV 236
Query: 237 LGTLAHAFSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIE 296
G LAHA++PTDGRLH D + W V ++S D+E+V +HEIGH+LGL HS+IE
Sbjct: 237 GGVLAHAYAPTDGRLHFDGDDAWSV------GAISGYFDVETVALHEIGHILGLQHSTIE 290
Query: 297 EAIMYPTISSRTKKVELESDDIEGIQMLY 325
EAIM+P+I K L DDI GI+ LY
Sbjct: 291 EAIMFPSIPEGVTK-GLHGDDIAGIKALY 318
>UniRef100_Q94LQ0 Putative metalloproteinase [Oryza sativa]
Length = 261
Score = 160 bits (406), Expect = 4e-38
Identities = 85/190 (44%), Positives = 119/190 (61%), Gaps = 14/190 (7%)
Query: 155 VAHYSFFPGQPRWPEGTQ--TLTYAFDPSENLD----DATKQVFANAFNQWSKVTTITFT 208
V+ Y+F+ G+PRW + LTYA ++ + DA VF +AF +W++V ++F
Sbjct: 45 VSRYAFWTGKPRWTRHGRPMVLTYAVSHTDAVGYLPGDAVLAVFRSAFARWAEVIPVSFA 104
Query: 209 EATSY-----SSSDIKIGFY-SGDHGDGEAFDGVLGTLAHAFSPTDGRLHLDKAEDWVVN 262
E T+ + +DI++GFY +G+HGDG FDG L AHA P DGR+ D AE W V
Sbjct: 105 EITTEDDAAAAEADIRVGFYGAGEHGDGHPFDGPLNVYAHATGPEDGRIDFDAAERWAV- 163
Query: 263 GDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEGIQ 322
D+ + AVDLE+V HEIGH LGL HS+ E ++MYP + +R +KV L DD+EGIQ
Sbjct: 164 -DLAADASPAAVDLETVATHEIGHALGLDHSTSESSVMYPYVGTRERKVRLTVDDVEGIQ 222
Query: 323 MLYGSNPNFT 332
LYG NP+F+
Sbjct: 223 ELYGVNPSFS 232
>UniRef100_P91953 Hatching enzyme precursor [Hemicentrotus pulcherrimus]
Length = 591
Score = 154 bits (389), Expect = 4e-36
Identities = 103/277 (37%), Positives = 129/277 (46%), Gaps = 46/277 (16%)
Query: 69 KLKNYFNHFGYIPNGPPSNFTDDFDEA---LESAVRTYQKNFNLNITGELDDATMNYIVK 125
KLK FGY+P G F EA SA+ YQ+N +N TG LD T +
Sbjct: 106 KLKANLEQFGYVPLG------STFGEANINYTSAILEYQQNGGINQTGILDAETAALLDT 159
Query: 126 PRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDPSEN-- 183
PRCGV DI+ T G WP +TY+F N
Sbjct: 160 PRCGVPDILPYVT------------------------GGIAWPRNV-AVTYSFGTLSNDL 194
Query: 184 LDDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHA 243
A K AF W V+++TF E SS DI+I F S +HGDG +FDG G LAHA
Sbjct: 195 SQTAIKNELRRAFQVWDDVSSLTFREVVDSSSVDIRIKFGSYEHGDGISFDGQGGVLAHA 254
Query: 244 FSPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPT 303
F P +G H D +E W + ++ +L V HE GH LGL HS ++ A+MYP
Sbjct: 255 FLPRNGDAHFDDSERWTI-------GTNSGTNLFQVAAHEFGHSLGLYHSDVQSALMYPY 307
Query: 304 ISSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSR 340
L+ DDI GI LYG N TG+TT T+R
Sbjct: 308 YRGYNPNFNLDRDDIAGITSLYGRN---TGSTTTTTR 341
>UniRef100_O60882 Matrix metalloproteinase-20 precursor [Homo sapiens]
Length = 483
Score = 149 bits (377), Expect = 9e-35
Identities = 92/242 (38%), Positives = 119/242 (49%), Gaps = 37/242 (15%)
Query: 100 VRTYQKNFNLNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYS 159
++ Q F L +TG+LD TMN I KPRCGV D VA+Y
Sbjct: 72 IKELQAFFGLQVTGKLDQTTMNVIKKPRCGVPD----------------------VANYR 109
Query: 160 FFPGQPRWPEGTQTLTYA-FDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDI 218
FPG+P+W + T T + + PS + + K V A WS ++F S +DI
Sbjct: 110 LFPGEPKWKKNTLTYRISKYTPSMSSVEVDKAV-EMALQAWSSAVPLSFVRINS-GEADI 167
Query: 219 KIGFYSGDHGDGEAFDGVLGTLAHAFSPTD---GRLHLDKAEDWVVNGDVTESSLSNAVD 275
I F +GDHGD FDG GTLAHAF+P + G H D AE W + +N +
Sbjct: 168 MISFENGDHGDSYPFDGPRGTLAHAFAPGEGLGGDTHFDNAEKWTMG--------TNGFN 219
Query: 276 LESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTK-KVELESDDIEGIQMLYGSNPNFTGT 334
L +V HE GH LGL HS+ A+MYPT + L DD++GIQ LYG F G
Sbjct: 220 LFTVAAHEFGHALGLAHSTDPSALMYPTYKYKNPYGFHLPKDDVKGIQALYGPRKVFLGK 279
Query: 335 TT 336
T
Sbjct: 280 PT 281
>UniRef100_P57748 Matrix metalloproteinase-20 precursor [Mus musculus]
Length = 482
Score = 149 bits (375), Expect = 2e-34
Identities = 91/242 (37%), Positives = 120/242 (48%), Gaps = 37/242 (15%)
Query: 100 VRTYQKNFNLNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYS 159
++ Q F L +TG+LD TMN I KPRCGV D VA+Y
Sbjct: 71 IKELQIFFGLKVTGKLDQNTMNVIKKPRCGVPD----------------------VANYR 108
Query: 160 FFPGQPRWPEGTQTLTYA-FDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDI 218
FPG+P+W + T + + PS + + K + A + WS + F S +DI
Sbjct: 109 LFPGEPKWKKNILTYRISKYTPSMSPTEVDKAI-QMALHAWSTAVPLNFVRINS-GEADI 166
Query: 219 KIGFYSGDHGDGEAFDGVLGTLAHAFSPTD---GRLHLDKAEDWVVNGDVTESSLSNAVD 275
I F +GDHGD FDG GTLAHAF+P + G H D AE W + +N +
Sbjct: 167 MISFETGDHGDSYPFDGPRGTLAHAFAPGEGLGGDTHFDNAEKWTMG--------TNGFN 218
Query: 276 LESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTK-KVELESDDIEGIQMLYGSNPNFTGT 334
L +V HE GH LGLGHS+ A+MYPT + + L DD++GIQ LYG F G
Sbjct: 219 LFTVAAHEFGHALGLGHSTDPSALMYPTYKYQNPYRFHLPKDDVKGIQALYGPRKIFPGK 278
Query: 335 TT 336
T
Sbjct: 279 PT 280
>UniRef100_UPI000036F042 UPI000036F042 UniRef100 entry
Length = 483
Score = 147 bits (372), Expect = 3e-34
Identities = 91/242 (37%), Positives = 118/242 (48%), Gaps = 37/242 (15%)
Query: 100 VRTYQKNFNLNITGELDDATMNYIVKPRCGVADIINGTTSMNSGKFNSSSTNFHTVAHYS 159
++ Q F L +TG+LD TMN I KPRCGV D VA+Y
Sbjct: 72 IKELQAFFGLQVTGKLDQTTMNVIKKPRCGVPD----------------------VANYR 109
Query: 160 FFPGQPRWPEGTQTLTYA-FDPSENLDDATKQVFANAFNQWSKVTTITFTEATSYSSSDI 218
FPG+P+W + T + + PS + + K V A WS ++F S +DI
Sbjct: 110 LFPGEPKWKKNILTYRISKYTPSMSSVEVDKAV-EMALQAWSSAVPLSFVRINS-GEADI 167
Query: 219 KIGFYSGDHGDGEAFDGVLGTLAHAFSPTD---GRLHLDKAEDWVVNGDVTESSLSNAVD 275
I F +GDHGD FDG GTLAHAF+P + G H D AE W + +N +
Sbjct: 168 MISFENGDHGDSYPFDGPRGTLAHAFAPGEGLGGDTHFDNAEKWTMG--------TNGFN 219
Query: 276 LESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTK-KVELESDDIEGIQMLYGSNPNFTGT 334
L +V HE GH LGL HS+ A+MYPT + L DD++GIQ LYG F G
Sbjct: 220 LFTVAAHEFGHALGLAHSTDPSALMYPTYKYKNPYGFHLPKDDVKGIQALYGPRKAFLGK 279
Query: 335 TT 336
T
Sbjct: 280 PT 281
>UniRef100_Q94LQ1 Putative metalloproteinase [Oryza sativa]
Length = 289
Score = 147 bits (371), Expect = 5e-34
Identities = 82/192 (42%), Positives = 109/192 (56%), Gaps = 19/192 (9%)
Query: 158 YSFFPGQPRWPEGTQTLTYAFDPSENLD----DATKQVFANAFNQWSKVTTITFTEATSY 213
++FFPG+PRW + LTYA P+ D A + +AF +W+ V + F EA Y
Sbjct: 45 FTFFPGKPRWTRPDRVLTYAVSPTATADHLPPSAVRAALRSAFARWADVIPMRFLEAERY 104
Query: 214 SSSDIKIGFYSGDHG------------DGEAFDGVLGTLAHAFSPT-DGRLHLDKAEDWV 260
++DIK+GFY G D + D G LAH+ P G++HL A W
Sbjct: 105 DAADIKVGFYLYTDGRCDGCACIDSDDDDDDGDDCEGVLAHSSMPEKSGQIHLHAAHRWT 164
Query: 261 VNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTISSRTKKVELESDDIEG 320
VN + L AVDLESV HEIGH+LGL HSS ++MYP IS R +KV L +DD+ G
Sbjct: 165 VNLAADTAPL--AVDLESVAAHEIGHVLGLDHSSSRSSMMYPFISCRERKVRLTTDDVHG 222
Query: 321 IQMLYGSNPNFT 332
IQ LYG+NP+F+
Sbjct: 223 IQELYGANPHFS 234
>UniRef100_P22757 Hatching enzyme precursor [Paracentrotus lividus]
Length = 587
Score = 146 bits (369), Expect = 8e-34
Identities = 101/276 (36%), Positives = 132/276 (47%), Gaps = 44/276 (15%)
Query: 70 LKNYFNHFGYIPNGPPSNFTDDFDEA---LESAVRTYQKNFNLNITGELDDATMNYIVKP 126
LK + FGY P G F EA SA+ +Q++ +N TG LD T + P
Sbjct: 104 LKAHLEKFGYTPPG------STFGEANLNYTSAILDFQEHGGINQTGILDADTAELLSTP 157
Query: 127 RCGVADIINGTTSMNSGKFNSSSTNFHTVAHYSFFPGQPRWPEGTQTLTYAFDP--SENL 184
RCGV D++ TS + N Q +TY+F S+
Sbjct: 158 RCGVPDVLPFVTSSITWSRN-------------------------QPVTYSFGALTSDLN 192
Query: 185 DDATKQVFANAFNQWSKVTTITFTEATSYSSSDIKIGFYSGDHGDGEAFDGVLGTLAHAF 244
+ K AF W V+ ++F E +S DI+I F S DHGDG +FDG G LAHAF
Sbjct: 193 QNDVKDEIRRAFRVWDDVSGLSFREVPDTTSVDIRIKFGSYDHGDGISFDGRGGVLAHAF 252
Query: 245 SPTDGRLHLDKAEDWVVNGDVTESSLSNAVDLESVVVHEIGHLLGLGHSSIEEAIMYPTI 304
P +G H D +E W TE + S +L V HE GH LGL HS++ A+MYP
Sbjct: 253 LPRNGDAHFDDSETW------TEGTRS-GTNLFQVAAHEFGHSLGLYHSTVRSALMYPYY 305
Query: 305 SSRTKKVELESDDIEGIQMLYGSNPNFTGTTTATSR 340
L++DDI GI+ LYGSN +GTTT T R
Sbjct: 306 QGYVPNFRLDNDDIAGIRSLYGSNSG-SGTTTTTRR 340
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 671,161,863
Number of Sequences: 2790947
Number of extensions: 30348273
Number of successful extensions: 78260
Number of sequences better than 10.0: 645
Number of HSP's better than 10.0 without gapping: 507
Number of HSP's successfully gapped in prelim test: 138
Number of HSP's that attempted gapping in prelim test: 76134
Number of HSP's gapped (non-prelim): 812
length of query: 368
length of database: 848,049,833
effective HSP length: 129
effective length of query: 239
effective length of database: 488,017,670
effective search space: 116636223130
effective search space used: 116636223130
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144345.3


