

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144343.7 - phase: 0
(480 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
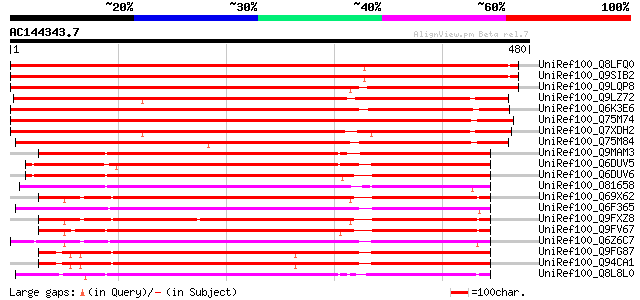
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LFQ0 Putative fatty acid elongase [Arabidopsis thali... 645 0.0
UniRef100_Q9SIB2 Putative fatty acid elongase [Arabidopsis thali... 644 0.0
UniRef100_Q9LQP8 Hypothetical protein At1g07720; F24B9.18 [Arabi... 639 0.0
UniRef100_Q9LZ72 Putative fatty acid elongase [Arabidopsis thali... 498 e-139
UniRef100_Q6K3E6 Putative fatty acid elongase 3-ketoacyl-CoA syn... 485 e-135
UniRef100_Q75M74 Putative fatty acid elongase [Oryza sativa] 484 e-135
UniRef100_Q7XDH2 Putative fatty acid elongase 3-ketoacyl-CoA syn... 483 e-135
UniRef100_Q75M84 Putative fatty acid elongase [Oryza sativa] 407 e-112
UniRef100_Q9MAM3 T25K16.11 [Arabidopsis thaliana] 355 2e-96
UniRef100_Q6DUV5 Fatty acid elongase 3-ketoacyl-CoA synthase [Br... 353 5e-96
UniRef100_Q6DUV6 Fatty acid elongase 3-ketoacyl-CoA synthase [Br... 348 1e-94
UniRef100_O81658 Senescence-associated protein 15 [Hemerocallis ... 345 2e-93
UniRef100_Q69X62 Putative beta-ketoacyl-CoA synthase [Oryza sativa] 334 3e-90
UniRef100_Q6F365 Putative beta-ketoacyl synthase [Oryza sativa] 333 5e-90
UniRef100_Q9FXZ8 Putative fatty acid elongase [Zea mays] 330 7e-89
UniRef100_Q9FV67 Fatty acid elongase 1-like protein [Limnanthes ... 328 3e-88
UniRef100_Q6Z6C7 Putative beta-ketoacyl-CoA-synthase [Oryza sativa] 327 4e-88
UniRef100_Q9FG87 Beta-ketoacyl-CoA synthase [Arabidopsis thaliana] 324 4e-87
UniRef100_Q94CA1 Putative beta-ketoacyl-CoA synthase [Arabidopsi... 324 4e-87
UniRef100_Q8L8L0 Fatty acid condensing enzyme CUT1, putative [Ar... 323 7e-87
>UniRef100_Q8LFQ0 Putative fatty acid elongase [Arabidopsis thaliana]
Length = 476
Score = 645 bits (1664), Expect = 0.0
Identities = 308/472 (65%), Positives = 383/472 (80%), Gaps = 3/472 (0%)
Query: 1 MEFLTLLFIFTTMHLCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNE 60
M+ L L F +L F IWK D ++D++CYIL+YQC+KP+D+R + + G+II N+
Sbjct: 1 MDLLFLFFSLLLSYLFFKIWKLIDSKQDKDCYILDYQCHKPTDDRMVSTQFSGEIIYRNQ 60
Query: 61 SIGLDECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNK 120
++GL E KF LKA+V+SGIGE+TYAPR EGR P+L+DG+ EMEEF DSI KLL +
Sbjct: 61 NLGLTEYKFLLKAIVSSGIGEQTYAPRLVFEGREERPSLQDGISEMEEFYVDSIGKLLER 120
Query: 121 SGISPSEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQN 180
+ ISP +IDILVVNVS+ SS PSL SRIINHYKMR+D+K +NL+GMGCSASLIS+D+++N
Sbjct: 121 NQISPKDIDILVVNVSMLSSTPSLASRIINHYKMRDDVKVFNLTGMGCSASLISVDIVKN 180
Query: 181 IFKSEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKL 240
IFKS NK AL+ TSESLS NWYSG NRSMILANCLFR+GGCAILLTNKRSL+ KA+ KL
Sbjct: 181 IFKSYANKLALVATSESLSPNWYSGNNRSMILANCLFRSGGCAILLTNKRSLRKKAMFKL 240
Query: 241 KCLVRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARE 300
KC+VRTHHGAR++++NCC Q EDEQG++GFYL K+LPKAATRAF DNL+V++PKILP E
Sbjct: 241 KCMVRTHHGAREESYNCCIQAEDEQGRVGFYLGKNLPKAATRAFVDNLKVITPKILPVTE 300
Query: 301 LLRYMLMSLIKKVIKYFAPKSAARWF--GTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGM 358
L+R+ML LIKK+ P + GT K+ +NFKTG++HF +HTGGKAVIDGIG
Sbjct: 301 LIRFMLKLLIKKIKIRQNPSXGSTNLPPGTPLKAGINFKTGIEHFCIHTGGKAVIDGIGH 360
Query: 359 SLDLNDYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSC 418
SLDLN+YD+EPARM+LHRFGNTSASSLWYVL YMEAKKRLK+G+RV MIS GAGFKCNSC
Sbjct: 361 SLDLNEYDIEPARMTLHRFGNTSASSLWYVLAYMEAKKRLKRGDRVFMISFGAGFKCNSC 420
Query: 419 YWEVMRDVVVGDDRNNVWDDCIDYYPPHSLANPFMEKYGWINNIQDPSTFKL 470
WEV+RD+ G+ + NVW+ CID YPP S+ NP++EK+GWI + +DP TFK+
Sbjct: 421 VWEVVRDLTGGESKGNVWNHCIDDYPPKSILNPYLEKFGWIQD-EDPDTFKV 471
>UniRef100_Q9SIB2 Putative fatty acid elongase [Arabidopsis thaliana]
Length = 476
Score = 644 bits (1660), Expect = 0.0
Identities = 307/472 (65%), Positives = 383/472 (81%), Gaps = 3/472 (0%)
Query: 1 MEFLTLLFIFTTMHLCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNE 60
M+ L L F +L F IWK D ++D++CYIL+YQC+KP+D+R + + G+II N+
Sbjct: 1 MDLLFLFFSLLLSYLFFKIWKLIDSKQDKDCYILDYQCHKPTDDRMVSTQFSGEIIYRNQ 60
Query: 61 SIGLDECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNK 120
++GL E KF LKA+V+SGIGE+TYAPR EGR P+L+DG+ EMEEF DSI KLL +
Sbjct: 61 NLGLTEYKFLLKAIVSSGIGEQTYAPRLVFEGREERPSLQDGISEMEEFYVDSIGKLLER 120
Query: 121 SGISPSEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQN 180
+ ISP +IDILVVNVS+ SS PSL SRIINHYKMR+D+K +NL+GMGCSASLIS+D+++N
Sbjct: 121 NQISPKDIDILVVNVSMLSSTPSLASRIINHYKMRDDVKVFNLTGMGCSASLISVDIVKN 180
Query: 181 IFKSEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKL 240
IFKS NK AL+ TSESLS NWYSG NRSMILANCLFR+GGCAILLTNKRSL+ KA+ KL
Sbjct: 181 IFKSYANKLALVATSESLSPNWYSGNNRSMILANCLFRSGGCAILLTNKRSLRKKAMFKL 240
Query: 241 KCLVRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARE 300
KC+VRTHHGAR++++NCC Q EDEQG++GFYL K+LPKAATRAF +NL+V++PKILP E
Sbjct: 241 KCMVRTHHGAREESYNCCIQAEDEQGRVGFYLGKNLPKAATRAFVENLKVITPKILPVTE 300
Query: 301 LLRYMLMSLIKKVIKYFAPKSAARWF--GTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGM 358
L+R+ML LIKK+ P + GT K+ +NFKTG++HF +HTGGKAVIDGIG
Sbjct: 301 LIRFMLKLLIKKIKIRQNPSKGSTNLPPGTPLKAGINFKTGIEHFCIHTGGKAVIDGIGH 360
Query: 359 SLDLNDYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSC 418
SLDLN+YD+EPARM+LHRFGNTSASSLWYVL YMEAKKRLK+G+RV MIS GAGFKCNSC
Sbjct: 361 SLDLNEYDIEPARMTLHRFGNTSASSLWYVLAYMEAKKRLKRGDRVFMISFGAGFKCNSC 420
Query: 419 YWEVMRDVVVGDDRNNVWDDCIDYYPPHSLANPFMEKYGWINNIQDPSTFKL 470
WEV+RD+ G+ + NVW+ CID YPP S+ NP++EK+GWI + +DP TFK+
Sbjct: 421 VWEVVRDLTGGESKGNVWNHCIDDYPPKSILNPYLEKFGWIQD-EDPDTFKV 471
>UniRef100_Q9LQP8 Hypothetical protein At1g07720; F24B9.18 [Arabidopsis thaliana]
Length = 478
Score = 639 bits (1648), Expect = 0.0
Identities = 310/480 (64%), Positives = 379/480 (78%), Gaps = 17/480 (3%)
Query: 1 MEFLTLLFIFTTMHLCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNE 60
M+ L +L +L F IWK+ D +RDQ CYIL+YQC+KPSD+R + + G II N+
Sbjct: 1 MDLLVMLLSLLVSYLIFKIWKRIDSKRDQNCYILDYQCHKPSDDRMVNTQFSGDIILRNK 60
Query: 61 SIGLDECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNK 120
+ L+E KF LKA+V+SGIGE+TYAPR F EGR PTL+DG+ EMEEF D+I K+L +
Sbjct: 61 HLRLNEYKFLLKAIVSSGIGEQTYAPRLFFEGREQRPTLQDGLSEMEEFYIDTIEKVLKR 120
Query: 121 SGISPSEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQN 180
+ ISPSEIDILVVNVS+ +S PSL++RIINHYKMREDIK +NL+ MGCSAS+IS+D+++N
Sbjct: 121 NKISPSEIDILVVNVSMLNSTPSLSARIINHYKMREDIKVFNLTAMGCSASVISIDIVKN 180
Query: 181 IFKSEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKL 240
IFK+ KNK AL++TSESLS NWYSG NRSMILANCLFR+GGCA+LLTNKRSL +A+ KL
Sbjct: 181 IFKTYKNKLALVVTSESLSPNWYSGNNRSMILANCLFRSGGCAVLLTNKRSLSRRAMFKL 240
Query: 241 KCLVRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARE 300
+CLVRTHHGARDD+ N C QKEDE G +G +L K LPKAATRAF DNL+V++PKILP E
Sbjct: 241 RCLVRTHHGARDDSFNACVQKEDELGHIGVHLDKTLPKAATRAFIDNLKVITPKILPVTE 300
Query: 301 LLRYMLMSLIKKV----------IKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGK 350
LLR+ML L+KK+ + APK+ K+ +NFKTG+DHF +HTGGK
Sbjct: 301 LLRFMLCLLLKKLRSSPSKGSTNVTQAAPKAGV-------KAGINFKTGIDHFCIHTGGK 353
Query: 351 AVIDGIGMSLDLNDYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLG 410
AVID IG SLDLN+YDLEPARM+LHRFGNTSASSLWYVLGYMEAKKRLK+G+RV MIS G
Sbjct: 354 AVIDAIGYSLDLNEYDLEPARMTLHRFGNTSASSLWYVLGYMEAKKRLKRGDRVFMISFG 413
Query: 411 AGFKCNSCYWEVMRDVVVGDDRNNVWDDCIDYYPPHSLANPFMEKYGWINNIQDPSTFKL 470
AGFKCNSC WEV+RD+ VG+ NVW+ CI+ YPP S+ NPF EKYGWI+ +DP TFK+
Sbjct: 414 AGFKCNSCVWEVVRDLNVGEAVGNVWNHCINQYPPKSILNPFFEKYGWIHEEEDPDTFKM 473
>UniRef100_Q9LZ72 Putative fatty acid elongase [Arabidopsis thaliana]
Length = 464
Score = 498 bits (1282), Expect = e-139
Identities = 237/461 (51%), Positives = 341/461 (73%), Gaps = 14/461 (3%)
Query: 4 LTLLFIFTTMHLCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNESIG 63
L+ L + +T+ + ++ F +R + CY+L+Y+CYK +ERKL + C K++ N+++G
Sbjct: 6 LSSLLLLSTLFVFYIFKFVFKRRNQRNCYMLHYECYKGMEERKLDTETCAKVVQRNKNLG 65
Query: 64 LDECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKS-- 121
L+E +F L+ + +SGIGEETY PRN +EGR +PTL D EM+E D++ KL +K+
Sbjct: 66 LEEYRFLLRTMASSGIGEETYGPRNVLEGREDSPTLLDAHSEMDEIMFDTLDKLFHKTKG 125
Query: 122 GISPSEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNI 181
ISPS+IDILVVNVSLF+ PSLTSR+IN YKMREDIK+YNLSG+GCSAS+IS+D++Q +
Sbjct: 126 SISPSDIDILVVNVSLFAPSPSLTSRVINRYKMREDIKSYNLSGLGCSASVISIDIVQRM 185
Query: 182 FKSEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLK 241
F++ +N AL++++E++ +WY G +RSM+L+NCLFR GG ++LLTN K++A++KL
Sbjct: 186 FETRENALALVVSTETMGPHWYCGKDRSMMLSNCLFRAGGSSVLLTNAARFKNQALMKLV 245
Query: 242 CLVRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPAREL 301
+VR H G+ D+A++CC Q ED G GF L K L KAA RA T NL+VL P++LP +EL
Sbjct: 246 TVVRAHVGSDDEAYSCCIQMEDRDGHPGFLLTKYLKKAAARALTKNLQVLLPRVLPVKEL 305
Query: 302 LRYMLMSLIKKVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLD 361
+RY ++ +K+ +++A+ SS +N KTG+ HF +H GG+A+I+G+G SL
Sbjct: 306 IRYAIVRALKR-------RTSAKREPASSGIGLNLKTGLQHFCIHPGGRAIIEGVGKSLG 358
Query: 362 LNDYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWE 421
L ++D+EPARM+LHRFGNTS+ LWYVLGYMEAK RLKKGE++LM+S+GAGF+ N+C WE
Sbjct: 359 LTEFDIEPARMALHRFGNTSSGGLWYVLGYMEAKNRLKKGEKILMMSMGAGFESNNCVWE 418
Query: 422 VMRDVVVGDDRNNVWDDCIDYYPPHS-LANPFMEKYGWINN 461
V++D+ D NVW+D +D YP S + NPF+EKY WIN+
Sbjct: 419 VLKDL----DDKNVWEDSVDRYPELSRIPNPFVEKYDWIND 455
>UniRef100_Q6K3E6 Putative fatty acid elongase 3-ketoacyl-CoA synthase 1 [Oryza
sativa]
Length = 463
Score = 485 bits (1248), Expect = e-135
Identities = 231/463 (49%), Positives = 325/463 (69%), Gaps = 13/463 (2%)
Query: 1 MEFLTLLFIFTTMHLC-FLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNN 59
M+ L L+ + H ++ W+ +RR CY+L+Y C+KPSD+RK+ + G II N
Sbjct: 1 MDLLPLVTLLLLAHAAAWVAWQAAARRRRATCYLLDYACHKPSDDRKVTTELAGAIIERN 60
Query: 60 ESIGLDECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLN 119
+ +GL E +F LK +VNSGIGE TY+PRN ++ R PTL D ++EM++F +D++ +L
Sbjct: 61 KRLGLPEYRFLLKVIVNSGIGEHTYSPRNVLDAREDCPTLRDALDEMDDFFDDAVAAVLA 120
Query: 120 KSGISPSEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQ 179
++ +SP ++D+LV+NV FS PSL R++ + +R+D+ AYNLSGMGCSA L+S+D+ +
Sbjct: 121 RAAVSPRDVDLLVINVGSFSPSPSLADRVVRRFGLRDDVMAYNLSGMGCSAGLVSVDLAR 180
Query: 180 NIFKSEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILK 239
N+ + AL+LTSES + NWY+G ++SM+L NCLFR GG A LLTN + + +A ++
Sbjct: 181 NVMLTRPRTMALVLTSESCAPNWYTGTDKSMMLGNCLFRCGGAAALLTNDPAFRSRAKME 240
Query: 240 LKCLVRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPAR 299
L+CLVR H GA DDAH +ED G+LG L+K LPKAA RAFT+NL+ L+P+ILPA
Sbjct: 241 LRCLVRAHIGAHDDAHAAAVHREDADGRLGVSLSKALPKAAVRAFTENLQRLAPRILPAG 300
Query: 300 ELLRYMLMSLIKKVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMS 359
EL R+ L++K+++ A AA + +NFKTGVDHF LH GG AVI+ + S
Sbjct: 301 ELARFAARLLLRKLLRRKAAAGAA--------AKINFKTGVDHFCLHPGGTAVIEAVRKS 352
Query: 360 LDLNDYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCY 419
L L+ YD+EPARM+LHR+GNTSASSLWYVL YMEAK+RL G+RVLM++ G+GFKCNS Y
Sbjct: 353 LGLDSYDVEPARMALHRWGNTSASSLWYVLSYMEAKRRLNAGDRVLMVTFGSGFKCNSSY 412
Query: 420 WEVMRDVVVGDDRNNVWDDCIDYYPPHSLANPFMEKYGWINNI 462
W V +D+ W+DCI YPP +L NP+MEK+GW+N++
Sbjct: 413 WVVTKDLADA----GAWEDCIHDYPPANLVNPYMEKFGWVNDL 451
>UniRef100_Q75M74 Putative fatty acid elongase [Oryza sativa]
Length = 467
Score = 484 bits (1245), Expect = e-135
Identities = 237/468 (50%), Positives = 322/468 (68%), Gaps = 6/468 (1%)
Query: 1 MEFLTLLFIFTTMH-LCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNN 59
ME L L+ + H + +L W +RR CY+L+Y C+KPSD+RK+ + G +I +
Sbjct: 1 MELLALVTVLLLAHAVAYLAWTAAARRRQSRCYLLDYVCHKPSDDRKVSTEAAGAVIERS 60
Query: 60 ESIGLDECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLN 119
+ + L E +F L+ +V SGIGEETYAPRN ++GR PT D + EME+F DSI +L
Sbjct: 61 KRLSLPEYRFLLRVIVRSGIGEETYAPRNVLDGREGEPTHGDSLGEMEDFFGDSIAELFA 120
Query: 120 KSGISPSEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQ 179
++G P ++D+LVVN S+FS PSL S I++ Y MRED+ AY+L+GMGCSA LISLD+ +
Sbjct: 121 RTGFGPRDVDVLVVNASMFSPDPSLASMIVHRYGMREDVAAYSLAGMGCSAGLISLDLAR 180
Query: 180 NIFKSEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILK 239
N + AL++++ES++ NWY+G ++SM+LANCLFR GG ++L+TN L+ +A ++
Sbjct: 181 NTLATRPRALALVVSTESIAPNWYTGTDKSMMLANCLFRCGGASVLVTNDPVLRGRAKME 240
Query: 240 LKCLVRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPAR 299
L CLVR + A DDAH C Q+ED+ G +G L+K LPKAA RAF NLR L+P+ILP
Sbjct: 241 LGCLVRANIAANDDAHACALQREDDDGTVGISLSKALPKAAVRAFAANLRRLAPRILPIT 300
Query: 300 ELLRYMLMSLI-KKVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGM 358
EL R+ LI KK+++ A + A +NFKTGVDHF LH GG AVI+ +
Sbjct: 301 ELARFAAQLLITKKLLRRRATAATATKHTGGDGPRINFKTGVDHFCLHPGGTAVIEAVKR 360
Query: 359 SLDLNDYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSC 418
SL L+D D+EPARM+LHR+GNTSASSLWYVL YMEAK RL++G++VLM++ G+GFKCNSC
Sbjct: 361 SLGLDDDDVEPARMTLHRWGNTSASSLWYVLSYMEAKGRLRRGDKVLMVTFGSGFKCNSC 420
Query: 419 YWEVMRDVVVGDDRNNVWDDCIDYYPPHSLANPFMEKYGWINNIQDPS 466
WEV D+ W DCID YPP + ANP+MEKY WIN++ S
Sbjct: 421 VWEVTGDMA----DKGAWADCIDAYPPENTANPYMEKYSWINDVDGDS 464
>UniRef100_Q7XDH2 Putative fatty acid elongase 3-ketoacyl-CoA synthase [Oryza sativa]
Length = 465
Score = 483 bits (1244), Expect = e-135
Identities = 242/478 (50%), Positives = 328/478 (67%), Gaps = 28/478 (5%)
Query: 1 MEFLTLLFIFTTMHLCFLIWKKFDQR-RDQECYILNYQCYKPSDERKLGSVQCGKIIGNN 59
ME T+L + + ++ + R R + CY+L+Y CYK +D+RKL + CG+II N
Sbjct: 1 MEVATMLTLTLLAYSAAMLARLLVARSRRRRCYLLDYVCYKATDDRKLPTDLCGEIIQRN 60
Query: 60 ESIGLDECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPT-LEDGVEEMEEFCNDSITKLL 118
+ +GL+E KF LK +VNSGIGEETY PRN I G P L +G+EEM+E + + +L
Sbjct: 61 KLLGLEEYKFLLKVIVNSGIGEETYGPRNIIGGGDARPDRLAEGMEEMDETFHAVLDELF 120
Query: 119 NKS------GISPSEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASL 172
+S G+ P+++D+LVVNVS+FS PSL++R++ Y +RED+K YNL+GMGCSA+L
Sbjct: 121 ARSSAAGGGGVRPADVDLLVVNVSMFSPAPSLSARVVRRYNLREDVKVYNLTGMGCSATL 180
Query: 173 ISLDVIQNIFKSEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSL 232
I+LD++ N ++ NK AL++TSES++ NWY+G RS +L NCLFR+GGCA LTN
Sbjct: 181 IALDLVNNFLRTHANKVALVMTSESIAPNWYAGNKRSFMLGNCLFRSGGCAYFLTNDPRH 240
Query: 233 KHKAILKLKCLVRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLS 292
+ A L+L+ LVRTH GA DDA++C Q ED+ G+ GF+L KDLP+AA AF NLR+L+
Sbjct: 241 RRHAKLRLRHLVRTHTGASDDAYSCALQMEDDAGRPGFHLGKDLPRAAVHAFVKNLRLLA 300
Query: 293 PKILPARELLRYMLMSLIKKVIKYFAPKSAARWFGTSSKSP------VNFKTGVDHFVLH 346
P++LP ELLR + + S+ R G SP + K GVDHF +H
Sbjct: 301 PRVLPLPELLRLAFATFL----------SSGRRSGGKKTSPSQQPLTIRMKAGVDHFCVH 350
Query: 347 TGGKAVIDGIGMSLDLNDYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLM 406
TGG AVIDG+G L L ++DLEP+RM+LHRFGNTSASS+WYVLGYMEAK+RL+ G+RVLM
Sbjct: 351 TGGAAVIDGVGKGLTLTEHDLEPSRMTLHRFGNTSASSVWYVLGYMEAKRRLRPGDRVLM 410
Query: 407 ISLGAGFKCNSCYWEVMRDVVVGDDRNNVWDDCIDYYPPHSLANPFMEKYGWINNIQD 464
++ GAGFKCNSC W V + V VW DCID+YPP LANPFMEKYG++ ++ +
Sbjct: 411 LTFGAGFKCNSCVWTVEKPV----SDAGVWKDCIDHYPPKELANPFMEKYGFVKDMMN 464
>UniRef100_Q75M84 Putative fatty acid elongase [Oryza sativa]
Length = 466
Score = 407 bits (1045), Expect = e-112
Identities = 201/458 (43%), Positives = 299/458 (64%), Gaps = 13/458 (2%)
Query: 6 LLFIFTTMHLCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNESIGLD 65
+LF+ + + + W +RR CY+L+Y CYKP+D+ L + I+ NE +G+
Sbjct: 10 VLFLPPPLAVAHMAWTAASRRRGMRCYLLDYVCYKPADDLTLTTELACAIVQRNERLGIP 69
Query: 66 ECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISP 125
E +F ++ + +G+G+ TYAPRN ++GR D V+EM+ + ++ +LL ++G+
Sbjct: 70 EFRFLVRLISRTGLGDRTYAPRNLLDGREELAAQRDSVDEMDACFDGAVPELLARTGLRA 129
Query: 126 SEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIF--K 183
++D+LVVNV+ F P L SR++ Y MRED+ AYNLSGMGCSA+L+++DV +N +
Sbjct: 130 RDVDVLVVNVNGFFPEPCLASRVVRRYGMREDVAAYNLSGMGCSATLVAVDVGRNAMRAR 189
Query: 184 SEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCL 243
S + AL++++ESL+ +WY+G R+M+LA CLFR GG A+LL+N + + +A ++L+ L
Sbjct: 190 SPRPVVALVVSTESLAPHWYAGKERTMMLAQCLFRCGGAAVLLSNDPAHRGRAKMELRRL 249
Query: 244 VRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLR 303
VR+ A DDA++C Q+ED+ G G ++K LPKAA RAF NL+ L P++LPA E+ R
Sbjct: 250 VRSTTAASDDAYSCIMQREDDDGLRGVSISKALPKAALRAFAANLQRLLPRVLPAMEIAR 309
Query: 304 YMLMSLIKKVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLN 363
+ +++ R +K +N K GVDH LH GG AVID + S L
Sbjct: 310 LAADLAWQNLLQ-------RRRHRGQTKLKINLKAGVDHICLHAGGVAVIDAVKKSFGLE 362
Query: 364 DYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVM 423
+ D+EP RM+LHR+GNTSASS+WYVL YMEAK RL++G++VLM++ G+GFKCNSC WEV
Sbjct: 363 ERDVEPLRMTLHRWGNTSASSVWYVLSYMEAKGRLRRGDKVLMVTFGSGFKCNSCVWEVA 422
Query: 424 RDVVVGDDRNNVWDDCIDYYPPHSLANPFMEKYGWINN 461
D+ W DCID YPP S +PF+EK+ WIN+
Sbjct: 423 GDMA----DKGAWADCIDAYPPESKPSPFLEKFAWIND 456
>UniRef100_Q9MAM3 T25K16.11 [Arabidopsis thaliana]
Length = 528
Score = 355 bits (911), Expect = 2e-96
Identities = 181/419 (43%), Positives = 261/419 (62%), Gaps = 14/419 (3%)
Query: 27 RDQECYILNYQCYKPSDERKLGSVQCGKIIGNNESIGLDECKFFLKAVVNSGIGEETYAP 86
R + Y++++ CYKP DERK+ + N S D +F + +G+G+ETY P
Sbjct: 118 RSKPVYLVDFSCYKPEDERKISVDSFLTMTEENGSFTDDTVQFQQRISNRAGLGDETYLP 177
Query: 87 RNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISPSEIDILVVNVSLFSSVPSLTS 146
R I + + E E ++ L K+GI P+E+ IL+VN SLF+ PSL++
Sbjct: 178 RG-ITSTPPKLNMSEARAEAEAVMFGALDSLFEKTGIKPAEVGILIVNCSLFNPTPSLSA 236
Query: 147 RIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSEKNKYALLLTSESLSTNWYSGI 206
I+NHYKMREDIK+YNL GMGCSA LIS+D+ N+ K+ N YA+++++E+++ NWY G
Sbjct: 237 MIVNHYKMREDIKSYNLGGMGCSAGLISIDLANNLLKANPNSYAVVVSTENITLNWYFGN 296
Query: 207 NRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVRTHHGARDDAHNCCSQKEDEQG 266
+RSM+L NC+FR GG AILL+N+R + K+ L +VRTH G+ D +NC QKEDE+G
Sbjct: 297 DRSMLLCNCIFRMGGAAILLSNRRQDRKKSKYSLVNVVRTHKGSDDKNYNCVYQKEDERG 356
Query: 267 KLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRYMLMSLIKKVIKYFAPKSAARWF 326
+G LA++L A A N+ L P +LP E L + L+SL+K+ + F
Sbjct: 357 TIGVSLARELMSVAGDALKTNITTLGPMVLPLSEQLMF-LISLVKR-----------KMF 404
Query: 327 GTSSKSPV-NFKTGVDHFVLHTGGKAVIDGIGMSLDLNDYDLEPARMSLHRFGNTSASSL 385
K + +FK +HF +H GG+AV+D + +LDL D+ +EP+RM+LHRFGNTS+SSL
Sbjct: 405 KLKVKPYIPDFKLAFEHFCIHAGGRAVLDEVQKNLDLKDWHMEPSRMTLHRFGNTSSSSL 464
Query: 386 WYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRDVVVGDDRNNVWDDCIDYYP 444
WY + Y EAK R+K G+R+ I+ G+GFKCNS W+ +R V + N W ID YP
Sbjct: 465 WYEMAYTEAKGRVKAGDRLWQIAFGSGFKCNSAVWKALRPVSTEEMTGNAWAGSIDQYP 523
>UniRef100_Q6DUV5 Fatty acid elongase 3-ketoacyl-CoA synthase [Brassica napus]
Length = 528
Score = 353 bits (907), Expect = 5e-96
Identities = 185/433 (42%), Positives = 269/433 (61%), Gaps = 19/433 (4%)
Query: 15 LCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNESIGLDECKFFLKAV 74
LCF++ + R + Y++++ CYKP DERK+ K+ N + D +F +
Sbjct: 107 LCFVLII-YVTNRSKPVYLVDFSCYKPEDERKMSVDSFLKMTEQNGAFTDDTVQFQQRIS 165
Query: 75 VNSGIGEETYAPRNFIEGRTVNP---TLEDGVEEMEEFCNDSITKLLNKSGISPSEIDIL 131
+G+G+ETY PR G T NP + + E E ++ L K+GI P+EI IL
Sbjct: 166 NRAGLGDETYFPR----GITSNPPKLNMSEARAEAEAVMFGALDSLFEKTGIKPAEIGIL 221
Query: 132 VVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSEKNKYAL 191
+VN SLFS PSL++ I+N YKMR+DIK+YNL GMGCSA LIS+D+ N+ K+ N YA+
Sbjct: 222 IVNCSLFSPTPSLSAMIVNRYKMRQDIKSYNLGGMGCSAGLISIDLANNLLKANPNSYAV 281
Query: 192 LLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVRTHHGAR 251
++++E+++ NWY G +RSM+L NC+FR GG AILL+N+R + K+ +L +VRTH G+
Sbjct: 282 VVSTENITLNWYFGNDRSMLLCNCIFRMGGAAILLSNRRQDRSKSKYELVNVVRTHKGSD 341
Query: 252 DDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRYMLMSLIK 311
D +NC QKEDE+G +G LA++L A A N+ L P +LP E L + L+SL+K
Sbjct: 342 DKNYNCVYQKEDERGTIGVSLARELMSVAGDALKTNITTLGPMVLPLSEQLMF-LVSLVK 400
Query: 312 KVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLNDYDLEPAR 371
+ + K +FK +HF +H GG+AV+D + +LDL D+ +EP+R
Sbjct: 401 RKLLKLKVKPYI----------PDFKLAFEHFCIHAGGRAVLDEVQKNLDLKDWHMEPSR 450
Query: 372 MSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRDVVVGDD 431
M+LHRFGNTS+SSLWY + Y EAK R+K G+R+ I+ G+GFKCNS W+ +R V +
Sbjct: 451 MTLHRFGNTSSSSLWYEMAYTEAKGRVKAGDRLWQIAFGSGFKCNSAVWKALRVVSTEEL 510
Query: 432 RNNVWDDCIDYYP 444
N W I+ YP
Sbjct: 511 TGNAWAGSIENYP 523
>UniRef100_Q6DUV6 Fatty acid elongase 3-ketoacyl-CoA synthase [Brassica napus]
Length = 528
Score = 348 bits (894), Expect = 1e-94
Identities = 179/434 (41%), Positives = 265/434 (60%), Gaps = 21/434 (4%)
Query: 15 LCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNESIGLDECKFFLKAV 74
LCF++ + R + Y++++ CYKP DERK+ K+ N + D +F +
Sbjct: 107 LCFVLII-YVTNRSKPVYLVDFSCYKPEDERKMSVDSFLKMTEQNGAFTDDTVQFQQRIS 165
Query: 75 VNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISPSEIDILVVN 134
+G+G+ETY PR I + + E E ++ L K+GI P+E+ IL+V+
Sbjct: 166 NRAGLGDETYLPRG-ITSTPPKLNMSEARAEAEAVMFGALDSLFEKTGIKPAEVGILIVS 224
Query: 135 VSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSEKNKYALLLT 194
SLF+ PSL++ I+NHYKMREDIK+YNL GMGCSA LIS+D+ N+ K+ N YA++++
Sbjct: 225 CSLFNPTPSLSAMIVNHYKMREDIKSYNLGGMGCSAGLISIDLANNLLKANPNSYAVVVS 284
Query: 195 SESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVRTHHGARDDA 254
+E+++ NWY G +RSM+L NC+FR GG AILL+N+R + K+ +L +VRTH G+ D
Sbjct: 285 TENITLNWYFGNDRSMLLCNCIFRMGGAAILLSNRRQDRSKSKYELVNVVRTHKGSDDKN 344
Query: 255 HNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRYML----MSLI 310
+NC QKEDE+G +G LA++L A A N+ L P +LP L + + L+
Sbjct: 345 YNCVYQKEDERGTIGVSLARELMSVAGDALKTNITTLGPMVLPLSGQLMFSVSLVKRKLL 404
Query: 311 KKVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLNDYDLEPA 370
K +K + P +FK +HF +H GG+AV+D + +LDL D+ +EP+
Sbjct: 405 KLKVKPYIP---------------DFKLAFEHFCIHAGGRAVLDEVQKNLDLEDWHMEPS 449
Query: 371 RMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRDVVVGD 430
RM+LHRFGNTS+SSLWY + Y EAK R+K G+R+ I+ G+GFKCNS W+ +R V +
Sbjct: 450 RMTLHRFGNTSSSSLWYEMAYTEAKGRVKAGDRLWQIAFGSGFKCNSAVWKALRVVSTEE 509
Query: 431 DRNNVWDDCIDYYP 444
N W I+ YP
Sbjct: 510 LTGNAWAGSIENYP 523
>UniRef100_O81658 Senescence-associated protein 15 [Hemerocallis sp.]
Length = 517
Score = 345 bits (884), Expect = 2e-93
Identities = 185/443 (41%), Positives = 267/443 (59%), Gaps = 19/443 (4%)
Query: 10 FTTMHLCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNESIGLDECKF 69
F++ + L+ + RR + Y++++ CYKP DE K+ + ++ + + F
Sbjct: 84 FSSSSIALLLLGVYYTRRPRPVYLVDFACYKPEDEHKISNEGFLEMTESTTAFNDKSLDF 143
Query: 70 FLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISPS-EI 128
K V SG+G+ETY P I+ R ++ + E E + L +GI+PS +I
Sbjct: 144 QTKITVRSGLGDETYLPPG-IQARPPKLSMAEARLEAETVMFGCLDALFESTGINPSRDI 202
Query: 129 DILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSEKNK 188
IL+VN SLF+ PSL++ I+NHYKMRED+K++NL GMGCSA LIS+D+ +++ ++ N
Sbjct: 203 GILIVNCSLFNPTPSLSAMIVNHYKMREDVKSFNLGGMGCSAGLISIDLAKDMLQANPNS 262
Query: 189 YALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVRTHH 248
YAL+L++E+++ NWY G +RSM+L+NC+FR GG A+LL+NKR +A +L VRTH
Sbjct: 263 YALVLSTENITLNWYFGNDRSMLLSNCIFRMGGAAVLLSNKRKDAKRAKYRLLHTVRTHK 322
Query: 249 GARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRYMLMS 308
GA D +NC Q+EDE GK+G LAK+L A A N+ L P +LP E +++
Sbjct: 323 GADDSCYNCVYQREDEGGKVGVSLAKELMAVAGDALKTNITTLGPLVLPLTEQGKFLATL 382
Query: 309 LIKKVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLNDYDLE 368
+ +K++K G P NFK +HF +H GG+AV+D + +L L D+E
Sbjct: 383 ITRKLLKL---------KGVRPYIP-NFKRAFEHFCVHAGGRAVLDEVEKNLGLEKTDME 432
Query: 369 PARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRDV-- 426
+R LHRFGNTS+SSLWY L Y EAK R+ KG+RV I G+GFKCNS W+ M+DV
Sbjct: 433 ASRSVLHRFGNTSSSSLWYELAYNEAKGRVGKGDRVWQIGFGSGFKCNSAVWKAMKDVPA 492
Query: 427 ----VVGDDRN-NVWDDCIDYYP 444
G R N W DCID YP
Sbjct: 493 IDRTASGSSRMCNPWGDCIDRYP 515
>UniRef100_Q69X62 Putative beta-ketoacyl-CoA synthase [Oryza sativa]
Length = 519
Score = 334 bits (857), Expect = 3e-90
Identities = 174/425 (40%), Positives = 264/425 (61%), Gaps = 27/425 (6%)
Query: 27 RDQECYILNYQCYKPSDERKLGS---VQCGKIIGNNESIGLDECKFFLKAVVNSGIGEET 83
R + Y+L++ CYKP +RK ++C + G+ LD F K + SG+GE+T
Sbjct: 103 RPRPVYLLDFACYKPDPQRKCTRETFMRCSSLTGSFTDANLD---FQRKILERSGLGEDT 159
Query: 84 YAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISPSEIDILVVNVSLFSSVPS 143
Y P + NP +++ +E +I +LL K+G+ P +I ++VVN SLF+ PS
Sbjct: 160 YLPPAVLRVPP-NPCMDEARKEARTVMFGAIDQLLEKTGVKPKDIGVVVVNCSLFNPTPS 218
Query: 144 LTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSEKNKYALLLTSESLSTNWY 203
L++ ++NHYK+R ++ +YNL GMGCSA L+S+D+ +++ + N YAL+++ E+++ NWY
Sbjct: 219 LSAMVVNHYKLRGNVISYNLGGMGCSAGLLSVDLAKDLLQVHPNSYALVVSMENITLNWY 278
Query: 204 SGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVRTHHGARDDAHNCCSQKED 263
G NRSM+++NCLFR GG AILL+N+RS + ++ +L VRTH GA D C +Q+ED
Sbjct: 279 FGNNRSMLVSNCLFRMGGAAILLSNRRSDRRRSKYELVHTVRTHKGANDKCFGCVTQEED 338
Query: 264 EQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRYMLMSLIKKV----IKYFAP 319
E GK+G L+KDL A A N+ L P +LP E L +M + KKV IK + P
Sbjct: 339 EIGKIGVSLSKDLMAVAGDALKTNITTLGPLVLPLSEQLLFMATLVAKKVLKMKIKPYIP 398
Query: 320 KSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLNDYDLEPARMSLHRFGN 379
+FK +HF +H GG+AV+D + +L+L D+ +EP+RM+L+RFGN
Sbjct: 399 ---------------DFKLAFEHFCIHAGGRAVLDELEKNLELTDWHMEPSRMTLYRFGN 443
Query: 380 TSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRDVVVGDDRNNVWDDC 439
TS+SSLWY L Y EAK R++K +R+ I+ G+GFKCNS W+ ++ V ++ N W D
Sbjct: 444 TSSSSLWYELAYTEAKGRIRKRDRIWQIAFGSGFKCNSAVWKALQTVNPAKEK-NPWMDE 502
Query: 440 IDYYP 444
ID +P
Sbjct: 503 IDNFP 507
>UniRef100_Q6F365 Putative beta-ketoacyl synthase [Oryza sativa]
Length = 520
Score = 333 bits (855), Expect = 5e-90
Identities = 173/441 (39%), Positives = 262/441 (59%), Gaps = 14/441 (3%)
Query: 6 LLFIFTTMHLCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNESIGLD 65
L+ + L L+ + R + Y++++ CYKP DERK + + +
Sbjct: 83 LVSVLLCSTLLVLVSTAYFLTRPRPVYLVDFACYKPDDERKCSRARFMNCTERLGTFTPE 142
Query: 66 ECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISP 125
+F K + SG+GE+TY P + NP++ + +E E ++ +LL K+G++P
Sbjct: 143 NVEFQRKIIERSGLGEDTYLPEAVLN-IPPNPSMANARKEAEMVMYGALDELLAKTGVNP 201
Query: 126 SEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSE 185
+I ILVVN SLF+ PSL++ ++NHYK+R ++ +YNL GMGCSA LIS+D+ +++ +
Sbjct: 202 KDIGILVVNCSLFNPTPSLSAMVVNHYKLRGNVVSYNLGGMGCSAGLISIDLAKDLLQVY 261
Query: 186 KNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVR 245
N YA++++ E+++ NWY G +RSM+++NCLFR GG AILL+N+ S + ++ +L VR
Sbjct: 262 PNTYAVVISMENITLNWYFGNDRSMLVSNCLFRMGGAAILLSNRGSARRRSKYQLVHTVR 321
Query: 246 THHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRYM 305
TH GA D C +Q+ED GK G L+KDL A A N+ L P +LP E L +
Sbjct: 322 THRGADDRCFGCVTQREDADGKTGVSLSKDLMAVAGEALKTNITTLGPLVLPMSEQLLFF 381
Query: 306 LMSLIKKVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLNDY 365
+ +KV+K +FK +HF +H GG+AV+D + +L L+D+
Sbjct: 382 ATLVTRKVLKRKVKPYIP-----------DFKLAFEHFCIHAGGRAVLDELEKNLQLSDW 430
Query: 366 DLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRD 425
+EP+RM+LHRFGNTS+SSLWY L Y EAK R+KKG+R I+ G+GFKCNS W +R
Sbjct: 431 HMEPSRMTLHRFGNTSSSSLWYELAYAEAKGRIKKGDRTWQIAFGSGFKCNSAVWRALRS 490
Query: 426 VVVGDDRN--NVWDDCIDYYP 444
V + N N W D I +P
Sbjct: 491 VNPAKENNFTNPWIDEIHRFP 511
>UniRef100_Q9FXZ8 Putative fatty acid elongase [Zea mays]
Length = 513
Score = 330 bits (845), Expect = 7e-89
Identities = 177/425 (41%), Positives = 262/425 (61%), Gaps = 29/425 (6%)
Query: 27 RDQECYILNYQCYKPSDERKLGS---VQCGKIIGNNESIGLDECKFFLKAVVNSGIGEET 83
R + Y+L++ CYKP ERK + C K+ G+ L+ F K + SG+GE+T
Sbjct: 99 RPRPVYLLDFACYKPESERKCTRETFMHCSKLTGSFTDENLE---FQRKILERSGLGEDT 155
Query: 84 YAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISPSEIDILVVNVSLFSSVPS 143
Y P + NP +++ +E +I +LL K+G+ P +I +LVVN SLF+ PS
Sbjct: 156 YLPPAVLRVPP-NPCMDEARKEARAVMFGAIDQLLEKTGVRPKDIGVLVVNCSLFNPTPS 214
Query: 144 LTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSEKNKYALLLTSESLSTNWY 203
L++ ++NHYK+R +I +YNL GMGCSA LI D+ +++ + N YAL+++ E+++ NWY
Sbjct: 215 LSAMVVNHYKLRGNIVSYNLGGMGCSAGLI--DLAKDLLQVHPNSYALVISMENITLNWY 272
Query: 204 SGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVRTHHGARDDAHNCCSQKED 263
G NRSM+++NCLFR GG AILL+N+RS + ++ +L VRTH GA D C +Q+ED
Sbjct: 273 FGNNRSMLVSNCLFRMGGAAILLSNRRSDRRRSKYELVHTVRTHKGANDKCFGCVTQEED 332
Query: 264 EQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRYMLMSLIKKV----IKYFAP 319
E GK+G L+KDL A A N+ L P +LP E L +M + KKV IK + P
Sbjct: 333 EIGKIGVSLSKDLMAVAGDALKTNITTLGPLVLPLSEQLLFMGTLIAKKVLKMKIKPYIP 392
Query: 320 KSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLNDYDLEPARMSLHRFGN 379
+FK +HF +H GG+AV+D + +L+L D+ +EP+RM+L+RFGN
Sbjct: 393 ---------------DFKLAFEHFCIHAGGRAVLDELEKNLELTDWHMEPSRMTLYRFGN 437
Query: 380 TSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRDVVVGDDRNNVWDDC 439
TS+SSLWY L Y EAK R++K +R+ I+ G+GFKCNS W+ +R V ++ + W D
Sbjct: 438 TSSSSLWYELAYTEAKGRIRKRDRIWQIAFGSGFKCNSAVWKALRTVNPAKEK-SPWMDE 496
Query: 440 IDYYP 444
ID +P
Sbjct: 497 IDNFP 501
>UniRef100_Q9FV67 Fatty acid elongase 1-like protein [Limnanthes douglasii]
Length = 505
Score = 328 bits (840), Expect = 3e-88
Identities = 177/425 (41%), Positives = 260/425 (60%), Gaps = 27/425 (6%)
Query: 27 RDQECYILNYQCYKPSDERKLGS---VQCGKIIGNNESIGLDECKFFLKAVVNSGIGEET 83
R + Y++++ CYKP + RK ++CG+ +G S D F K V SG+G+ T
Sbjct: 97 RPRPVYLMDFACYKPDETRKSTREHFMKCGESLG---SFTEDNIDFQRKLVARSGLGDAT 153
Query: 84 YAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISPSEIDILVVNVSLFSSVPS 143
Y P I +P+++ E E +I +LL K+ ++P +I ILVVN SLFS PS
Sbjct: 154 YLPEA-IGTIPAHPSMKAARREAELVMFGAIDQLLEKTKVNPKDIGILVVNCSLFSPTPS 212
Query: 144 LTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSEKNKYALLLTSESLSTNWY 203
L+S I+NHYK+R +I +YNL GMGCSA LIS+D+ + + ++ N YAL++++E+++ NWY
Sbjct: 213 LSSMIVNHYKLRGNIISYNLGGMGCSAGLISVDLAKRLLETNPNTYALVMSTENITLNWY 272
Query: 204 SGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVRTHHGARDDAHNCCSQKED 263
G +RS +++NCLFR GG A+LL+NK S K ++ +L VR+H GA D+ + C Q+ED
Sbjct: 273 MGNDRSKLVSNCLFRMGGAAVLLSNKTSDKKRSKYQLVTTVRSHKGADDNCYGCIFQEED 332
Query: 264 EQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRY----MLMSLIKKVIKYFAP 319
GK+G L+K+L A A N+ L P +LP E L + + + KK IK + P
Sbjct: 333 SNGKIGVSLSKNLMAVAGDALKTNITTLGPLVLPMSEQLLFFATLVARKVFKKKIKPYIP 392
Query: 320 KSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLNDYDLEPARMSLHRFGN 379
+FK DHF +H GG+AV+D + +L L+ + LEP+RM+L+RFGN
Sbjct: 393 ---------------DFKLAFDHFCIHAGGRAVLDELEKNLQLSSWHLEPSRMTLYRFGN 437
Query: 380 TSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRDVVVGDDRNNVWDDC 439
TS+SSLWY L Y EAK R++KGERV I G+GFKCNS W+ ++ V ++ N W D
Sbjct: 438 TSSSSLWYELAYSEAKGRIRKGERVWQIGFGSGFKCNSAVWKALKSVDPKKEK-NPWMDE 496
Query: 440 IDYYP 444
I +P
Sbjct: 497 IHQFP 501
>UniRef100_Q6Z6C7 Putative beta-ketoacyl-CoA-synthase [Oryza sativa]
Length = 519
Score = 327 bits (839), Expect = 4e-88
Identities = 175/449 (38%), Positives = 271/449 (59%), Gaps = 20/449 (4%)
Query: 1 MEFLTLLFIFTTMHLCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGS---VQCGKIIG 57
+ F + + T L FL +F R + Y++++ CYKP ER+ ++C ++ G
Sbjct: 74 LRFNLVSVVACTTLLVFLSTVRF-LTRPRPVYLVDFACYKPPPERRCSRDAFMRCSRLAG 132
Query: 58 NNESIGLDECKFFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKL 117
+ LD F + V SG+G++TY P + NP++ + E E ++ L
Sbjct: 133 CFTAASLD---FQRRIVERSGLGDDTYLPAAVLR-EPPNPSMAEARREAEAVMFGAVDDL 188
Query: 118 LNKSGISPSEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDV 177
L K+G+S EI +LVVN SLF+ PSL++ ++NHYK+R +I +YNL GMGCSA L+++D+
Sbjct: 189 LAKTGVSAKEIGVLVVNCSLFNPTPSLSAMVVNHYKLRGNIVSYNLGGMGCSAGLLAIDL 248
Query: 178 IQNIFKSEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAI 237
+++ + +N YAL+++ E+++ NWYSG +RSM+++NCLFR GG AILL+N+ S + ++
Sbjct: 249 AKDLLQVHRNSYALVISMENITLNWYSGNDRSMLVSNCLFRMGGAAILLSNRWSERRRSK 308
Query: 238 LKLKCLVRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILP 297
+L VRTH G D C +Q+ED +G +G L+KDL A A N+ L P +LP
Sbjct: 309 YELVHTVRTHKGGDDKCFGCVTQEEDGEGNVGVALSKDLMAVAGDALKTNITTLGPLVLP 368
Query: 298 ARELLRYMLMSLIKKVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIG 357
E L +M + KK++K K +FK +HF +H GG+AV+D I
Sbjct: 369 LSEQLLFMATLVAKKLLKMKNVKPYI----------PDFKLAFEHFCVHAGGRAVLDEIE 418
Query: 358 MSLDLNDYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNS 417
+L L ++ +EP+RM+L+RFGNTS+SSLWY L Y EAK R+++ +RV I+ G+GFKCNS
Sbjct: 419 KNLSLGEWQMEPSRMTLYRFGNTSSSSLWYELAYSEAKGRVRRRDRVWQIAFGSGFKCNS 478
Query: 418 CYWEVMRDVVVGDD--RNNVWDDCIDYYP 444
W +R V ++ + N W D ID +P
Sbjct: 479 AVWRALRSVDPEEEALKKNPWMDEIDRFP 507
>UniRef100_Q9FG87 Beta-ketoacyl-CoA synthase [Arabidopsis thaliana]
Length = 529
Score = 324 bits (830), Expect = 4e-87
Identities = 172/427 (40%), Positives = 263/427 (61%), Gaps = 25/427 (5%)
Query: 27 RDQECYILNYQCYKPSDERKLGSVQCGK--IIGNNESIGL---DECKFFLKAVVNSGIGE 81
R + ++L++ CYKP S+ C + + ++ +G+ D F K + SG+G+
Sbjct: 107 RPRRVFLLDFSCYKPDP-----SLICTRETFMDRSQRVGIFTEDNLAFQQKILERSGLGQ 161
Query: 82 ETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISPSEIDILVVNVSLFSSV 141
+TY P + NP +E+ +E E +I +L K+G+ P +I ILVVN SLF+
Sbjct: 162 KTYFPEALLRVPP-NPCMEEARKEAETVMFGAIDAVLEKTGVKPKDIGILVVNCSLFNPT 220
Query: 142 PSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSEKNKYALLLTSESLSTN 201
PSL++ I+N YK+R +I +YNL GMGCSA LIS+D+ + + + + N YAL++++E+++ N
Sbjct: 221 PSLSAMIVNKYKLRGNILSYNLGGMGCSAGLISIDLAKQMLQVQPNSYALVVSTENITLN 280
Query: 202 WYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVRTHHGARDDAHNCCSQK 261
WY G +RSM+L+NC+FR GG A+LL+N+ S + ++ +L VRTH GA D+A C Q+
Sbjct: 281 WYLGNDRSMLLSNCIFRMGGAAVLLSNRSSDRSRSKYQLIHTVRTHKGADDNAFGCVYQR 340
Query: 262 ED----EQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRYMLMSLIKKVIKYF 317
ED E GK+G L+K+L A A N+ L P +LP E L + + +KV K
Sbjct: 341 EDNNAEETGKIGVSLSKNLMAIAGEALKTNITTLGPLVLPMSEQLLFFATLVARKVFKVK 400
Query: 318 APKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLNDYDLEPARMSLHRF 377
K +FK +HF +H GG+AV+D I +LDL+++ +EP+RM+L+RF
Sbjct: 401 KIKPYI----------PDFKLAFEHFCIHAGGRAVLDEIEKNLDLSEWHMEPSRMTLNRF 450
Query: 378 GNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRDVVVGDDRNNVWD 437
GNTS+SSLWY L Y EAK R+K+G+R I+ G+GFKCNS W+ +R + D++ N W
Sbjct: 451 GNTSSSSLWYELAYSEAKGRIKRGDRTWQIAFGSGFKCNSAVWKALRTIDPMDEKTNPWI 510
Query: 438 DCIDYYP 444
D ID +P
Sbjct: 511 DEIDDFP 517
>UniRef100_Q94CA1 Putative beta-ketoacyl-CoA synthase [Arabidopsis thaliana]
Length = 529
Score = 324 bits (830), Expect = 4e-87
Identities = 172/427 (40%), Positives = 263/427 (61%), Gaps = 25/427 (5%)
Query: 27 RDQECYILNYQCYKPSDERKLGSVQCGK--IIGNNESIGL---DECKFFLKAVVNSGIGE 81
R + ++L++ CYKP S+ C + + ++ +G+ D F K + SG+G+
Sbjct: 107 RPRRVFLLDFSCYKPDP-----SLICTRETFMDRSQRVGIFTEDNLAFQQKILERSGLGQ 161
Query: 82 ETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSGISPSEIDILVVNVSLFSSV 141
+TY P + NP +E+ +E E +I +L K+G+ P +I ILVVN SLF+
Sbjct: 162 KTYFPEALLRVPP-NPCMEEARKEAETVMFGAIDAVLEKTGVKPKDIGILVVNCSLFNPT 220
Query: 142 PSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIFKSEKNKYALLLTSESLSTN 201
PSL++ I+N YK+R +I +YNL GMGCSA LIS+D+ + + + + N YAL++++E+++ N
Sbjct: 221 PSLSAMIVNKYKLRGNILSYNLGGMGCSAGLISIDLAKQMLQVQPNSYALVVSTENITLN 280
Query: 202 WYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKCLVRTHHGARDDAHNCCSQK 261
WY G +RSM+L+NC+FR GG A+LL+N+ S + ++ +L VRTH GA D+A C Q+
Sbjct: 281 WYLGNDRSMLLSNCIFRMGGAAVLLSNRSSDRSRSKYQLIHTVRTHKGADDNAFGCVYQR 340
Query: 262 ED----EQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELLRYMLMSLIKKVIKYF 317
ED E GK+G L+K+L A A N+ L P +LP E L + + +KV K
Sbjct: 341 EDNNAEEAGKIGVSLSKNLMAIAGEALKTNITTLGPLVLPMSEQLLFFATLVARKVFKVK 400
Query: 318 APKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDLNDYDLEPARMSLHRF 377
K +FK +HF +H GG+AV+D I +LDL+++ +EP+RM+L+RF
Sbjct: 401 KIKPYI----------PDFKLAFEHFCIHAGGRAVLDEIEKNLDLSEWHMEPSRMTLNRF 450
Query: 378 GNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEVMRDVVVGDDRNNVWD 437
GNTS+SSLWY L Y EAK R+K+G+R I+ G+GFKCNS W+ +R + D++ N W
Sbjct: 451 GNTSSSSLWYELAYSEAKGRIKRGDRTWQIAFGSGFKCNSAVWKALRTIDPMDEKTNPWI 510
Query: 438 DCIDYYP 444
D ID +P
Sbjct: 511 DEIDDFP 517
>UniRef100_Q8L8L0 Fatty acid condensing enzyme CUT1, putative [Arabidopsis thaliana]
Length = 492
Score = 323 bits (828), Expect = 7e-87
Identities = 184/442 (41%), Positives = 264/442 (59%), Gaps = 20/442 (4%)
Query: 6 LLFIFTTMHLCFLIWKKFDQRRDQECYILNYQCYKPSDERKLGSVQCGKIIGNNESIGLD 65
LL I + L + + + + Y+++Y CYKP + V + ++ I D
Sbjct: 59 LLHILCSSFLIIFVSTVYFMSKPRTVYLVDYSCYKPPVTCR---VPFSSFMEHSRLILKD 115
Query: 66 ECK---FFLKAVVNSGIGEETYAPRNFIEGRTVNPTLEDGVEEMEEFCNDSITKLLNKSG 122
K F ++ + SG+GEET P I PT+E E + ++ L +G
Sbjct: 116 NPKSVEFQMRILERSGLGEETCLPPA-IHYIPPTPTMESARNEAQMVIFTAMEDLFKNTG 174
Query: 123 ISPSEIDILVVNVSLFSSVPSLTSRIINHYKMREDIKAYNLSGMGCSASLISLDVIQNIF 182
+ P +IDIL+VN SLFS PSL++ IIN YK+R +IK+YNLSGMGCSASLIS+DV +N+
Sbjct: 175 LKPKDIDILIVNCSLFSPTPSLSAMIINKYKLRSNIKSYNLSGMGCSASLISVDVARNLL 234
Query: 183 KSEKNKYALLLTSESLSTNWYSGINRSMILANCLFRTGGCAILLTNKRSLKHKAILKLKC 242
+ N A+++++E ++ N+Y G R+M+L NCLFR GG AILL+N+RS + +A KL
Sbjct: 235 QVHPNSNAIIISTEIITPNYYKGNERAMLLPNCLFRMGGAAILLSNRRSDRWRAKYKLCH 294
Query: 243 LVRTHHGARDDAHNCCSQKEDEQGKLGFYLAKDLPKAATRAFTDNLRVLSPKILPARELL 302
LVRTH GA D ++NC ++ED+ G +G L+KDL A A N+ + P +LPA E L
Sbjct: 295 LVRTHRGADDKSYNCVMEQEDKNGNVGINLSKDLMTIAGEALKANITTIGPLVLPASEQL 354
Query: 303 RYMLMSLIKKVIKYFAPKSAARWFGTSSKSPVNFKTGVDHFVLHTGGKAVIDGIGMSLDL 362
+ L SLI + K F PK W +FK +HF +H GG+AVID + +L L
Sbjct: 355 LF-LSSLIGR--KIFNPK----W----KPYIPDFKQAFEHFCIHAGGRAVIDELQKNLQL 403
Query: 363 NDYDLEPARMSLHRFGNTSASSLWYVLGYMEAKKRLKKGERVLMISLGAGFKCNSCYWEV 422
+ +E +RM+LHRFGNTS+SSLWY L Y+EA+ R+K+ +RV I+ G+GFKCNS W+
Sbjct: 404 SGEHVEASRMTLHRFGNTSSSSLWYELSYIEAQGRMKRNDRVWQIAFGSGFKCNSAVWKC 463
Query: 423 MRDVVVGDDRNNVWDDCIDYYP 444
R + D W DCI+ YP
Sbjct: 464 NRTIKTPTD--GAWSDCIERYP 483
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 819,931,878
Number of Sequences: 2790947
Number of extensions: 35006840
Number of successful extensions: 81407
Number of sequences better than 10.0: 839
Number of HSP's better than 10.0 without gapping: 575
Number of HSP's successfully gapped in prelim test: 264
Number of HSP's that attempted gapping in prelim test: 80267
Number of HSP's gapped (non-prelim): 1045
length of query: 480
length of database: 848,049,833
effective HSP length: 131
effective length of query: 349
effective length of database: 482,435,776
effective search space: 168370085824
effective search space used: 168370085824
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC144343.7


