

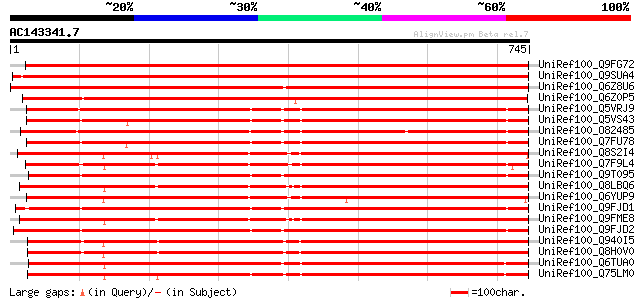
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.7 + phase: 0
(745 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FG72 Sexual differentiation process protein ISP4-lik... 1067 0.0
UniRef100_Q9SUA4 Isp4 like protein [Arabidopsis thaliana] 1060 0.0
UniRef100_Q6Z8U6 Putative glutathione transporter [Oryza sativa] 1046 0.0
UniRef100_Q6Z0P5 Putative sexual differentiation process protein... 987 0.0
UniRef100_Q5VRJ9 Putative isp4 protein [Oryza sativa] 848 0.0
UniRef100_Q5VS43 Putative sexual differentiation process protein... 848 0.0
UniRef100_O82485 T12H20.7 protein [Arabidopsis thaliana] 843 0.0
UniRef100_Q7FU78 Glutathione transporter [Oryza sativa] 823 0.0
UniRef100_Q8S2I4 Isp4 protein-like [Oryza sativa] 808 0.0
UniRef100_Q7F9L4 OSJNBa0006A01.13 protein [Oryza sativa] 797 0.0
UniRef100_Q9T095 Hypothetical protein AT4g27730 [Arabidopsis tha... 794 0.0
UniRef100_Q8LBQ6 Isp4-like protein [Arabidopsis thaliana] 794 0.0
UniRef100_Q6YUP9 Putative sexual differentiation process protein... 793 0.0
UniRef100_Q9FJD1 Isp4 protein [Arabidopsis thaliana] 793 0.0
UniRef100_Q9FME8 Isp4-like protein [Arabidopsis thaliana] 793 0.0
UniRef100_Q9FJD2 Isp4 protein [Arabidopsis thaliana] 790 0.0
UniRef100_Q940I5 Hypothetical protein Z97341.3 [Arabidopsis thal... 790 0.0
UniRef100_Q8H0V0 Isp4 like protein [Arabidopsis thaliana] 788 0.0
UniRef100_Q6TUA0 Putative glutathione transporter [Zea mays] 774 0.0
UniRef100_Q75LM0 Putative oligopeptide transporter protein [Oryz... 764 0.0
>UniRef100_Q9FG72 Sexual differentiation process protein ISP4-like [Arabidopsis
thaliana]
Length = 755
Score = 1067 bits (2759), Expect = 0.0
Identities = 488/723 (67%), Positives = 595/723 (81%), Gaps = 1/723 (0%)
Query: 23 NDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQ 82
ND+PIE+VRLTVPITDDP+ P LTFRTW LGL SCILLAFVNQF G+R+N L ++SV+AQ
Sbjct: 32 NDNPIEEVRLTVPITDDPTLPVLTFRTWTLGLFSCILLAFVNQFFGFRSNQLWVSSVAAQ 91
Query: 83 IIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSGVYAINII 142
I+ LPLGKLMA TLPTK P T WS+S NPGPF++KEHVLITIFA++G+ GVYA +II
Sbjct: 92 IVTLPLGKLMAKTLPTKKFGFPGTNWSWSFNPGPFNMKEHVLITIFANTGAGGVYATSII 151
Query: 143 TIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAFH 202
TIVKAFY+R ++ AA LL +TQ+LGYGWAGIFR+FLVDSPYMWWP +LVQVSLFRA H
Sbjct: 152 TIVKAFYNRQLNVAAAMLLTQTTQLLGYGWAGIFRKFLVDSPYMWWPSNLVQVSLFRALH 211
Query: 203 EKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGSGM 262
EKE +G +R +FF +VF SFAYYIIPGY F +IS +SFVC IWK S+TAQ +GSG+
Sbjct: 212 EKEDLQKGQQTRFRFFIIVFCVSFAYYIIPGYLFPSISAISFVCWIWKSSVTAQIVGSGL 271
Query: 263 KGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNLYDAKKFP 322
KGLGIGSFGLDW+TVAGFLGSPLAVP FAI N GFF+++Y+++PI YW N YDA+KFP
Sbjct: 272 KGLGIGSFGLDWSTVAGFLGSPLAVPFFAIANFFGGFFIFLYIVLPIFYWTNAYDAQKFP 331
Query: 323 LISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTATIS 382
+SHTFD TG TYN+TRILN + FDI++++Y+ YSK+YLSV FA YG F +L ATIS
Sbjct: 332 FYTSHTFDQTGHTYNITRILNEKNFDINLDAYNGYSKLYLSVMFALLYGLSFGSLCATIS 391
Query: 383 HVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMALLAC 442
HV L+ G+ I MWKK T+ K++ GDVH+R+MKKNY+ VP+WWF+ +L++ AL AC
Sbjct: 392 HVALYDGKFIWGMWKKAKTATKDKYGDVHSRLMKKNYQSVPQWWFIAVLVISFAFALYAC 451
Query: 443 EGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKPLAN 502
EGF KQLQLPWWG++L+ IAL FTLPIGVI+ATTN + GLNVITEL+IG++YPGKPLAN
Sbjct: 452 EGFDKQLQLPWWGLILACAIALFFTLPIGVIQATTNQQMGLNVITELIIGYLYPGKPLAN 511
Query: 503 VAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSI 562
VAFKTYG+ISM QAL F+GDFKLGHYMKI P+SMFIVQLV TVVAS+V FGT WWL+TS+
Sbjct: 512 VAFKTYGYISMSQALYFVGDFKLGHYMKIPPRSMFIVQLVATVVASTVCFGTTWWLITSV 571
Query: 563 ENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIAPV 622
ENIC+ LLP GSPWTCPGD+VFYNASIIWGV+GP RMFTK+G+YP +NWFFLIGL+APV
Sbjct: 572 ENICNVDLLPVGSPWTCPGDEVFYNASIIWGVIGPGRMFTKEGIYPGMNWFFLIGLLAPV 631
Query: 623 PVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKAWW 682
P W LS KFP +KW++ I++P+I + S +P ++V+Y +W IVG+ FN+Y++R+FK WW
Sbjct: 632 PFWYLSKKFPEKKWLKQIHVPLIFSAVSAMPQAKAVHYWSWAIVGVVFNYYIFRRFKTWW 691
Query: 683 ARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLE-PDHCPLAKCPTAPGVHAEG 741
ARH YILSA LDAG A +G+L++F+ Q+ I P WWGLE DHCPLA CP A GV EG
Sbjct: 692 ARHNYILSAALDAGTAIMGVLIFFAFQNNDISLPDWWGLENSDHCPLAHCPLAKGVVVEG 751
Query: 742 CPV 744
CPV
Sbjct: 752 CPV 754
>UniRef100_Q9SUA4 Isp4 like protein [Arabidopsis thaliana]
Length = 753
Score = 1060 bits (2740), Expect = 0.0
Identities = 487/741 (65%), Positives = 597/741 (79%), Gaps = 3/741 (0%)
Query: 5 RVSQNTVIEDAEIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVN 64
+V VI D E E NDSPIE+VRLTVPITDDPS P LTFRTW LG+ SC++LAFVN
Sbjct: 14 KVESKIVIADEE--EEDENDSPIEEVRLTVPITDDPSLPVLTFRTWFLGMVSCVVLAFVN 71
Query: 65 QFLGYRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVL 124
F GYR+NPL ++SV AQII LPLGKLMA TLPT +++P T WS SLNPGPF++KEHVL
Sbjct: 72 NFFGYRSNPLTVSSVVAQIITLPLGKLMATTLPTTKLRLPGTNWSCSLNPGPFNMKEHVL 131
Query: 125 ITIFASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSP 184
ITIFA++G+ G YA +I+TIVKAFYHR+++P AA LL +TQ+LGYGWAG+FR++LVDSP
Sbjct: 132 ITIFANTGAGGAYATSILTIVKAFYHRNLNPAAAMLLVQTTQLLGYGWAGMFRKYLVDSP 191
Query: 185 YMWWPESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSF 244
YMWWP +LVQVSLFRA HEKE++ G ++L+FF +VF SF YYI+PGY F +IS +SF
Sbjct: 192 YMWWPANLVQVSLFRALHEKEEKREGKQTKLRFFLIVFFLSFTYYIVPGYLFPSISYLSF 251
Query: 245 VCLIWKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMY 304
VC IW S+TAQQIGSG+ GLGIGSFGLDW+TVAGFLGSPLAVP FAI N GF ++ Y
Sbjct: 252 VCWIWTRSVTAQQIGSGLHGLGIGSFGLDWSTVAGFLGSPLAVPFFAIANSFGGFIIFFY 311
Query: 305 VLIPISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSV 364
+++PI YW+N Y+AKKFP +SH FD TG YN TRILN +TF+ID+ +Y +YSK+YLS+
Sbjct: 312 IILPIFYWSNAYEAKKFPFYTSHPFDHTGQRYNTTRILNQKTFNIDLPAYESYSKLYLSI 371
Query: 365 AFAFEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPE 424
FA YG F LTATISHV LF G+ I ++WKK T + K++ GDVHTR+MKKNY++VP+
Sbjct: 372 LFALIYGLSFGTLTATISHVALFDGKFIWELWKKATLTTKDKFGDVHTRLMKKNYKEVPQ 431
Query: 425 WWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLN 484
WWFV +L ++AL ACEGFGKQLQLPWWG+LL+ IA FTLPIGVI ATTN R GLN
Sbjct: 432 WWFVAVLAASFVLALYACEGFGKQLQLPWWGLLLACAIAFTFTLPIGVILATTNQRMGLN 491
Query: 485 VITELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGT 544
VI+EL+IGF+YPGKPLANVAFKTYG +S+ QAL F+GDFKLGHYMKI P+SMFIVQLV T
Sbjct: 492 VISELIIGFLYPGKPLANVAFKTYGSVSIAQALYFVGDFKLGHYMKIPPRSMFIVQLVAT 551
Query: 545 VVASSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKD 604
+VAS+V FGT WWLL+S+ENIC+ +LPK SPWTCPGD VFYNASIIWG++GP RMFT
Sbjct: 552 IVASTVSFGTTWWLLSSVENICNTDMLPKSSPWTCPGDVVFYNASIIWGIIGPGRMFTSK 611
Query: 605 GVYPELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWG 664
G+YP +NWFFLIG +APVPVW + KFP +KWI I+IP+I +GA+ +P ++V+Y +W
Sbjct: 612 GIYPGMNWFFLIGFLAPVPVWFFARKFPEKKWIHQIHIPLIFSGANVMPMAKAVHYWSWF 671
Query: 665 IVGIFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLE-P 723
VGI FN+Y++R++K WWARH YILSA LDAG A +G+L+YF+LQ+ I P WWG E
Sbjct: 672 AVGIVFNYYIFRRYKGWWARHNYILSAALDAGTAVMGVLIYFALQNNNISLPDWWGNENT 731
Query: 724 DHCPLAKCPTAPGVHAEGCPV 744
DHCPLA CPT G+ A+GCPV
Sbjct: 732 DHCPLANCPTEKGIVAKGCPV 752
>UniRef100_Q6Z8U6 Putative glutathione transporter [Oryza sativa]
Length = 752
Score = 1046 bits (2704), Expect = 0.0
Identities = 483/745 (64%), Positives = 596/745 (79%), Gaps = 5/745 (0%)
Query: 1 MSSSRVSQNTVIEDAEIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILL 60
+ R S+N D + +E ++D PIE+VRLTVPITDDP+ PALTFRTW+LGL SC +L
Sbjct: 11 LEGHRSSENPESRDEKTEE-EVDDCPIEEVRLTVPITDDPALPALTFRTWLLGLISCAML 69
Query: 61 AFVNQFLGYRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLK 120
AF NQF GYR NPL I+S+S QI+ LPLGKLMAA LP K +V T WSFSLNPGPF+LK
Sbjct: 70 AFSNQFFGYRQNPLYISSLSVQIVVLPLGKLMAACLPKKVFRVKGTAWSFSLNPGPFNLK 129
Query: 121 EHVLITIFASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFL 180
EHVLITIFA++GS+ VYA+ IITIVKAFY R IHP+AA LL +TQ++GYGWAG+FR+FL
Sbjct: 130 EHVLITIFANTGSNSVYAVGIITIVKAFYRREIHPLAAMLLTQTTQLMGYGWAGLFRKFL 189
Query: 181 VDSPYMWWPESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAIS 240
VDSPYMWWP +LVQVSLFRA HEKEKRP+GGT+RLQFF V + SFAYYI+P Y F IS
Sbjct: 190 VDSPYMWWPSNLVQVSLFRALHEKEKRPKGGTTRLQFFLTVLITSFAYYIVPNYLFPTIS 249
Query: 241 TVSFVCLIWKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFF 300
T+S VCL+WK+S+TAQQIGSG+ GLG+GSFGLDW TVAGFLG+PL+ P FAI+NIMAGFF
Sbjct: 250 TISVVCLVWKKSVTAQQIGSGVYGLGVGSFGLDWATVAGFLGTPLSTPAFAIVNIMAGFF 309
Query: 301 LYMYVLIPISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKI 360
L +YV++P +YW + Y AK+FP+ISSH F + G+ Y+V ++L+T TF+ Y KI
Sbjct: 310 LIVYVIVPAAYWADAYGAKRFPIISSHVFSANGSRYDVNQVLDTATFEFSQAGYDAAGKI 369
Query: 361 YLSVAFAFEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYE 420
LS+ FAF YG FA L AT+SHV LFHG I W++T ++ Q GDVHTR+MK+NY
Sbjct: 370 NLSIFFAFTYGLSFATLAATLSHVALFHGGSI---WRQTKAAVSGQGGDVHTRLMKRNYA 426
Query: 421 QVPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIR 480
VP+WWF +L+ ++ +++ CEGFG+QLQLP+WG+LL+ +A FTLPIG+I ATTN +
Sbjct: 427 AVPQWWFQVMLVAVLGLSVFTCEGFGQQLQLPYWGVLLAAGLAFFFTLPIGIITATTNQQ 486
Query: 481 SGLNVITELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQ 540
GLNVITEL+IG++YPG+PLANVAFKTYG+ISM QA+ FL DFKLGHYMKI P+SMFIVQ
Sbjct: 487 PGLNVITELIIGYLYPGRPLANVAFKTYGYISMSQAIMFLQDFKLGHYMKIPPRSMFIVQ 546
Query: 541 LVGTVVASSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRM 600
LVGTV+ASSV+FGT+WWLL S+ NICD + LP+GSPWTCPGDDVF+NASIIWGVVGP RM
Sbjct: 547 LVGTVLASSVYFGTSWWLLESVSNICDPAKLPEGSPWTCPGDDVFFNASIIWGVVGPLRM 606
Query: 601 FTKDGVYPELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNY 660
F + G+Y ++N+FFL G +APVPVW LS FP + WI +N+P+++ +PP RSVNY
Sbjct: 607 FGRLGLYAKMNYFFLAGALAPVPVWALSRAFPGRAWIGLVNMPVLLGATGMMPPARSVNY 666
Query: 661 ITWGIVGIFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWG 720
+ WG VG+ FN+ VYR++K WWARH Y+LSAGLDAGVAF+G+L Y LQS GI G WWG
Sbjct: 667 LMWGAVGLAFNYVVYRRYKGWWARHNYVLSAGLDAGVAFMGILSYAVLQSRGINGVNWWG 726
Query: 721 LE-PDHCPLAKCPTAPGVHAEGCPV 744
L+ DHC LA+CPTAPGV A GCPV
Sbjct: 727 LQVDDHCALARCPTAPGVSAPGCPV 751
>UniRef100_Q6Z0P5 Putative sexual differentiation process protein isp4 [Oryza sativa]
Length = 733
Score = 987 bits (2551), Expect = 0.0
Identities = 458/730 (62%), Positives = 571/730 (77%), Gaps = 7/730 (0%)
Query: 19 EYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITS 78
E +ND PIE+VR TVP+TDDPS+P LTFR WVLGL+SC+ LAFVN+F YRT L I +
Sbjct: 2 EEEVNDHPIEEVRNTVPVTDDPSEPCLTFRVWVLGLSSCVFLAFVNEFFMYRTTQLSIGT 61
Query: 79 VSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSGVYA 138
V QII LP+G+LMA+TLP + ++V WSFSLNPGPFSLKEH LI IFA +G+SGVYA
Sbjct: 62 VVVQIITLPIGRLMASTLPARRLRVG--GWSFSLNPGPFSLKEHCLIIIFAGAGASGVYA 119
Query: 139 INIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLF 198
+NII IVK FY R I P AA LLA +TQ+LGYGWAG+FR++LVDS YMWWP +LVQV+LF
Sbjct: 120 MNIIAIVKVFYKRQISPYAAMLLAQTTQLLGYGWAGLFRKYLVDSAYMWWPSNLVQVTLF 179
Query: 199 RAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQI 258
RA HE+EKR +G +RLQFF +V SFAYYI+P Y F AIST+S +C ++++S+TAQQI
Sbjct: 180 RAMHEEEKRNKGQLTRLQFFIMVMTCSFAYYIVPSYLFPAISTISVLCWLYRDSVTAQQI 239
Query: 259 GSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNLYDA 318
GSG GLG+GSFGLDWNTV GFLG+PLA P FAI N+MAGF L YV +PI YW + Y+A
Sbjct: 240 GSGASGLGVGSFGLDWNTVVGFLGNPLASPAFAIFNVMAGFALSTYVAVPILYWTDTYNA 299
Query: 319 KKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALT 378
K+FPL+SSH F++ G Y+ RIL+ TF +++ Y Y +I LS+ FA YG FA L
Sbjct: 300 KRFPLVSSHVFNAAGGRYDTARILDPATFTLNLREYDAYGRINLSILFAINYGIGFAGLM 359
Query: 379 ATISHVVLFHGEMIVQMWKKTTTSLKNQLG----DVHTRIMKKNYEQVPEWWFVTILILM 434
+T+SHV L+HG+ I +W+K T N G DVHTRIMK+NY+ VP+WWF +L ++
Sbjct: 360 STLSHVALYHGKDIWGLWRKATAEQANGGGKERQDVHTRIMKRNYKAVPQWWFHLMLAIV 419
Query: 435 VMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFI 494
+ ++L CEGFG+QLQLP+WG+LL+ IA FTLPIGVI ATTN++ GLN+ITEL+IG++
Sbjct: 420 MALSLYTCEGFGRQLQLPYWGLLLACAIAFTFTLPIGVISATTNMQPGLNIITELIIGYL 479
Query: 495 YPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGT 554
YPGKPLANV FKTYG+ISM QAL F+ DFKLGHYMKI P+SMF+VQL GTVVAS+VHF T
Sbjct: 480 YPGKPLANVVFKTYGYISMTQALTFVSDFKLGHYMKIPPRSMFMVQLAGTVVASTVHFAT 539
Query: 555 AWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFF 614
AWWLLT++ NICD LP GSPWTCPG+DVFYNASIIWGVVGP RMF + G Y ++N+FF
Sbjct: 540 AWWLLTTVRNICDVDSLPLGSPWTCPGEDVFYNASIIWGVVGPLRMFGRLGNYWQMNYFF 599
Query: 615 LIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYV 674
L+G++APVPVWLLS ++P ++ IN+P+++AGAS + P RSVN++ WG+VG FN V
Sbjct: 600 LVGVLAPVPVWLLSRRYPRSALLRDINLPLVLAGASGLLPARSVNFVMWGLVGFVFNHVV 659
Query: 675 YRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWW-GLEPDHCPLAKCPT 733
YR+ +AWW RH Y+L+AGLDAGVAF+G+L + SL + IYG WW G DHCPLA CPT
Sbjct: 660 YRRCRAWWMRHNYVLAAGLDAGVAFMGVLTFVSLGYFDIYGVQWWGGAADDHCPLASCPT 719
Query: 734 APGVHAEGCP 743
APGV A GCP
Sbjct: 720 APGVFARGCP 729
>UniRef100_Q5VRJ9 Putative isp4 protein [Oryza sativa]
Length = 760
Score = 848 bits (2192), Expect = 0.0
Identities = 386/721 (53%), Positives = 529/721 (72%), Gaps = 7/721 (0%)
Query: 24 DSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQI 83
+SP+EQV LTVP+ DDP+ P LTFR WVLG ASC++L+F+N F YR PL +T++SAQI
Sbjct: 44 NSPVEQVALTVPVGDDPATPVLTFRIWVLGTASCVVLSFLNTFFWYRKEPLTVTAISAQI 103
Query: 84 IALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSGVYAINIIT 143
+PLG+LMAA LP + + W F+LNPGPF++KEHVLITIFA++G+ V+AIN+IT
Sbjct: 104 AVVPLGRLMAAALPER-VFFRGRPWEFTLNPGPFNVKEHVLITIFANAGAGSVFAINVIT 162
Query: 144 IVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAFHE 203
V+ FY + I + L+ L++Q+LG+GWAGIFRR+LV+ MWWP +LVQVSLFRA HE
Sbjct: 163 AVRVFYGKRISFFVSLLVVLTSQVLGFGWAGIFRRYLVEPAAMWWPSNLVQVSLFRALHE 222
Query: 204 KEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGSGMK 263
KE+R +GG +R QFF V FV SFAYYI PGY FQ ++++S++C I+ +S+ AQQ+GSG+
Sbjct: 223 KERRSKGGMTRTQFFLVAFVCSFAYYIFPGYLFQMLTSLSWICWIFPKSVLAQQLGSGLH 282
Query: 264 GLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNLYDAKKFPL 323
GLGIG+ GLDW++++ +LGSPLA P FA NI AGFF+Y+YV+ PI+YW NLY A+ FP+
Sbjct: 283 GLGIGAIGLDWSSISSYLGSPLASPWFATANIAAGFFIYIYVITPIAYWINLYKAQNFPI 342
Query: 324 ISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTATISH 383
S F TG YN++ I++++ F D ++Y +Y+S F+ YG FA LTAT+ H
Sbjct: 343 FSDGLFTVTGQKYNISTIIDSQ-FHFDTKAYEKNGPLYISTFFSISYGLGFACLTATVVH 401
Query: 384 VVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMALLACE 443
V+LFHG ++W+ + ++ +++ D+HT++MK+ Y+QVPEWWF++ILI V + + CE
Sbjct: 402 VLLFHGS---EIWQLSRSAFQDKKMDIHTKLMKR-YKQVPEWWFISILIASVAITMFTCE 457
Query: 444 GFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKPLANV 503
+ +QLQLPWWG+LL+ +A+ FTLPIG++ ATTN GLN+ITE ++G++YPG+P+AN+
Sbjct: 458 YYIEQLQLPWWGVLLACALAIFFTLPIGIVTATTNQTPGLNIITEYIMGYLYPGRPVANM 517
Query: 504 AFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIE 563
FK YG+I QAL FL DFKLGHYMKI P++MF+ Q+VGT++A+ V+ GTAWWL+ +I
Sbjct: 518 CFKVYGYIGPQQALAFLQDFKLGHYMKIPPRTMFMAQVVGTLIAAFVYLGTAWWLMDTIP 577
Query: 564 NICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIAPVP 623
NIC+ LLP GSPWTCP D +FY+AS+IWG++GP+R+F G Y +NWFFL G IAP+
Sbjct: 578 NICNTELLPPGSPWTCPYDHLFYDASVIWGLIGPRRIFGDLGTYSAVNWFFLGGAIAPLL 637
Query: 624 VWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKAWWA 683
VW FP QKWI +N+P++IAG S +PP SVNY W V + VY+ + WW
Sbjct: 638 VWFAHKAFPGQKWILLVNMPVLIAGISQMPPATSVNYTAWIFVAFLSGYVVYKYRRDWWE 697
Query: 684 RHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAPGVHAEGCP 743
RH Y+LS LDAG+AF+ +LLY L I WWG + D CPLA CP A G+ +GCP
Sbjct: 698 RHNYLLSGALDAGLAFMAVLLYLCLGLEKI-SLNWWGNDLDGCPLASCPIAEGITVQGCP 756
Query: 744 V 744
V
Sbjct: 757 V 757
>UniRef100_Q5VS43 Putative sexual differentiation process protein isp4 [Oryza sativa]
Length = 763
Score = 848 bits (2192), Expect = 0.0
Identities = 397/726 (54%), Positives = 524/726 (71%), Gaps = 12/726 (1%)
Query: 24 DSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQI 83
+SPIEQV LTVP++D+P P LTFR WVLG ASC +L+F+NQF YR PL IT++SAQI
Sbjct: 43 NSPIEQVALTVPVSDEPETPVLTFRMWVLGTASCAVLSFLNQFFWYRKEPLTITAISAQI 102
Query: 84 IALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSGVYAINIIT 143
+PLG+LMAA LP + W F+LNPGPF++KEHVLITIFA+SG+ VYAI++IT
Sbjct: 103 AVVPLGRLMAAALPERAF-FRGRPWEFTLNPGPFNVKEHVLITIFANSGAGTVYAIHVIT 161
Query: 144 IVKAFYHRSIHPVAAYLLALSTQM-----LGYGWAGIFRRFLVDSPYMWWPESLVQVSLF 198
V+ FY + I + L+ L+TQ+ LG+GWAGIFRR+LV+ MWWP +LVQVSLF
Sbjct: 162 AVRVFYGKHISFFVSLLVVLTTQVVDDQVLGFGWAGIFRRYLVEPAAMWWPSNLVQVSLF 221
Query: 199 RAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQI 258
RA HEKE R +GG +R QFF V F+ SFAYYI PGY FQ ++++S++C ++ S+ AQQ+
Sbjct: 222 RALHEKEARSKGGLTRNQFFLVAFICSFAYYIFPGYLFQMLTSLSWICWVFPHSVLAQQL 281
Query: 259 GSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNLYDA 318
GSG+ GLGIG+ GLDW+TV+ +LGSPLA P FA N+ AGFF MY++ PI+YW N Y A
Sbjct: 282 GSGLSGLGIGAIGLDWSTVSSYLGSPLASPWFATANVAAGFFFIMYIITPIAYWFNFYKA 341
Query: 319 KKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALT 378
+ FP+ S F STG YN++ I+++ F D ++Y +YLS FA YG FA+LT
Sbjct: 342 QNFPIFSDGLFTSTGQKYNISSIVDSH-FHFDTKAYEKNGPLYLSTFFAVTYGVGFASLT 400
Query: 379 ATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMA 438
ATI HV+LFHG ++W+ + ++ + + D+HT++M++ Y+QVPEWWFV ILI + +
Sbjct: 401 ATIVHVLLFHGS---EIWQLSKSAFQEKRMDIHTKLMRR-YKQVPEWWFVCILIANIAVT 456
Query: 439 LLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGK 498
+ ACE + +QLQLPWWG+LL+ IA FTLPIG+I ATTN GLN+ITE ++G++YPG+
Sbjct: 457 IFACEYYIEQLQLPWWGVLLACAIAFFFTLPIGIITATTNQTPGLNIITEYIMGYLYPGR 516
Query: 499 PLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWL 558
P+AN+ FK YG+ISM QAL FL DFKLGHYMKI P++MF+ Q+VGT++A+ V+ GTAWWL
Sbjct: 517 PVANMCFKVYGYISMSQALTFLQDFKLGHYMKIPPRTMFMAQVVGTLIAAFVYIGTAWWL 576
Query: 559 LTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGL 618
+ +I NIC+ LLP SPWTCPGD VFY+AS+IWG++ P+R+F G Y +NWFFL G
Sbjct: 577 METIPNICNTELLPSDSPWTCPGDHVFYDASVIWGLISPRRIFGDLGTYSAVNWFFLGGA 636
Query: 619 IAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKF 678
IAPV VW FPNQ WI IN+P++I +PP +VNY TW +VG + VYR
Sbjct: 637 IAPVLVWFAHKAFPNQNWILLINMPVLIGATGQMPPATAVNYTTWILVGFLSGYVVYRYR 696
Query: 679 KAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAPGVH 738
+ WW RH Y+LS LDAG+AF+ +L+Y L I WWG + D CPLA CPTA GV
Sbjct: 697 RDWWERHNYLLSGALDAGLAFMAVLIYLCLGLENI-SLNWWGNDLDGCPLASCPTAKGVV 755
Query: 739 AEGCPV 744
+GCPV
Sbjct: 756 VDGCPV 761
>UniRef100_O82485 T12H20.7 protein [Arabidopsis thaliana]
Length = 766
Score = 843 bits (2178), Expect = 0.0
Identities = 395/729 (54%), Positives = 529/729 (72%), Gaps = 10/729 (1%)
Query: 16 EIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLK 75
E +E +SPI QV LTVP TDDPS P LTFR WVLG SCILL+F+NQF YRT PL
Sbjct: 46 ENEEEEEENSPIRQVALTVPTTDDPSLPVLTFRMWVLGTLSCILLSFLNQFFWYRTEPLT 105
Query: 76 ITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSG 135
I+++SAQI +PLG+LMAA + T + + W F+LNPGPF++KEHVLITIFA++G+
Sbjct: 106 ISAISAQIAVVPLGRLMAAKI-TDRVFFQGSKWQFTLNPGPFNVKEHVLITIFANAGAGS 164
Query: 136 VYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQV 195
VYAI+++T+VKAFY ++I ++++ ++TQ+LG+GWAGIFR++LV+ MWWP +LVQV
Sbjct: 165 VYAIHVVTVVKAFYMKNITFFVSFIVIVTTQVLGFGWAGIFRKYLVEPAAMWWPANLVQV 224
Query: 196 SLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITA 255
SLFRA HEKE+R +GG +R QFF + FV SFAYY+ PGY FQ ++++S+VC + S+ A
Sbjct: 225 SLFRALHEKEERTKGGLTRTQFFVIAFVCSFAYYVFPGYLFQIMTSLSWVCWFFPSSVMA 284
Query: 256 QQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNL 315
QQIGSG+ GLG+G+ GLDW+T++ +LGSPLA P FA N+ GF L +YVL+PI YW ++
Sbjct: 285 QQIGSGLHGLGVGAIGLDWSTISSYLGSPLASPWFATANVGVGFVLVIYVLVPICYWLDV 344
Query: 316 YDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFA 375
Y AK FP+ SS F S G+ YN+T I+++ F +D+ +Y +YL FA YG FA
Sbjct: 345 YKAKTFPIFSSSLFSSQGSKYNITSIIDS-NFHLDLPAYERQGPLYLCTFFAISYGVGFA 403
Query: 376 ALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMV 435
AL+ATI HV LFHG ++W+++ S K + DVH R+M++ Y+QVPEWWF IL+ V
Sbjct: 404 ALSATIMHVALFHGR---EIWEQSKESFKEKKLDVHARLMQR-YKQVPEWWFWCILVTNV 459
Query: 436 MMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIY 495
+ ACE + QLQLPWWG+LL+ T+A+IFTLPIG+I A TN GLN+ITE +IG+IY
Sbjct: 460 GATIFACEYYNDQLQLPWWGVLLACTVAIIFTLPIGIITAITNQAPGLNIITEYIIGYIY 519
Query: 496 PGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTA 555
PG P+AN+ FK YG+ISM QA+ FL DFKLGHYMKI P++MF+ Q+VGT+++ V+ TA
Sbjct: 520 PGYPVANMCFKVYGYISMQQAITFLQDFKLGHYMKIPPRTMFMAQIVGTLISCFVYLTTA 579
Query: 556 WWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFL 615
WWL+ +I NICD S WTCP D VFY+AS+IWG++GP+R+F G+Y +NWFFL
Sbjct: 580 WWLMETIPNICDS---VTNSVWTCPSDKVFYDASVIWGLIGPRRIFGDLGLYKSVNWFFL 636
Query: 616 IGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVY 675
+G IAP+ VWL S FP Q+WI+ IN+P++I+ S +PP +VNY TW + G F V+
Sbjct: 637 VGAIAPILVWLASRMFPRQEWIKLINMPVLISATSSMPPATAVNYTTWVLAGFLSGFVVF 696
Query: 676 RKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAP 735
R W R+ Y+LS LDAG+AF+G+LLY L + WWG E D CPLA CPTAP
Sbjct: 697 RYRPNLWQRYNYVLSGALDAGLAFMGVLLYMCLGLENV-SLDWWGNELDGCPLASCPTAP 755
Query: 736 GVHAEGCPV 744
G+ EGCP+
Sbjct: 756 GIIVEGCPL 764
>UniRef100_Q7FU78 Glutathione transporter [Oryza sativa]
Length = 766
Score = 823 bits (2127), Expect = 0.0
Identities = 383/724 (52%), Positives = 514/724 (70%), Gaps = 10/724 (1%)
Query: 24 DSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQI 83
+SP+E V LTVP++D+P P LTFR WVLG ASC +L+F+NQF YR PL IT++SAQI
Sbjct: 48 NSPVELVALTVPVSDEPETPVLTFRMWVLGTASCAVLSFLNQFFWYRKEPLTITAISAQI 107
Query: 84 IALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSGVYAINIIT 143
+PLG+LMAATLP W F+LNPGPF++KEHVLITIFA+SG+ VYAI++IT
Sbjct: 108 AVVPLGRLMAATLPEHAF-FRGRPWEFTLNPGPFNVKEHVLITIFANSGAGTVYAIHVIT 166
Query: 144 IVKAFYHRSIHPVAAYLLALSTQ---MLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRA 200
V+ FY +++ + L+ L+TQ MLG+GWAGIFRR+LV+ MWWP +LVQVSLF A
Sbjct: 167 GVRVFYGKTLSFFISLLVVLTTQYHQMLGFGWAGIFRRYLVEPASMWWPSNLVQVSLFSA 226
Query: 201 FHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGS 260
HEKE R +GG +R QFF V FV SFAYYI PGY FQ ++++S++C ++ S+ AQQ+GS
Sbjct: 227 LHEKEARRKGGLTRNQFFLVAFVCSFAYYIFPGYLFQMLTSLSWICWVFPSSVLAQQLGS 286
Query: 261 GMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNLYDAKK 320
G++GLG+G+ GLDW++++ +LGSPLA P FA +N+ GFF+ MY++ PI+YW N Y A+
Sbjct: 287 GLRGLGVGAIGLDWSSISSYLGSPLASPWFATVNVGVGFFIVMYIITPIAYWFNFYKAQN 346
Query: 321 FPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTAT 380
FP+ S F STG YNV+ I+++ F D ++Y +YLS + YG FA L AT
Sbjct: 347 FPIFSDGLFTSTGQKYNVSSIVDSH-FHFDTKAYEKNGPLYLSTSLLVTYGVGFATLAAT 405
Query: 381 ISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMALL 440
I H +LFHG ++W + ++ + + D+HT++M++ Y+QVPEWWF+ ILI + +
Sbjct: 406 IVHALLFHGS---EIWLLSKSAFQEKRMDIHTKLMRR-YKQVPEWWFICILIANIGTTIF 461
Query: 441 ACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKPL 500
ACE + ++LQLPWWG+L + +IA FTLPIG+I+ATTN GLNVITE +IG++YPG+P+
Sbjct: 462 ACEYYNEELQLPWWGVLFACSIAFFFTLPIGIIKATTNQTPGLNVITEYIIGYLYPGRPV 521
Query: 501 ANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLT 560
AN+ FK YG+ISM QAL FL DFKLGHYMKI P++MF+ Q+VGT +A+ V+ GTAWWL+
Sbjct: 522 ANMCFKVYGYISMKQALAFLEDFKLGHYMKIPPRTMFMAQVVGTSIAAFVYIGTAWWLME 581
Query: 561 SIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIA 620
+I NIC+ LLP SPWTCPGD VFY+AS+ WG++ P+R+F G Y LNWFFL G IA
Sbjct: 582 TIPNICNTELLPSDSPWTCPGDHVFYDASVTWGLISPRRIFGDLGTYSALNWFFLCGAIA 641
Query: 621 PVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKA 680
P+ VW FP Q WI I P++I +PP +VNY TW +VG + VYR +
Sbjct: 642 PLLVWFAHKTFPGQNWILLIKTPVLIGATFQMPPATAVNYTTWILVGFLSGYVVYRYRRD 701
Query: 681 WWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAPGVHAE 740
WW RH Y+LS LDAG+AF+ +L+Y L I WWG + D CPLA CPTA G+ +
Sbjct: 702 WWERHNYLLSGALDAGLAFMAVLIYLCLGLEDI-SLNWWGNDLDGCPLASCPTAKGIVVK 760
Query: 741 GCPV 744
GC V
Sbjct: 761 GCAV 764
>UniRef100_Q8S2I4 Isp4 protein-like [Oryza sativa]
Length = 755
Score = 808 bits (2086), Expect = 0.0
Identities = 394/752 (52%), Positives = 529/752 (69%), Gaps = 26/752 (3%)
Query: 12 IEDAEIDEYSIND--SPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGY 69
+E + E +++D SPIE+VRLTVP+TDD S P TFR W LGL SC+L++F+NQF Y
Sbjct: 9 VERGVVAEGALDDDASPIEEVRLTVPVTDDSSLPVWTFRMWTLGLLSCVLMSFLNQFFSY 68
Query: 70 RTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWS--FSLNPGPFSLKEHVLITI 127
RT PL +T ++ Q+ +LPLG ++A LP + + P SLNPGPF++KEHVL++I
Sbjct: 69 RTEPLIVTQITVQVASLPLGHILARVLPRRKFKAPALLGGGECSLNPGPFNMKEHVLVSI 128
Query: 128 FASSG----SSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDS 183
FA++G S YA+ I+ I++AFY RSI A+LL +TQ+LGYGWAG+ RR++V+
Sbjct: 129 FANAGCAFGSGSAYAVMIVDIIRAFYGRSISLFPAWLLITTTQVLGYGWAGLMRRYVVEP 188
Query: 184 PYMWWPESLVQVSLFRAFH----EKEKRPRG---GTSRLQFFFVVFVASFAYYIIPGYFF 236
MWWP +LVQVSLFRA H EKE+ G G S+ +FF + SF +Y +PGY F
Sbjct: 189 AQMWWPGTLVQVSLFRALHGKGEEKEENKEGSGGGMSQAKFFLIALACSFLWYAVPGYLF 248
Query: 237 QAISTVSFVCLIWKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIM 296
+++VS+VC I+ +S+TAQQ+GSGMKGLG+G+F LDW V+ FL SPL P FA NI+
Sbjct: 249 PTLTSVSWVCWIFSKSVTAQQLGSGMKGLGLGAFTLDWTAVSAFLYSPLISPFFATANIL 308
Query: 297 AGFFLYMYVLIPISYWN-NLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYS 355
AG+ L MYV++P+SYW +LY+A++FP+ SSH F +TG+TY++T I+N + F+IDM+ Y
Sbjct: 309 AGYVLLMYVVVPVSYWGLDLYNARRFPIFSSHLFTATGSTYDITAIVN-DRFEIDMDGYH 367
Query: 356 NYSKIYLSVAFAFEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIM 415
+I +S FA YG FA + AT++HV LFHG+ I + ++ + + DVHTR+M
Sbjct: 368 RMGRINMSTFFALSYGLGFATIAATVTHVALFHGKEIYRRFRAS----QRDKPDVHTRLM 423
Query: 416 KKNYEQVPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEA 475
K +Y VP WWF +L L + ++LL C +QLPWWG+L + +A +FTLPI +I A
Sbjct: 424 K-SYRDVPSWWFYAMLALSMAVSLLLCTVLRSAVQLPWWGLLFACAMAFVFTLPISIITA 482
Query: 476 TTNIRSGLNVITELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKS 535
TTN GLN+ITE VIG + PGKP+ANV FK YG++SM QA+ FL DFKLGHYMKI PKS
Sbjct: 483 TTNQTPGLNIITEYVIGLMLPGKPIANVCFKAYGYMSMSQAVSFLSDFKLGHYMKIPPKS 542
Query: 536 MFIVQLVGTVVASSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVV 595
MF+V+LVGTVVAS+V+ A+WLL SI NIC ++LLP SPWTCP D VF++AS+IWG+V
Sbjct: 543 MFLVKLVGTVVASTVNLVVAYWLLGSIPNICQDALLPADSPWTCPNDRVFFDASVIWGLV 602
Query: 596 GPKRMFTKDGVYPELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPV 655
GP+R+F G Y LNWFFL G + PV V+LL FP++ WI IN+P++I S +PP
Sbjct: 603 GPRRIFGPLGNYGALNWFFLAGAVGPVIVYLLHRAFPSKTWIPMINLPVLIGATSYMPPA 662
Query: 656 RSVNYITWGIVGIFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYG 715
+VNY +W I+GI FNF+V+R K WW R+ YILSA LDAGVAF+ +LLYFSL S
Sbjct: 663 TAVNYNSWLIIGIIFNFFVFRYRKLWWKRYNYILSAALDAGVAFMAVLLYFSL-SMENRS 721
Query: 716 PTWWGLEPDHCPLAKCPTAPGVHAEG---CPV 744
+WWG +HCPLA CPTA G++ CPV
Sbjct: 722 ISWWGTAGEHCPLASCPTAKGINLGADSVCPV 753
>UniRef100_Q7F9L4 OSJNBa0006A01.13 protein [Oryza sativa]
Length = 737
Score = 797 bits (2059), Expect = 0.0
Identities = 378/733 (51%), Positives = 515/733 (69%), Gaps = 25/733 (3%)
Query: 23 NDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQ 82
+ SP+EQVRLTVP TDDPS P TFR W +GL SC +L+++NQF YR+ P+ IT ++ Q
Sbjct: 18 DQSPVEQVRLTVPTTDDPSLPVWTFRMWTIGLLSCAMLSYINQFFSYRSEPIVITQITVQ 77
Query: 83 IIALPLGKLMAATLPTKPIQVPFTTWS--FSLNPGPFSLKEHVLITIFASSGSS----GV 136
+ ALP+G +A LP + FT + SLNPGPF++KEHVLI+IFA++G++ G
Sbjct: 78 VAALPIGHFLARVLPKRK----FTVFGRECSLNPGPFNVKEHVLISIFANAGAAFGNGGA 133
Query: 137 YAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVS 196
YAI+II I+KAFYHRSI + LL ++TQ+LGYGWAG+ R+++V+ +MWWP+SLVQVS
Sbjct: 134 YAIDIINIIKAFYHRSISFPTSLLLVITTQVLGYGWAGLMRKYVVEPAHMWWPQSLVQVS 193
Query: 197 LFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQ 256
L RA HEKE +R +FF + + S A+Y++PGY F + VS++C + S+T Q
Sbjct: 194 LLRALHEKENLRM---TRAKFFLIALICSAAWYVVPGYLFPTVGAVSWLCWAFPRSVTMQ 250
Query: 257 QIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWN-NL 315
QIGSGM GLG+G+F LDW TV FLGSPL P FAI+N+ GF L +YV++PI+YW NL
Sbjct: 251 QIGSGMSGLGVGAFTLDWATVVSFLGSPLVYPFFAIVNVWVGFVLLVYVMLPIAYWVLNL 310
Query: 316 YDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFA 375
Y A FP S+ FD TG Y ++ I+N + F++D ++Y+ KI+LS+ FA YG FA
Sbjct: 311 YQASTFPFFSASLFDHTGEEYRISEIVN-DRFELDTDAYARQGKIHLSLFFATSYGLGFA 369
Query: 376 ALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMV 435
+ AT+SHV LF+G + + +++ + DVHTR+M++ Y+ +P WWF +L L +
Sbjct: 370 TIAATLSHVTLFYGTEMYRRFRQAA----RENPDVHTRLMRR-YDDIPNWWFYGMLALAM 424
Query: 436 MMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIY 495
+ ALL C F ++QLPWW +L ++ +A FTLPI VI ATTN GLN+ITE V+G I
Sbjct: 425 VAALLLCTVFKDEVQLPWWALLCAVAVAAFFTLPISVITATTNTTPGLNIITEYVMGLIM 484
Query: 496 PGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTA 555
PGKP+ANV FK YG+ISM QA+ FL DFKLGHYMKI P+SMF+VQ +GT+VA +V+ A
Sbjct: 485 PGKPIANVCFKVYGYISMNQAVSFLTDFKLGHYMKIPPRSMFLVQFIGTIVAGTVNMSVA 544
Query: 556 WWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFL 615
WWLL+++ +ICD+ LP+GSPWTCPG VF++AS+IWG+VGP+R+F G Y LNWFFL
Sbjct: 545 WWLLSTVPHICDKKHLPEGSPWTCPGSRVFFDASVIWGLVGPRRIFGPLGYYGALNWFFL 604
Query: 616 IGLIAPVPVWLLSLKFPNQK-WIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYV 674
GL P VWLL+ P WI+ I++P+++ +++PP ++NY W VG FN+ V
Sbjct: 605 GGLAGPAVVWLLARALPRHAGWIRLIHLPVLLGATANMPPASTLNYTAWCSVGAVFNYLV 664
Query: 675 YRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWG---LEPDHCPLAKC 731
+R+ KAWW R+ Y+LSA +DAGVA +G+L+YF L S GI P WWG + DHC L+ C
Sbjct: 665 FRRRKAWWQRYNYVLSAAMDAGVAIMGVLIYFCLSSRGI-TPDWWGNSDINIDHCDLSTC 723
Query: 732 PTAPGVHAEGCPV 744
PTA GV EGCPV
Sbjct: 724 PTAKGVIVEGCPV 736
>UniRef100_Q9T095 Hypothetical protein AT4g27730 [Arabidopsis thaliana]
Length = 736
Score = 794 bits (2051), Expect = 0.0
Identities = 380/721 (52%), Positives = 510/721 (70%), Gaps = 9/721 (1%)
Query: 27 IEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQIIAL 86
+ +V LTVP TDD + P LTFR WVLG+ +CI+L+F+NQF YRT PL IT +SAQI +
Sbjct: 21 VPEVELTVPKTDDSTLPVLTFRMWVLGIGACIVLSFINQFFWYRTMPLSITGISAQIAVV 80
Query: 87 PLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSGSSGVYAINIITIVK 146
PLG LMA LPTK + T + F+LNPG F++KEHVLITIFA+SG+ VYA +I++ +K
Sbjct: 81 PLGHLMARVLPTKRF-LEGTRFQFTLNPGAFNVKEHVLITIFANSGAGSVYATHILSAIK 139
Query: 147 AFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAFHEKEK 206
+Y RS+ + A+L+ ++TQ+LG+GWAG+FR+ LV+ MWWP +LVQVSLF A HEKEK
Sbjct: 140 LYYKRSLPFLPAFLVMITTQILGFGWAGLFRKHLVEPGEMWWPSNLVQVSLFGALHEKEK 199
Query: 207 RPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGSGMKGLG 266
+ RGG SR QFF +V VASFAYYI PGY F ++++S+VC + +SI Q+GSG GLG
Sbjct: 200 KSRGGMSRTQFFLIVLVASFAYYIFPGYLFTMLTSISWVCWLNPKSILVNQLGSGEHGLG 259
Query: 267 IGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWNNLYDAKKFPLISS 326
IGS G DW T++ +LGSPLA P FA +N+ GF L MY++ P+ YW N+YDAK FP+ SS
Sbjct: 260 IGSIGFDWVTISAYLGSPLASPLFASVNVAIGFVLVMYIVTPVCYWLNIYDAKTFPIFSS 319
Query: 327 HTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTATISHVVL 386
F G+ Y+V I++++ F +D YS I +S FA YG FA L+ATI HV++
Sbjct: 320 QLFMGNGSRYDVLSIIDSK-FHLDRVVYSRTGSINMSTFFAVTYGLGFATLSATIVHVLV 378
Query: 387 FHGEMIVQMWKKTTTSL-KNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMALLACEGF 445
F+G +WK+T + KN+ D+HTRIMKKNY +VP WWF+ IL+L + + + +
Sbjct: 379 FNGS---DLWKQTRGAFQKNKKMDIHTRIMKKNYREVPLWWFLVILLLNIALIMFISVHY 435
Query: 446 GKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKPLANVAF 505
+QLPWWG+LL+ IA+ FT IGVI ATTN GLN+ITE VIG+IYP +P+AN+ F
Sbjct: 436 NATVQLPWWGVLLACAIAISFTPLIGVIAATTNQAPGLNIITEYVIGYIYPERPVANMCF 495
Query: 506 KTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIENI 565
K YG+ISM QAL F+ DFKLGHYMKI P+SMF+ Q+ GT+VA V+ GTAWWL+ I ++
Sbjct: 496 KVYGYISMTQALTFISDFKLGHYMKIPPRSMFMAQVAGTLVAVVVYTGTAWWLMEEIPHL 555
Query: 566 CDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIAPVPVW 625
CD SLLP S WTCP D VF++AS+IWG+VGP+R+F G Y +NWFFL+G IAP+ VW
Sbjct: 556 CDTSLLPSDSQWTCPMDRVFFDASVIWGLVGPRRVFGDLGEYSNVNWFFLVGAIAPLLVW 615
Query: 626 LLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKAWWARH 685
L + FP Q WI I+IP+++ + +PP +VN+ +W IV F ++++ + WW ++
Sbjct: 616 LATKMFPAQTWIAKIHIPVLVGATAMMPPATAVNFTSWLIVAFIFGHFIFKYRRVWWTKY 675
Query: 686 TYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGL--EPDHCPLAKCPTAPGVHAEGCP 743
Y+LS GLDAG AF+ +LL+ +L GI WWG + D CPLA CPTA GV +GCP
Sbjct: 676 NYVLSGGLDAGSAFMTILLFLALGRKGI-EVQWWGNSGDRDTCPLASCPTAKGVVVKGCP 734
Query: 744 V 744
V
Sbjct: 735 V 735
>UniRef100_Q8LBQ6 Isp4-like protein [Arabidopsis thaliana]
Length = 729
Score = 794 bits (2050), Expect = 0.0
Identities = 373/737 (50%), Positives = 518/737 (69%), Gaps = 17/737 (2%)
Query: 15 AEIDEYSIND-SPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNP 73
A DE+S D SPIE+VRLTV TDDP+ P TFR W LGL SC LL+F+NQF YRT P
Sbjct: 2 ATADEFSDEDTSPIEEVRLTVTNTDDPTLPVWTFRMWFLGLISCSLLSFLNQFFSYRTEP 61
Query: 74 LKITSVSAQIIALPLGKLMAATLPTKPIQVPFT-TWSFSLNPGPFSLKEHVLITIFASSG 132
L IT ++ Q+ LP+G +A LP +P + FSLNPGPF++KEHVLI+IFA++G
Sbjct: 62 LVITQITVQVATLPIGHFLAKVLPKTRFGLPGCGSARFSLNPGPFNMKEHVLISIFANAG 121
Query: 133 SS----GVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWW 188
S+ YA+ IITI+KAFY RSI +A +LL ++TQ+LGYGWAG+ R+++V+ +MWW
Sbjct: 122 SAFGSGSAYAVGIITIIKAFYGRSISFIAGWLLIITTQVLGYGWAGLLRKYVVEPAHMWW 181
Query: 189 PESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLI 248
P +LVQVSLFRA HEK+ + +R +FF + V SF +YI+PGY F ++++S+VC
Sbjct: 182 PSTLVQVSLFRALHEKDDQRM---TRAKFFVIALVCSFGWYIVPGYLFTTLTSISWVCWA 238
Query: 249 WKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIP 308
+ S+TAQQIGSGM+GLG+G+F LDW VA FL SPL P FAI N+ G+ L +Y+++P
Sbjct: 239 FPRSVTAQQIGSGMRGLGLGAFTLDWTAVASFLFSPLISPFFAIANVFIGYVLLIYLVLP 298
Query: 309 ISYWN-NLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFA 367
++YW + Y+A +FP+ SSH F S G TY++ I+N + F++D+ Y +I LS+ FA
Sbjct: 299 LAYWGFDSYNATRFPIFSSHLFTSVGKTYDIPAIVN-DNFELDLAKYEQQGRINLSMFFA 357
Query: 368 FEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWF 427
YG FA + +T++HV LF+G+ I + ++ S K + D+HTR+MK+ Y+ +P WWF
Sbjct: 358 LTYGLGFATIASTLTHVALFYGKEISERFR---VSYKGK-EDIHTRLMKR-YKDIPSWWF 412
Query: 428 VTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVIT 487
++L ++++L C ++Q+PWWG++ + +A +FTLPI +I ATTN GLN+IT
Sbjct: 413 YSMLAATLLISLALCVFLNDEVQMPWWGLVFASAMAFVFTLPISIITATTNQTPGLNIIT 472
Query: 488 ELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVA 547
E +G IYPG+P+ANV FK YG++SM QA+ FL DFKLGHYMKI P+SMF+VQ +GT++A
Sbjct: 473 EYAMGLIYPGRPIANVCFKVYGYMSMAQAVSFLNDFKLGHYMKIPPRSMFLVQFIGTILA 532
Query: 548 SSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVY 607
+++ AWW L SI+NIC E LLP SPWTCPGD VF++AS+IWG+VGPKR+F G Y
Sbjct: 533 GTINITVAWWQLNSIKNICQEELLPPNSPWTCPGDRVFFDASVIWGLVGPKRIFGSQGNY 592
Query: 608 PELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVG 667
+NWFFL G + PV VW L FP + WI +N+P+++ + +PP +VNY +W +VG
Sbjct: 593 AAMNWFFLGGALGPVIVWSLHKAFPKRSWIPLVNLPVLLGATAMMPPATAVNYNSWILVG 652
Query: 668 IFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCP 727
FN +V+R K+WW R+ Y+LSA +DAGVAF+ +LLYFS+ WWG +HC
Sbjct: 653 TIFNLFVFRYRKSWWQRYNYVLSAAMDAGVAFMAVLLYFSV-GMEEKSLDWWGTRGEHCD 711
Query: 728 LAKCPTAPGVHAEGCPV 744
LAKCPTA GV +GCPV
Sbjct: 712 LAKCPTARGVIVDGCPV 728
>UniRef100_Q6YUP9 Putative sexual differentiation process protein isp4 [Oryza sativa]
Length = 751
Score = 793 bits (2049), Expect = 0.0
Identities = 381/735 (51%), Positives = 517/735 (69%), Gaps = 23/735 (3%)
Query: 25 SPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQII 84
SPIE+VRLTVP DD + P TFR W +GL SC LL+F+NQF YRT PL +T ++ Q+
Sbjct: 24 SPIEEVRLTVPAGDDTALPVWTFRMWSIGLLSCALLSFLNQFFSYRTEPLIVTQITVQVA 83
Query: 85 ALPLGKLMAATLPTKPIQVPFTTWS--FSLNPGPFSLKEHVLITIFASSG----SSGVYA 138
+LP+G +A LP + + P +SLNPGPF++KEHVLI+IFA++G + YA
Sbjct: 84 SLPVGHFLARVLPRRAFRAPALLGGGEWSLNPGPFNMKEHVLISIFANAGCAFGNGNAYA 143
Query: 139 INIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLF 198
+ I+ I++AFY RSI VAA+LL ++TQ+LGYGWAG+ R+F+V+ +MWWP +LVQVSLF
Sbjct: 144 VMIVDIIRAFYKRSISFVAAWLLIITTQVLGYGWAGLMRKFVVEPAHMWWPGTLVQVSLF 203
Query: 199 RAFHEKEKRPRGGT--SRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQ 256
RA HEK++ P G SR +FF V + SFA+Y +PGY F ++++S+VC ++ +S+TAQ
Sbjct: 204 RALHEKDELPHGSRQISRSKFFLVALICSFAWYAVPGYLFPTLTSISWVCWVFSKSVTAQ 263
Query: 257 QIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWN-NL 315
Q+GSG+KGLG+G+F LDW ++ FL SPL P FA NI GF L++YVL+PI+YW +L
Sbjct: 264 QLGSGLKGLGVGAFSLDWTAISSFLFSPLISPFFATANIFVGFVLFLYVLVPIAYWGFDL 323
Query: 316 YDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFA 375
Y+AK FP+ SSH F S G +Y++T I+N + F++D+++Y+ +I LSV FA YG FA
Sbjct: 324 YNAKTFPIFSSHLFMSNGTSYDITAIVN-DKFELDIDAYNKLGRINLSVFFALAYGLSFA 382
Query: 376 ALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMV 435
+ +T++HV LF+G+ I ++ + + + D+HTR+MKK Y+ +P WWF +++ L +
Sbjct: 383 TIASTVTHVGLFYGKEIYHRFRAS----QKEKPDIHTRLMKK-YDDIPVWWFYSLMALSM 437
Query: 436 MMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRS---GLNVITELVIG 492
+AL+ C ++QLPWWG+L + +A IFTLPI +I ATTN S GLNVITE IG
Sbjct: 438 TVALILCTVLKHEVQLPWWGLLFACGMAFIFTLPISIISATTNQASYTPGLNVITEYAIG 497
Query: 493 FIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHF 552
I PG P+ANV FK YG++SM QA+ FL DFKLGHYMKI PKSMF+VQ +GT+VA +V+
Sbjct: 498 LIIPGHPIANVCFKVYGYMSMSQAIAFLSDFKLGHYMKIPPKSMFLVQFIGTIVAGTVNL 557
Query: 553 GTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNW 612
G AWWLL SI +IC +SL P SPWTCP D VF++AS+IWG++GP R+F G Y LNW
Sbjct: 558 GVAWWLLGSIHDICQDSL-PADSPWTCPNDRVFFDASVIWGLIGPIRIFGPHGNYSALNW 616
Query: 613 FFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNF 672
FFLIG PV V++ FPN+KWI N+P++I + +PP +VNY +W + G FNF
Sbjct: 617 FFLIGAAGPVIVYIFHKMFPNKKWITLTNLPVLIGATASMPPATAVNYNSWLLFGTIFNF 676
Query: 673 YVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCP 732
+V+R K WW R+ YILSA LDAGVAF+ +LLYFSL S WWG +HCPLA CP
Sbjct: 677 FVFRYRKKWWERYNYILSAALDAGVAFMAVLLYFSL-SMENRSIDWWGTAGEHCPLATCP 735
Query: 733 TAPGVH---AEGCPV 744
TA GV CPV
Sbjct: 736 TAKGVDLGPTSVCPV 750
>UniRef100_Q9FJD1 Isp4 protein [Arabidopsis thaliana]
Length = 733
Score = 793 bits (2048), Expect = 0.0
Identities = 377/737 (51%), Positives = 519/737 (70%), Gaps = 11/737 (1%)
Query: 9 NTVIEDAEIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLG 68
+T+ E DE SI + QV LTVP TDDP+ P +TFR WVLG+ +C+LL+F+NQF
Sbjct: 6 DTISESECDDEISI----VPQVELTVPKTDDPTSPTVTFRMWVLGITACVLLSFLNQFFW 61
Query: 69 YRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIF 128
YRTNPL I+SVSAQI +P+G LMA LPT+ T WSF++NPGPFS KEHVLIT+F
Sbjct: 62 YRTNPLTISSVSAQIAVVPIGHLMAKVLPTRRF-FEGTRWSFTMNPGPFSTKEHVLITVF 120
Query: 129 ASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWW 188
A+SGS VYA +I++ VK +Y R + + A L+ ++TQ+LG+GWAG++R+ LV+ MWW
Sbjct: 121 ANSGSGAVYATHILSAVKLYYKRRLDFLPALLVMITTQVLGFGWAGLYRKHLVEPGEMWW 180
Query: 189 PESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLI 248
P +LVQVSLFRA HEKE + + G SR QFF + + SF+YY++PGY F ++TVS++C I
Sbjct: 181 PSNLVQVSLFRALHEKENKSKWGISRNQFFVITLITSFSYYLLPGYLFTVLTTVSWLCWI 240
Query: 249 WKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIP 308
+SI Q+GSG GLGIGSFGLDW+T+A +LGSPLA P FA NI AGFFL MYV+ P
Sbjct: 241 SPKSILVNQLGSGSAGLGIGSFGLDWSTIASYLGSPLASPFFASANIAAGFFLVMYVITP 300
Query: 309 ISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAF 368
+ Y+ +LY+AK FP+ S F ++G Y VT I++ F +D ++Y+ +++S FA
Sbjct: 301 LCYYLDLYNAKTFPIYSGKLFVASGKEYKVTSIIDAN-FRLDRQAYAETGPVHMSTFFAV 359
Query: 369 EYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSL-KNQLGDVHTRIMKKNYEQVPEWWF 427
YG FA L+A+I HV++F+G+ +W +T + KN+ D+HT+IMK+NY++VP WWF
Sbjct: 360 TYGLGFATLSASIFHVLIFNGK---DLWTQTRGAFGKNKKMDIHTKIMKRNYKEVPLWWF 416
Query: 428 VTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVIT 487
++I + + + + C + Q+QLPWWG L+ IA+ FT +GVI ATTN GLN+IT
Sbjct: 417 LSIFAVNLAVIVFICIYYKTQIQLPWWGAFLACLIAIFFTPLVGVIMATTNQAPGLNIIT 476
Query: 488 ELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVA 547
E +IG+ YP +P+AN+ FKTYG+ISM Q+L FL D KLG YMKI P++MF+ Q+VGT+VA
Sbjct: 477 EYIIGYAYPERPVANICFKTYGYISMSQSLTFLSDLKLGTYMKIPPRTMFMAQVVGTLVA 536
Query: 548 SSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVY 607
+ GTAWWL+ I N+CD +LLP GS WTCP D VF++AS+IWG+VGP+RMF G Y
Sbjct: 537 VIAYAGTAWWLMAEIPNLCDTNLLPPGSQWTCPSDRVFFDASVIWGLVGPRRMFGDLGEY 596
Query: 608 PELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVG 667
+NWFF+ G IAP V+L S FPN+KWI I+IP++I + +PP +VN+ +W ++
Sbjct: 597 SNINWFFVGGAIAPALVYLASRLFPNKKWISDIHIPVLIGATAIMPPASAVNFTSWLVMA 656
Query: 668 IFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCP 727
F +V++ + WW R+ Y+LS G+DAG F+ +LL+ +LQ I WWG + CP
Sbjct: 657 FVFGHFVFKYRREWWQRYNYVLSGGMDAGTGFMSVLLFLALQRSEI-AIDWWGNSGEGCP 715
Query: 728 LAKCPTAPGVHAEGCPV 744
+AKCPTA GV GCPV
Sbjct: 716 VAKCPTAKGVVVHGCPV 732
>UniRef100_Q9FME8 Isp4-like protein [Arabidopsis thaliana]
Length = 729
Score = 793 bits (2047), Expect = 0.0
Identities = 373/737 (50%), Positives = 517/737 (69%), Gaps = 17/737 (2%)
Query: 15 AEIDEYSIND-SPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNP 73
A DE+S D SPIE+VRLTV TDDP+ P TFR W LGL SC LL+F+NQF YRT P
Sbjct: 2 ATADEFSDEDTSPIEEVRLTVTNTDDPTLPVWTFRMWFLGLISCSLLSFLNQFFSYRTEP 61
Query: 74 LKITSVSAQIIALPLGKLMAATLPTKPIQVPFT-TWSFSLNPGPFSLKEHVLITIFASSG 132
L IT ++ Q+ LP+G +A LP +P + FSLNPGPF++KEHVLI+IFA++G
Sbjct: 62 LVITQITVQVATLPIGHFLAKVLPKTRFGLPGCGSARFSLNPGPFNMKEHVLISIFANAG 121
Query: 133 SS----GVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWW 188
S+ YA+ IITI+KAFY RSI +A +LL ++TQ+LGYGWAG+ R+++V+ +MWW
Sbjct: 122 SAFGSGSAYAVGIITIIKAFYGRSISFIAGWLLIITTQVLGYGWAGLLRKYVVEPAHMWW 181
Query: 189 PESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLI 248
P +LVQVSLFRA HEK+ + +R +FF + V SF +YI+PGY F ++++S+VC
Sbjct: 182 PSTLVQVSLFRALHEKDDQRM---TRAKFFVIALVCSFGWYIVPGYLFTTLTSISWVCWA 238
Query: 249 WKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIP 308
+ S+TAQQIGSGM+GLG+G+F LDW VA FL SPL P FAI N+ G+ L +Y ++P
Sbjct: 239 FPRSVTAQQIGSGMRGLGLGAFTLDWTAVASFLFSPLISPFFAIANVFIGYVLLIYFVLP 298
Query: 309 ISYWN-NLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFA 367
++YW + Y+A +FP+ SSH F S G TY++ I+N + F++D+ Y +I LS+ FA
Sbjct: 299 LAYWGFDSYNATRFPIFSSHLFTSVGNTYDIPAIVN-DNFELDLAKYEQQGRINLSMFFA 357
Query: 368 FEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWF 427
YG FA + +T++HV LF+G+ I + ++ S K + D+HTR+MK+ Y+ +P WWF
Sbjct: 358 LTYGLGFATIASTLTHVALFYGKEISERFR---VSYKGK-EDIHTRLMKR-YKDIPSWWF 412
Query: 428 VTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVIT 487
++L ++++L C ++Q+PWWG++ + +A +FTLPI +I ATTN GLN+IT
Sbjct: 413 YSMLAATLLISLALCVFLNDEVQMPWWGLVFASAMAFVFTLPISIITATTNQTPGLNIIT 472
Query: 488 ELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVA 547
E +G IYPG+P+ANV FK YG++SM QA+ FL DFKLGHYMKI P+SMF+VQ +GT++A
Sbjct: 473 EYAMGLIYPGRPIANVCFKVYGYMSMAQAVSFLNDFKLGHYMKIPPRSMFLVQFIGTILA 532
Query: 548 SSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVY 607
+++ AWW L SI+NIC E LLP SPWTCPGD VF++AS+IWG+VGPKR+F G Y
Sbjct: 533 GTINITVAWWQLNSIKNICQEELLPPNSPWTCPGDRVFFDASVIWGLVGPKRIFGSQGNY 592
Query: 608 PELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVG 667
+NWFFL G + PV VW L FP + WI +N+P+++ + +PP +VNY +W +VG
Sbjct: 593 AAMNWFFLGGALGPVIVWSLHKAFPKRSWIPLVNLPVLLGATAMMPPATAVNYNSWILVG 652
Query: 668 IFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCP 727
FN +V+R K+WW R+ Y+LSA +DAGVAF+ +LLYFS+ WWG +HC
Sbjct: 653 TIFNLFVFRYRKSWWQRYNYVLSAAMDAGVAFMAVLLYFSV-GMEEKSLDWWGTRGEHCD 711
Query: 728 LAKCPTAPGVHAEGCPV 744
LAKCPTA GV +GCPV
Sbjct: 712 LAKCPTARGVIVDGCPV 728
>UniRef100_Q9FJD2 Isp4 protein [Arabidopsis thaliana]
Length = 741
Score = 790 bits (2040), Expect = 0.0
Identities = 378/742 (50%), Positives = 516/742 (68%), Gaps = 9/742 (1%)
Query: 6 VSQNTVIEDAEIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQ 65
V ++ + + E ++ +E+V LTVP TDDP+ P LTFR W LGL +CI+L+F+NQ
Sbjct: 5 VEDSSRVMEIEGQNDDLDRCVVEEVELTVPKTDDPTLPVLTFRMWTLGLGACIILSFINQ 64
Query: 66 FLGYRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLI 125
F YR PL I+ +SAQI +PLG LMA LPT+ + + W FS+NPGPF++KEHVLI
Sbjct: 65 FFWYRQMPLTISGISAQIAVVPLGHLMAKVLPTRMF-LEGSKWEFSMNPGPFNVKEHVLI 123
Query: 126 TIFASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPY 185
TIFA+SG+ VYA +I++ +K +Y RS+ + A+LL ++TQ LG+GWAG+FR+ LV+
Sbjct: 124 TIFANSGAGTVYATHILSAIKLYYKRSLPFLPAFLLMITTQFLGFGWAGLFRKHLVEPGE 183
Query: 186 MWWPESLVQVSLFRAFHEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFV 245
MWWP +LVQVSLF A HEKEK+ +GG +R+QFF +V V SFAYYI+PGY F I+++S++
Sbjct: 184 MWWPSNLVQVSLFSALHEKEKKKKGGMTRIQFFLIVLVTSFAYYILPGYLFTMITSISWI 243
Query: 246 CLIWKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYV 305
C + +S+ Q+GSG +GLGIG+ G+DW T++ +LGSPLA P FA IN+ GF + MYV
Sbjct: 244 CWLGPKSVLVHQLGSGEQGLGIGAIGIDWATISSYLGSPLASPLFATINVTIGFVVIMYV 303
Query: 306 LIPISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVA 365
PI YW N+Y AK +P+ SS F G++Y+V I++ + F +D + Y+ I +S
Sbjct: 304 ATPICYWLNIYKAKTYPIFSSGLFMGNGSSYDVLSIIDKK-FHLDRDIYAKTGPINMSTF 362
Query: 366 FAFEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSL-KNQLGDVHTRIMKKNYEQVPE 424
FA YG FA L+ATI HV+LF+G +WK+T + +N+ D HTRIMKKNY +VP
Sbjct: 363 FAVTYGLGFATLSATIVHVLLFNGR---DLWKQTRGAFQRNKKMDFHTRIMKKNYREVPM 419
Query: 425 WWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLN 484
WWF IL+L + + + + +QLPWWG+LL+ IA+ FT IGVI ATTN GLN
Sbjct: 420 WWFYVILVLNIALIMFISFYYNATVQLPWWGVLLACAIAVFFTPLIGVIAATTNQEPGLN 479
Query: 485 VITELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGT 544
VITE VIG++YP +P+AN+ FK YG+ISM QAL F+ DFKLG YMKI P+SMF+ Q+VGT
Sbjct: 480 VITEYVIGYLYPERPVANMCFKVYGYISMTQALTFIQDFKLGLYMKIPPRSMFMAQVVGT 539
Query: 545 VVASSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKD 604
+V+ V+ GTAWWL+ I ++CD+SLLP S WTCP D VF++AS+IWG+VGP+RMF
Sbjct: 540 LVSVVVYTGTAWWLMVDIPHLCDKSLLPPDSEWTCPMDRVFFDASVIWGLVGPRRMFGNL 599
Query: 605 GVYPELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWG 664
G Y +NWFFL+G IAP VWL + FP KWI I+ P+I+ S +PP +VN+ +W
Sbjct: 600 GEYAAINWFFLVGAIAPFFVWLATKAFPAHKWISKIHFPVILGATSMMPPAMAVNFTSWC 659
Query: 665 IVGIFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPD 724
IV F ++Y+ + WW ++ Y+LS GLDAG AF+ +L++ S+ GI G WWG D
Sbjct: 660 IVAFVFGHFLYKYKRQWWKKYNYVLSGGLDAGTAFMTILIFLSVGRKGI-GLLWWGNADD 718
Query: 725 --HCPLAKCPTAPGVHAEGCPV 744
+C LA CPTA GV GCPV
Sbjct: 719 STNCSLASCPTAKGVIMHGCPV 740
>UniRef100_Q940I5 Hypothetical protein Z97341.3 [Arabidopsis thaliana]
Length = 737
Score = 790 bits (2039), Expect = 0.0
Identities = 374/724 (51%), Positives = 503/724 (68%), Gaps = 14/724 (1%)
Query: 26 PIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQIIA 85
P+E+V L VP TDDPS P +TFRTW LGL SC+LL F+N F YRT PL I+++ QI
Sbjct: 22 PVEEVALVVPETDDPSLPVMTFRTWFLGLTSCVLLIFLNTFFTYRTQPLTISAILMQIAV 81
Query: 86 LPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSG----SSGVYAINI 141
LP+GK MA TLPT + WSFSLNPGPF++KEHV+ITIFA+ G Y+I
Sbjct: 82 LPIGKFMARTLPTTSHNL--LGWSFSLNPGPFNIKEHVIITIFANCGVAYGGGDAYSIGA 139
Query: 142 ITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAF 201
IT++KA+Y +S+ + + L+TQ+LGYGWAGI RR+LVD MWWP +L QVSLFRA
Sbjct: 140 ITVMKAYYKQSLSFICGLFIVLTTQILGYGWAGILRRYLVDPVDMWWPSNLAQVSLFRAL 199
Query: 202 HEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGSG 261
HEKE + +G T R++FF V ASF YY +PGY F ++ S+VC W SITAQQ+GSG
Sbjct: 200 HEKENKSKGLT-RMKFFLVALGASFIYYALPGYLFPILTFFSWVCWAWPNSITAQQVGSG 258
Query: 262 MKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWN-NLYDAKK 320
GLG+G+F LDW ++ + GSPL P +I+N+ GF +++Y+++P+ YW N +DA+K
Sbjct: 259 YHGLGVGAFTLDWAGISAYHGSPLVAPWSSILNVGVGFIMFIYIIVPVCYWKFNTFDARK 318
Query: 321 FPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTAT 380
FP+ S+ F ++G Y+ T+IL T FD+D+ +Y+NY K+YLS FA G FA TAT
Sbjct: 319 FPIFSNQLFTTSGQKYDTTKIL-TPQFDLDIGAYNNYGKLYLSPLFALSIGSGFARFTAT 377
Query: 381 ISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMALL 440
++HV LF+G I WK+T +++ D+H ++M+ +Y++VPEWWF +L V M+LL
Sbjct: 378 LTHVALFNGRDI---WKQTWSAVNTTKLDIHGKLMQ-SYKKVPEWWFYILLAGSVAMSLL 433
Query: 441 ACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKPL 500
+ + +QLPWWG+L + +A I TLPIGVI+ATTN + G ++I + +IG+I PGKP+
Sbjct: 434 MSFVWKESVQLPWWGMLFAFALAFIVTLPIGVIQATTNQQPGYDIIGQFIIGYILPGKPI 493
Query: 501 ANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLT 560
AN+ FK YG IS V AL FL D KLGHYMKI P+ M+ QLVGTVVA V+ G AWW+L
Sbjct: 494 ANLIFKIYGRISTVHALSFLADLKLGHYMKIPPRCMYTAQLVGTVVAGVVNLGVAWWMLE 553
Query: 561 SIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIA 620
SI++ICD SPWTCP V ++AS+IWG++GP+R+F G+Y L W FLIG +
Sbjct: 554 SIQDICDIEGDHPNSPWTCPKYRVTFDASVIWGLIGPRRLFGPGGMYRNLVWLFLIGAVL 613
Query: 621 PVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKA 680
PVPVW LS FPN+KWI INIP+I G + +PP N +W + G FN++V+ K
Sbjct: 614 PVPVWALSKIFPNKKWIPLINIPVISYGFAGMPPATPTNIASWLVTGTIFNYFVFNYHKR 673
Query: 681 WWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAPGVHAE 740
WW ++ Y+LSA LDAG AF+G+LL+F+LQ+ G + WWG E DHCPLA CPTAPG+ A+
Sbjct: 674 WWQKYNYVLSAALDAGTAFMGVLLFFALQNAG-HDLKWWGTEVDHCPLASCPTAPGIKAK 732
Query: 741 GCPV 744
GCPV
Sbjct: 733 GCPV 736
>UniRef100_Q8H0V0 Isp4 like protein [Arabidopsis thaliana]
Length = 737
Score = 788 bits (2034), Expect = 0.0
Identities = 373/724 (51%), Positives = 502/724 (68%), Gaps = 14/724 (1%)
Query: 26 PIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQIIA 85
P+E+V L VP TDDPS P +TFR W LGL SC+LL F+N F YRT PL I+++ QI
Sbjct: 22 PVEEVALVVPETDDPSLPVMTFRAWFLGLTSCVLLIFLNTFFTYRTQPLTISAILMQIAV 81
Query: 86 LPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSG----SSGVYAINI 141
LP+GK MA TLPT + WSFSLNPGPF++KEHV+ITIFA+ G Y+I
Sbjct: 82 LPIGKFMARTLPTTSHNL--LGWSFSLNPGPFNIKEHVIITIFANCGVAYGGGDAYSIGA 139
Query: 142 ITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAF 201
IT++KA+Y +S+ + + L+TQ+LGYGWAGI RR+LVD MWWP +L QVSLFRA
Sbjct: 140 ITVMKAYYKQSLSFICGLFIVLTTQILGYGWAGILRRYLVDPVDMWWPSNLAQVSLFRAL 199
Query: 202 HEKEKRPRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIGSG 261
HEKE + +G T R++FF V ASF YY +PGY F ++ S+VC W SITAQQ+GSG
Sbjct: 200 HEKENKSKGLT-RMKFFLVALGASFIYYALPGYLFPILTFFSWVCWAWPNSITAQQVGSG 258
Query: 262 MKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWN-NLYDAKK 320
GLG+G+F LDW ++ + GSPL P +I+N+ GF +++Y+++P+ YW N +DA+K
Sbjct: 259 YHGLGVGAFTLDWAGISAYHGSPLVAPWSSILNVGVGFIMFIYIIVPVCYWKFNTFDARK 318
Query: 321 FPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALTAT 380
FP+ S+ F ++G Y+ T+IL T FD+D+ +Y+NY K+YLS FA G FA TAT
Sbjct: 319 FPIFSNQLFTTSGQKYDTTKIL-TPQFDLDIGAYNNYGKLYLSPLFALSIGSGFARFTAT 377
Query: 381 ISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMALL 440
++HV LF+G I WK+T +++ D+H ++M+ +Y++VPEWWF +L V M+LL
Sbjct: 378 LTHVALFNGRDI---WKQTWSAVNTTKLDIHGKLMQ-SYKKVPEWWFYILLAGSVAMSLL 433
Query: 441 ACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGKPL 500
+ + +QLPWWG+L + +A I TLPIGVI+ATTN + G ++I + +IG+I PGKP+
Sbjct: 434 MSFVWKESVQLPWWGMLFAFALAFIVTLPIGVIQATTNQQPGYDIIGQFIIGYILPGKPI 493
Query: 501 ANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWLLT 560
AN+ FK YG IS V AL FL D KLGHYMKI P+ M+ QLVGTVVA V+ G AWW+L
Sbjct: 494 ANLIFKIYGRISTVHALSFLADLKLGHYMKIPPRCMYTAQLVGTVVAGVVNLGVAWWMLE 553
Query: 561 SIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGLIA 620
SI++ICD SPWTCP V ++AS+IWG++GP+R+F G+Y L W FLIG +
Sbjct: 554 SIQDICDIEGDHPNSPWTCPKYRVTFDASVIWGLIGPRRLFGPGGMYRNLVWLFLIGAVL 613
Query: 621 PVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKFKA 680
PVPVW LS FPN+KWI INIP+I G + +PP N +W + G FN++V+ K
Sbjct: 614 PVPVWALSKIFPNKKWIPLINIPVISYGFAGMPPATPTNIASWLVTGTIFNYFVFNYHKR 673
Query: 681 WWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAPGVHAE 740
WW ++ Y+LSA LDAG AF+G+LL+F+LQ+ G + WWG E DHCPLA CPTAPG+ A+
Sbjct: 674 WWQKYNYVLSAALDAGTAFMGVLLFFALQNAG-HDLKWWGTEVDHCPLASCPTAPGIKAK 732
Query: 741 GCPV 744
GCPV
Sbjct: 733 GCPV 736
>UniRef100_Q6TUA0 Putative glutathione transporter [Zea mays]
Length = 721
Score = 774 bits (1999), Expect = 0.0
Identities = 360/726 (49%), Positives = 501/726 (68%), Gaps = 14/726 (1%)
Query: 27 IEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQIIAL 86
+E+V L VP TDDPS P +TFR W LGL SC++L F+N F YRT PL I+ + AQI+ L
Sbjct: 1 VEEVALVVPETDDPSMPVMTFRAWALGLGSCVVLIFLNTFFTYRTQPLTISGILAQILVL 60
Query: 87 PLGKLMAATLPTKPIQV-PFTTWSFSLNPGPFSLKEHVLITIFASSGSS----GVYAINI 141
P G+ +AA LP + +++ SF+LNPGPF++KEHV+ITIFA+ G S Y+I
Sbjct: 61 PAGRFLAAVLPDREVRILGGRLGSFNLNPGPFNVKEHVIITIFANCGVSYGGGDAYSIGA 120
Query: 142 ITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRAF 201
IT++KA+Y +++ A L+ L+TQ+LGYGWAG+ RR+LVD MWWP +L QVSLFRA
Sbjct: 121 ITVMKAYYKQTLSFACALLIVLTTQILGYGWAGLLRRYLVDPAEMWWPSNLAQVSLFRAL 180
Query: 202 HEKEKR--PRGGTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESITAQQIG 259
HEKE+ G SR++FF +VF ASFAYY +PGY ++ S+ C W SITAQQ+G
Sbjct: 181 HEKEEEGGKSRGPSRMRFFLIVFFASFAYYALPGYLLPILTFFSWACWAWPHSITAQQVG 240
Query: 260 SGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWN-NLYDA 318
SG GLG+G+F LDW ++ + GSPL P +I N GF +++YV++P+ YW N +DA
Sbjct: 241 SGYHGLGVGAFTLDWAGISAYHGSPLVAPWASIANTAVGFVMFIYVIVPLCYWQFNTFDA 300
Query: 319 KKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFCFAALT 378
++FP+ S+ F ++G Y+ T++L T+ FD+++ +Y +Y K+YLS FA G F T
Sbjct: 301 RRFPIFSNQLFTASGQKYDTTKVL-TKDFDLNVAAYDSYGKLYLSPLFAISIGSGFLRFT 359
Query: 379 ATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWFVTILILMVMMA 438
ATI HV+LFHG MWK++ +++ DVH ++M++ Y QVP+WWF+ +L+ V+++
Sbjct: 360 ATIVHVLLFHGS---DMWKQSKSAMNAVKPDVHAKLMQR-YRQVPQWWFLMLLLGSVVVS 415
Query: 439 LLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVIGFIYPGK 498
LL + +++QLPWWG+L + +A + TLPIGVI+ATTN + G ++I + +IG+ PGK
Sbjct: 416 LLMSFVWKEEMQLPWWGMLFAFALAFVVTLPIGVIQATTNQQPGYDIIAQFMIGYALPGK 475
Query: 499 PLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVHFGTAWWL 558
P+AN+ FK YG IS V AL FL D KLGHYMKI P+ M+ QLVGTVVA V+ AWW+
Sbjct: 476 PIANLLFKIYGRISTVHALSFLADLKLGHYMKIPPRCMYTAQLVGTVVAGVVNLAVAWWM 535
Query: 559 LTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELNWFFLIGL 618
L +IENICD L SPWTCP V ++AS+IWG++GP R+F + G+Y L W FL+G
Sbjct: 536 LDNIENICDVEALHPDSPWTCPKYRVTFDASVIWGLIGPGRLFGQHGLYRNLVWLFLVGA 595
Query: 619 IAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFNFYVYRKF 678
+ PVPVWLLS FP +KWI IN+P+I G + +PP N TW + G FN++V+R
Sbjct: 596 VLPVPVWLLSRAFPEKKWIALINVPVISYGFAGMPPATPTNIATWLVTGTIFNYFVFRYR 655
Query: 679 KAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKCPTAPGVH 738
K WW ++ Y+LSA LDAG AF+G+L++F+LQ+ + WWG E DHCPLA CP APG+
Sbjct: 656 KGWWQKYNYVLSAALDAGTAFMGVLIFFALQN-AHHELKWWGTEVDHCPLASCPPAPGIA 714
Query: 739 AEGCPV 744
+GCPV
Sbjct: 715 VKGCPV 720
>UniRef100_Q75LM0 Putative oligopeptide transporter protein [Oryza sativa]
Length = 757
Score = 764 bits (1973), Expect = 0.0
Identities = 358/733 (48%), Positives = 503/733 (67%), Gaps = 20/733 (2%)
Query: 26 PIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTNPLKITSVSAQIIA 85
P+E+V L VP TDDP+ P +TFR W LGLASC++L F+N F YRT PL I+ + AQI+
Sbjct: 30 PVEEVALVVPETDDPTTPVMTFRAWTLGLASCVVLIFLNTFFTYRTQPLTISGILAQILV 89
Query: 86 LPLGKLMAATLPTKPIQV-PFTTWSFSLNPGPFSLKEHVLITIFASSGSS----GVYAIN 140
LP G+ MAA LP++ +++ SF+LNPGPF++KEHV+ITIFA+ G S Y+I
Sbjct: 90 LPAGQFMAAVLPSREVRLLGGRLGSFNLNPGPFNIKEHVIITIFANCGVSYGGGDAYSIG 149
Query: 141 IITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESLVQVSLFRA 200
IT++KA+Y +S+ + A L+ L+TQ+LGYGWAG+ RR+LVD MWWP +L QVSLFRA
Sbjct: 150 AITVMKAYYKQSLSFLCALLIVLTTQILGYGWAGMLRRYLVDPADMWWPSNLAQVSLFRA 209
Query: 201 FHEKEKRPRG------GTSRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLIWKESIT 254
HEKE G G +R++FF + F ASFAYY +PGY ++ S+ C W SIT
Sbjct: 210 LHEKEGGDGGKGSSSRGPTRMRFFLIFFFASFAYYALPGYLLPILTFFSWACWAWPHSIT 269
Query: 255 AQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIPISYWN- 313
AQQ+GSG GLG+G+F LDW ++ + GSPL P +I N AGF +++Y+++P+ YW
Sbjct: 270 AQQVGSGYHGLGVGAFTLDWAGISAYHGSPLVAPWSSIANTAAGFVMFIYLIVPLCYWKF 329
Query: 314 NLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDIDMESYSNYSKIYLSVAFAFEYGFC 373
+ +DA+KFP+ S+ F ++G Y+ T++L E FD+++ +Y +Y K+YLS FA G
Sbjct: 330 DTFDARKFPIFSNQLFTASGQKYDTTKVLTRE-FDLNVAAYESYGKLYLSPLFAISIGSG 388
Query: 374 FAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLG--DVHTRIMKKNYEQVPEWWFVTIL 431
F TATI HV LFHG I W+++ +++ + DVH ++M++ Y+QVP+WWF+ +L
Sbjct: 389 FLRFTATIVHVALFHGGDI---WRQSRSAMSSAAAKMDVHAKLMRR-YKQVPQWWFLVLL 444
Query: 432 ILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITELVI 491
+ V ++L+ + +++QLPWWG+L + +A + TLPIGVI+ATTN + G ++I + +I
Sbjct: 445 VGSVAVSLVMSFVYREEVQLPWWGMLFAFALAFVVTLPIGVIQATTNQQPGYDIIAQFMI 504
Query: 492 GFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVASSVH 551
G+ PGKP+AN+ FK YG IS V AL FL D KLGHYMKI P+ M+ QLVGTVVA V+
Sbjct: 505 GYALPGKPIANLLFKIYGRISTVHALSFLADLKLGHYMKIPPRCMYTAQLVGTVVAGVVN 564
Query: 552 FGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVYPELN 611
AWW+L SI+NICD L SPWTCP V ++AS+IWG++GP R+F + G+Y L
Sbjct: 565 LAVAWWMLGSIDNICDVEALHPDSPWTCPKYRVTFDASVIWGLIGPARLFGRHGLYRNLV 624
Query: 612 WFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVGIFFN 671
W FL G + PVPVWLLS FP +KWI IN+P+I G + +PP N +W + G FN
Sbjct: 625 WLFLAGAVLPVPVWLLSRAFPEKKWIALINVPVISYGFAGMPPATPTNIASWLVTGTIFN 684
Query: 672 FYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWGLEPDHCPLAKC 731
++V++ K WW ++ Y+LSA LDAG AF+G+L++F+LQ+ + WWG DHCPLA C
Sbjct: 685 YFVFKYRKGWWQKYNYVLSAALDAGTAFMGVLIFFALQN-AHHELKWWGTAVDHCPLASC 743
Query: 732 PTAPGVHAEGCPV 744
PTAPG+ +GCPV
Sbjct: 744 PTAPGIAVKGCPV 756
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.141 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,270,805,522
Number of Sequences: 2790947
Number of extensions: 55451416
Number of successful extensions: 173608
Number of sequences better than 10.0: 183
Number of HSP's better than 10.0 without gapping: 129
Number of HSP's successfully gapped in prelim test: 54
Number of HSP's that attempted gapping in prelim test: 172473
Number of HSP's gapped (non-prelim): 241
length of query: 745
length of database: 848,049,833
effective HSP length: 135
effective length of query: 610
effective length of database: 471,271,988
effective search space: 287475912680
effective search space used: 287475912680
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 79 (35.0 bits)
Medicago: description of AC143341.7


