

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143338.3 - phase: 0
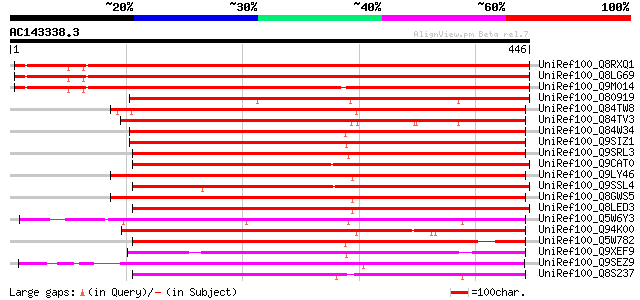
(446 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8RXQ1 Hypothetical protein At5g01620 [Arabidopsis tha... 649 0.0
UniRef100_Q8LG69 Hypothetical protein [Arabidopsis thaliana] 644 0.0
UniRef100_Q9M014 Hypothetical protein F7A7_140 [Arabidopsis thal... 639 0.0
UniRef100_O80919 Expressed protein [Arabidopsis thaliana] 397 e-109
UniRef100_Q84TW8 Hypothetical protein OSJNBa0094J08.9 [Oryza sat... 390 e-107
UniRef100_Q84TV3 Hypothetical protein OSJNBa0094J08.14 [Oryza sa... 385 e-105
UniRef100_Q84W34 Hypothetical protein At2g40320 [Arabidopsis tha... 375 e-102
UniRef100_Q9SIZ1 Hypothetical protein At2g40320 [Arabidopsis tha... 374 e-102
UniRef100_Q9SRL3 F9F8.15 protein [Arabidopsis thaliana] 371 e-101
UniRef100_Q9CAT0 Hypothetical protein T18K17.20 [Arabidopsis tha... 361 2e-98
UniRef100_Q9LY46 Hypothetical protein F27K19_170 [Arabidopsis th... 353 7e-96
UniRef100_Q9SSL4 F3N23.34 protein [Arabidopsis thaliana] 352 2e-95
UniRef100_Q8GWS5 Hypothetical protein At3g55990/F27K19_170 [Arab... 351 2e-95
UniRef100_Q8LED3 Hypothetical protein [Arabidopsis thaliana] 348 2e-94
UniRef100_Q5W6Y3 Hypothetical protein OSJNBa0036C12.14 [Oryza sa... 344 3e-93
UniRef100_Q94K00 Hypothetical protein At2g40150; T7M7.4 [Arabido... 340 5e-92
UniRef100_Q5W782 Hypothetical protein OJ1537_B05.7 [Oryza sativa] 329 1e-88
UniRef100_Q9XEF9 Hypothetical protein T07M07.22 [Arabidopsis tha... 327 4e-88
UniRef100_Q9SEZ9 At2g40160/T7M7.25 [Arabidopsis thaliana] 315 2e-84
UniRef100_Q8S237 P0446G04.14 protein [Oryza sativa] 305 2e-81
>UniRef100_Q8RXQ1 Hypothetical protein At5g01620 [Arabidopsis thaliana]
Length = 449
Score = 649 bits (1673), Expect = 0.0
Identities = 305/448 (68%), Positives = 354/448 (78%), Gaps = 8/448 (1%)
Query: 5 KWNKKKVPIFPFIVLLLLVFIAFSTLHNGSTIHKFHENPLHIHQE--QASSTTYVKPNLP 62
+W++KK + P LL ++ + F L N +I + H + Q + S+ +VKPN+P
Sbjct: 4 RWSRKKSRL-PLAGLLFILVVTFMILFNERSIQQIHHHAASHTQNLREPSTFDFVKPNVP 62
Query: 63 --NHLKKSQEILDRYSRCNSTVGYSGRKIARRGGSKSSSNRRVSSES-CDVFSGKWVFDN 119
N+L + E+LDR+S+CNST YSG+KI + + E CDVFSGKWVFDN
Sbjct: 63 RINYLG-AHEVLDRFSKCNSTKEYSGKKIGWVDPFEDHPGQVTKEEQKCDVFSGKWVFDN 121
Query: 120 AS-YPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWNVKEMWEKLRGKRL 178
+S YPL+ ES CPYMSDQLAC KHGR DL YQHWRWQPH C+LKRWN EMWEKLRGKRL
Sbjct: 122 SSSYPLHKESQCPYMSDQLACQKHGRKDLEYQHWRWQPHACNLKRWNAIEMWEKLRGKRL 181
Query: 179 MFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNATVEFLWAPLLAESN 238
MFVGDSLNRGQWISMVCLLQS IP DK+SMSPNAHLTIFRAE+YNATVEFLWAPLL ESN
Sbjct: 182 MFVGDSLNRGQWISMVCLLQSVIPRDKQSMSPNAHLTIFRAEDYNATVEFLWAPLLVESN 241
Query: 239 SDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQGPVKLLWTDEENGACEEL 298
SDDPVNHRL ERIIRPDSVLKHAS W+HADIL+FNTYLWWRQ VKL W+ EE G+CEE+
Sbjct: 242 SDDPVNHRLSERIIRPDSVLKHASKWQHADILIFNTYLWWRQDSVKLRWSSEEKGSCEEV 301
Query: 299 DGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREWNPESEGNCYGEKKPIDV 358
ME+AM +W DWV++ VDP KKRVFFVTMSPTH WSREWNP SEGNCYGEKKPI+
Sbjct: 302 KSAEGMEMAMDSWGDWVANNVDPNKKRVFFVTMSPTHQWSREWNPGSEGNCYGEKKPIEE 361
Query: 359 ESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLS 418
ESYWGSGSD+PTM V+ +L L KVSV+N+TQLSEYRKDGHPS++RKFWEPL ++L
Sbjct: 362 ESYWGSGSDIPTMRMVKRVLERLGPKVSVINITQLSEYRKDGHPSVYRKFWEPLNEDRLK 421
Query: 419 NPSSYSDCIHWCLPGVPDTWNELLFHFL 446
NP+SYSDC HWC+PGVPD WN+LLFHFL
Sbjct: 422 NPASYSDCTHWCVPGVPDVWNQLLFHFL 449
>UniRef100_Q8LG69 Hypothetical protein [Arabidopsis thaliana]
Length = 449
Score = 644 bits (1660), Expect = 0.0
Identities = 303/448 (67%), Positives = 353/448 (78%), Gaps = 8/448 (1%)
Query: 5 KWNKKKVPIFPFIVLLLLVFIAFSTLHNGSTIHKFHENPLHIHQE--QASSTTYVKPNLP 62
+W++KK + P LL ++ + F L N +I + H + Q + S+ +VKPN+P
Sbjct: 4 RWSRKKSRL-PLAGLLFILVVTFMILFNERSIQQIHHHAASHTQNLREPSTFDFVKPNVP 62
Query: 63 --NHLKKSQEILDRYSRCNSTVGYSGRKIARRGGSKSSSNRRVSSES-CDVFSGKWVFDN 119
N+L + E+L R+S+CNST YSG+KI + + E CDVFSGKWVFDN
Sbjct: 63 RINYLG-AHEVLGRFSKCNSTKEYSGKKIGCVDPFEDHPGQVTKEEQKCDVFSGKWVFDN 121
Query: 120 AS-YPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWNVKEMWEKLRGKRL 178
+S YPL+ ES CPYMSDQLAC KHGR DL YQHWRWQPH C+LKRWNV EMWEKLRGKRL
Sbjct: 122 SSSYPLHKESQCPYMSDQLACQKHGRKDLEYQHWRWQPHACNLKRWNVIEMWEKLRGKRL 181
Query: 179 MFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNATVEFLWAPLLAESN 238
MFVGDSLNR QWISMVCLL+S IP DK+SMSPNAHLTIFRAE+YNATVEFLWAPLL ESN
Sbjct: 182 MFVGDSLNRSQWISMVCLLRSVIPRDKQSMSPNAHLTIFRAEDYNATVEFLWAPLLVESN 241
Query: 239 SDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQGPVKLLWTDEENGACEEL 298
SDDPVNHRL ERIIRPDSVLKHAS W+HADIL+FNTYLWWRQ VKL W+ EE G+CEE+
Sbjct: 242 SDDPVNHRLSERIIRPDSVLKHASKWQHADILIFNTYLWWRQDSVKLRWSSEEKGSCEEV 301
Query: 299 DGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREWNPESEGNCYGEKKPIDV 358
ME+AM +W DWV++ VDP KKRVFFVTMSPTH WSREWNP SEGNCYGEKKPI+
Sbjct: 302 KSAEGMEMAMDSWGDWVANNVDPNKKRVFFVTMSPTHQWSREWNPGSEGNCYGEKKPIEE 361
Query: 359 ESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLS 418
ESYWGSGSD+PTM V+ +L L KVSV+N+TQLSEYRKDGHPS++RKFWEPL ++L
Sbjct: 362 ESYWGSGSDIPTMRMVKRVLERLGPKVSVINITQLSEYRKDGHPSVYRKFWEPLNEDRLK 421
Query: 419 NPSSYSDCIHWCLPGVPDTWNELLFHFL 446
NP+SYSDC HWC+PGVPD WN+LLFHFL
Sbjct: 422 NPASYSDCTHWCVPGVPDVWNQLLFHFL 449
>UniRef100_Q9M014 Hypothetical protein F7A7_140 [Arabidopsis thaliana]
Length = 446
Score = 639 bits (1647), Expect = 0.0
Identities = 304/448 (67%), Positives = 352/448 (77%), Gaps = 11/448 (2%)
Query: 5 KWNKKKVPIFPFIVLLLLVFIAFSTLHNGSTIHKFHENPLHIHQE--QASSTTYVKPNLP 62
+W++KK + P LL ++ + F L N +I + H + Q + S+ +VKPN+P
Sbjct: 4 RWSRKKSRL-PLAGLLFILVVTFMILFNERSIQQIHHHAASHTQNLREPSTFDFVKPNVP 62
Query: 63 --NHLKKSQEILDRYSRCNSTVGYSGRKIARRGGSKSSSNRRVSSES-CDVFSGKWVFDN 119
N+L + E+LDR+S+CNST YSG+KI + + E CDVFSGKWVFDN
Sbjct: 63 RINYLG-AHEVLDRFSKCNSTKEYSGKKIGWVDPFEDHPGQVTKEEQKCDVFSGKWVFDN 121
Query: 120 AS-YPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWNVKEMWEKLRGKRL 178
+S YPL+ ES CPYMSDQLAC KHGR DL YQHWRWQPH C+LKRWN EMWEKLRGKRL
Sbjct: 122 SSSYPLHKESQCPYMSDQLACQKHGRKDLEYQHWRWQPHACNLKRWNAIEMWEKLRGKRL 181
Query: 179 MFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNATVEFLWAPLLAESN 238
MFVGDSLNRGQWISMVCLLQS IP DK+SMSPNAHLTIFRAE+YNATVEFLWAPLL ESN
Sbjct: 182 MFVGDSLNRGQWISMVCLLQSVIPRDKQSMSPNAHLTIFRAEDYNATVEFLWAPLLVESN 241
Query: 239 SDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQGPVKLLWTDEENGACEEL 298
SDDPVNHRL ERIIRPDSVLKHAS W+HADIL+FNTYLWWRQ VKL EE G+CEE+
Sbjct: 242 SDDPVNHRLSERIIRPDSVLKHASKWQHADILIFNTYLWWRQDSVKL---REEKGSCEEV 298
Query: 299 DGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREWNPESEGNCYGEKKPIDV 358
ME+AM +W DWV++ VDP KKRVFFVTMSPTH WSREWNP SEGNCYGEKKPI+
Sbjct: 299 KSAEGMEMAMDSWGDWVANNVDPNKKRVFFVTMSPTHQWSREWNPGSEGNCYGEKKPIEE 358
Query: 359 ESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLS 418
ESYWGSGSD+PTM V+ +L L KVSV+N+TQLSEYRKDGHPS++RKFWEPL ++L
Sbjct: 359 ESYWGSGSDIPTMRMVKRVLERLGPKVSVINITQLSEYRKDGHPSVYRKFWEPLNEDRLK 418
Query: 419 NPSSYSDCIHWCLPGVPDTWNELLFHFL 446
NP+SYSDC HWC+PGVPD WN+LLFHFL
Sbjct: 419 NPASYSDCTHWCVPGVPDVWNQLLFHFL 446
>UniRef100_O80919 Expressed protein [Arabidopsis thaliana]
Length = 410
Score = 397 bits (1019), Expect = e-109
Identities = 183/350 (52%), Positives = 243/350 (69%), Gaps = 7/350 (2%)
Query: 104 SSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKR 163
S C++F GKWVFDN SYPLY E DC +MSDQLAC K GR DL Y+ WRWQPH CDL R
Sbjct: 55 SGRECNLFEGKWVFDNVSYPLYKEEDCKFMSDQLACEKFGRKDLSYKFWRWQPHTCDLPR 114
Query: 164 WNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPN--AHLTIFRAEE 221
+N ++ E+LR KR+++VGDSLNRGQW+SMVC++ S I K N ++L F+A E
Sbjct: 115 FNGTKLLERLRNKRMVYVGDSLNRGQWVSMVCMVSSVITNPKAMYMHNNGSNLITFKALE 174
Query: 222 YNATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQG 281
YNAT+++ WAPLL ESNSDDP NHR +RI+R S+ KHA W ++DI+VFN+YLWWR
Sbjct: 175 YNATIDYYWAPLLVESNSDDPTNHRFPDRIVRIQSIEKHARHWTNSDIIVFNSYLWWRMP 234
Query: 282 PVKLLWTDEE--NGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSR 339
+K LW E +G +E++ E+A+ + W+ V+P ++FF++MSPTH +
Sbjct: 235 HIKSLWGSFEKLDGIYKEVEMVRVYEMALQTLSQWLEVHVNPNITKLFFMSMSPTHERAE 294
Query: 340 EWNPESEGNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSK---VSVLNVTQLSEY 396
EW NCYGE ID E Y G GSD M +EN+L L ++ + ++N+TQLSEY
Sbjct: 295 EWGGILNQNCYGEASLIDKEGYTGRGSDPKMMRVLENVLDGLKNRGLNMQMINITQLSEY 354
Query: 397 RKDGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLFHFL 446
RK+GHPSI+RK W ++ ++SNPSS +DCIHWCLPGVPD WNELL+ ++
Sbjct: 355 RKEGHPSIYRKQWGTVKENEISNPSSNADCIHWCLPGVPDVWNELLYAYI 404
>UniRef100_Q84TW8 Hypothetical protein OSJNBa0094J08.9 [Oryza sativa]
Length = 441
Score = 390 bits (1002), Expect = e-107
Identities = 187/376 (49%), Positives = 242/376 (63%), Gaps = 19/376 (5%)
Query: 87 RKIAR-------RGGSKSSSNRRV---------SSESCDVFSGKWVFDNASYPLYNESDC 130
R+IAR GG SS V ++ CDV G+WV+D A+ P Y E +C
Sbjct: 60 RQIARPRAPSHHHGGGSSSGGGDVVPPFAVGAAAAAGCDVGVGEWVYDEAARPWYEEEEC 119
Query: 131 PYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWNVKEMWEKLRGKRLMFVGDSLNRGQW 190
PY+ QL C HGR D YQHWRWQP C L +N M E LRGKR+MFVGDSLNRGQ+
Sbjct: 120 PYIQPQLTCQAHGRPDTAYQHWRWQPRGCSLPSFNATLMLEMLRGKRMMFVGDSLNRGQY 179
Query: 191 ISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNATVEFLWAPLLAESNSDDPVNHRLDER 250
+S+VCLL IP +SM LT+FRA+ YNAT+EF WAP LAESNSDD V HR+ +R
Sbjct: 180 VSLVCLLHRSIPESSKSMETFDSLTVFRAKNYNATIEFYWAPFLAESNSDDAVVHRIADR 239
Query: 251 IIRPDSVLKHASLWEHADILVFNTYLWWRQG-PVKLLWTDEENGACE--ELDGRGAMELA 307
I+R ++ KHA W+ ADILVFN+YLWW G +K+L E+ + + E++ A +
Sbjct: 240 IVRGTALEKHARFWKGADILVFNSYLWWMTGQKMKILQGSFEDKSKDIVEMETEEAYGMV 299
Query: 308 MGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREWNPESEGNCYGEKKPIDVESYWGSGSD 367
+ A WV + ++P RVFFVTMSPTH S++W +S+GNCY + PI SYWG G+
Sbjct: 300 LNAVVRWVENNMNPRNSRVFFVTMSPTHTRSKDWGDDSDGNCYNQTTPIRDLSYWGPGTS 359
Query: 368 LPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSSYSDCI 427
M + + S+ V ++N+TQLSEYRKD H I++K W PL PEQ++NP SY+DC
Sbjct: 360 KGLMRVIGEVFSTSKVPVGIVNITQLSEYRKDAHTQIYKKQWNPLTPEQIANPKSYADCT 419
Query: 428 HWCLPGVPDTWNELLF 443
HWCLPG+ DTWNELL+
Sbjct: 420 HWCLPGLQDTWNELLY 435
>UniRef100_Q84TV3 Hypothetical protein OSJNBa0094J08.14 [Oryza sativa]
Length = 473
Score = 385 bits (990), Expect = e-105
Identities = 188/373 (50%), Positives = 248/373 (66%), Gaps = 25/373 (6%)
Query: 96 KSSSNRRVSSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQ 155
K S+ CD+FSG+WV+D A+YPLY ES C MS+Q AC K+GRTDL YQHWRWQ
Sbjct: 96 KLPSSSSSGGGECDLFSGRWVYDEAAYPLYRESACRVMSEQSACEKYGRTDLRYQHWRWQ 155
Query: 156 PHDCDLKRWNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLT 215
PH CDL R++ ++ KLR KRL+FVGDSLNR QW SM+CL+ + P S++ + LT
Sbjct: 156 PHGCDLPRFDAEKFLGKLRNKRLVFVGDSLNRNQWASMLCLIDTGAPELHTSINSSRSLT 215
Query: 216 IFRAEEYNATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTY 275
F+ EYNA+V+F W+PLL ESNSD P+ HR+ +R +R S+ KHA+ W +AD+LVFN+Y
Sbjct: 216 TFKIHEYNASVDFYWSPLLVESNSDHPLRHRVADRTVRAASINKHAAHWTNADVLVFNSY 275
Query: 276 LWWRQGPVKLLWTDEEN------GACEE---------LDGRGAMELAMGAWADWVSSKVD 320
LWW++ +K+LW +N A EE +D A ELA+ WADW+ VD
Sbjct: 276 LWWQRPAMKVLWGSFDNPAAVVAAAAEEGDEYAVSKVIDSLRAYELAVRTWADWMEFHVD 335
Query: 321 PLKKRVFFVTMSPTHLWSREWNPESE----GN--CYGEKKPIDVESYWG-SGSDLPTMSS 373
+ ++FF+TMSPTHL S EW + GN CYGE +PI E Y G SG+D+ +
Sbjct: 336 RARTQLFFMTMSPTHLRSDEWEDAAAAAAGGNHGCYGETEPIAAEEYRGTSGTDMAFARA 395
Query: 374 VENILSSLNSK---VSVLNVTQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWC 430
VE L + V ++NVT+LSE RKD HPS+ R++W+P+ EQ NPSSY+DCIHWC
Sbjct: 396 VEAEARRLGERSVAVRLINVTRLSERRKDAHPSVHRRYWDPVTDEQRRNPSSYADCIHWC 455
Query: 431 LPGVPDTWNELLF 443
LPGVPD WN+LL+
Sbjct: 456 LPGVPDVWNQLLY 468
>UniRef100_Q84W34 Hypothetical protein At2g40320 [Arabidopsis thaliana]
Length = 425
Score = 375 bits (964), Expect = e-102
Identities = 171/343 (49%), Positives = 224/343 (64%), Gaps = 3/343 (0%)
Query: 104 SSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKR 163
+ ESCDVFSGKWV D S PLY E +CPY+ QL C +HGR D YQ WRWQP+ CDL
Sbjct: 77 TEESCDVFSGKWVRDEVSRPLYEEWECPYIQPQLTCQEHGRPDKDYQFWRWQPNHCDLPS 136
Query: 164 WNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYN 223
+N M E LRGKR+M+VGDSLNRG ++SM+CLL IP D++S+ N LT+F A+EYN
Sbjct: 137 FNASLMLETLRGKRMMYVGDSLNRGMFVSMICLLHRLIPEDQKSIKTNGSLTVFTAKEYN 196
Query: 224 ATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQG-P 282
AT+EF WAP L ESNSDD + HR+ +R++R S+ KH W+ DI++FNTYLWW G
Sbjct: 197 ATIEFYWAPFLLESNSDDAIVHRISDRVVRKGSINKHGRHWKGVDIIIFNTYLWWMTGLK 256
Query: 283 VKLLW--TDEENGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSRE 340
+ +L D++ E+ A + M + WV + +D K RVFF +MSP H +
Sbjct: 257 MNILQGSFDDKEKNIVEVSTEDAYRMGMKSMLRWVKNNMDRKKTRVFFTSMSPAHAKGID 316
Query: 341 WNPESEGNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDG 400
W E NCY + I+ SYWGS M + + + +++LN+TQ+S YRKD
Sbjct: 317 WGGEPGQNCYNQTTLIEDPSYWGSDCRKSIMKVIGEVFGRSKTPITLLNITQMSNYRKDA 376
Query: 401 HPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
H SI++K W PL EQL NP+SY+DC+HWCLPG+ DTWNELLF
Sbjct: 377 HTSIYKKQWSPLTAEQLENPTSYADCVHWCLPGLQDTWNELLF 419
>UniRef100_Q9SIZ1 Hypothetical protein At2g40320 [Arabidopsis thaliana]
Length = 435
Score = 374 bits (960), Expect = e-102
Identities = 172/353 (48%), Positives = 225/353 (63%), Gaps = 13/353 (3%)
Query: 104 SSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKR 163
+ ESCDVFSGKWV D S PLY E +CPY+ QL C +HGR D YQ WRWQP+ CDL
Sbjct: 77 TEESCDVFSGKWVRDEVSRPLYEEWECPYIQPQLTCQEHGRPDKDYQFWRWQPNHCDLPS 136
Query: 164 WNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYN 223
+N M E LRGKR+M+VGDSLNRG ++SM+CLL IP D++S+ N LT+F A+EYN
Sbjct: 137 FNASLMLETLRGKRMMYVGDSLNRGMFVSMICLLHRLIPEDQKSIKTNGSLTVFTAKEYN 196
Query: 224 ATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQG-P 282
AT+EF WAP L ESNSDD + HR+ +R++R S+ KH W+ DI++FNTYLWW G
Sbjct: 197 ATIEFYWAPFLLESNSDDAIVHRISDRVVRKGSINKHGRHWKGVDIIIFNTYLWWMTGLK 256
Query: 283 VKLLWT------------DEENGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVT 330
+ +L D++ E+ A + M + WV + +D K RVFF +
Sbjct: 257 MNILEVSDIGGIYRQGSFDDKEKNIVEVSTEDAYRMGMKSMLRWVKNNMDRKKTRVFFTS 316
Query: 331 MSPTHLWSREWNPESEGNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSKVSVLNV 390
MSPTH +W E NCY + I+ SYWGS M + + + +++LN+
Sbjct: 317 MSPTHAKGIDWGGEPGQNCYNQTTLIEDPSYWGSDCRKSIMKVIGEVFGRSKTPITLLNI 376
Query: 391 TQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
TQ+S YRKD H SI++K W PL EQL NP+SY+DC+HWCLPG+ DTWNELLF
Sbjct: 377 TQMSNYRKDAHTSIYKKQWSPLTAEQLENPTSYADCVHWCLPGLQDTWNELLF 429
>UniRef100_Q9SRL3 F9F8.15 protein [Arabidopsis thaliana]
Length = 451
Score = 371 bits (952), Expect = e-101
Identities = 174/342 (50%), Positives = 220/342 (63%), Gaps = 4/342 (1%)
Query: 106 ESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWN 165
E CDVF G WV D ++ PLY ES+CPY+ QL C HGR D YQ WRW+P C L +N
Sbjct: 104 EGCDVFKGNWVKDWSTRPLYRESECPYIQPQLTCRTHGRPDSDYQSWRWRPDSCSLPSFN 163
Query: 166 VKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNAT 225
M E LRGK++MFVGDSLNRG ++S++CLL S+IP + +SM LT+F ++YNAT
Sbjct: 164 ATVMLESLRGKKMMFVGDSLNRGMYVSLICLLHSQIPENSKSMDTFGSLTVFSLKDYNAT 223
Query: 226 VEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQG-PVK 284
+EF WAP L ESNSD+ HR+ +RI+R S+ KH W ADI+VFNTYLWWR G +K
Sbjct: 224 IEFYWAPFLLESNSDNATVHRVSDRIVRKGSINKHGRHWRGADIVVFNTYLWWRTGFKMK 283
Query: 285 LLWTD--EENGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREWN 342
+L +E E++ A +A+ WV +DPLK RVFF TMSPTH +W
Sbjct: 284 ILEGSFKDEKKRIVEMESEDAYRMALKTMVKWVKKNMDPLKTRVFFATMSPTHYKGEDWG 343
Query: 343 PESEGNCYGEKKPIDVESYWGSGSDLPTMSSV-ENILSSLNSKVSVLNVTQLSEYRKDGH 401
E NCY + PI ++W S M + E + V+VLN+TQLS YRKD H
Sbjct: 344 GEQGKNCYNQTTPIQDMNHWPSDCSKTLMKVIGEELDQRAEFPVTVLNITQLSGYRKDAH 403
Query: 402 PSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
SI++K W PL EQL+NP+SYSDCIHWCLPG+ DTWNEL F
Sbjct: 404 TSIYKKQWSPLTKEQLANPASYSDCIHWCLPGLQDTWNELFF 445
>UniRef100_Q9CAT0 Hypothetical protein T18K17.20 [Arabidopsis thaliana]
Length = 403
Score = 361 bits (927), Expect = 2e-98
Identities = 164/342 (47%), Positives = 231/342 (66%), Gaps = 2/342 (0%)
Query: 106 ESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWN 165
ESC+VF G+WV+DN SYPLY E CPY+ Q C ++GR D YQ+WRW+P CDL R+N
Sbjct: 55 ESCNVFEGQWVWDNVSYPLYTEKSCPYLVKQTTCQRNGRPDSYYQNWRWKPSSCDLPRFN 114
Query: 166 VKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNAT 225
++ + LR KRLMF+GDS+ R + SMVC++QS IP K+S + IF+AEEYNA+
Sbjct: 115 ALKLLDVLRNKRLMFIGDSVQRSTFESMVCMVQSVIPEKKKSFHRIPPMKIFKAEEYNAS 174
Query: 226 VEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQGPVKL 285
+E+ WAP + ES SD NH + +R+++ D++ KH+ WE D+LVF +Y +G +
Sbjct: 175 IEYYWAPFIVESISDHATNHTVHKRLVKLDAIEKHSKSWEGVDVLVFESYK-SLEGKLIR 233
Query: 286 LWTDEENGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREWNPES 345
+ + E + A ++A+ WA W +K++ K++VFF +MSPTHLWS EWNP S
Sbjct: 234 ICRYGDTSEVREYNVTTAYKMALETWAKWFKTKINSEKQKVFFTSMSPTHLWSWEWNPGS 293
Query: 346 EGNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIF 405
+G CY E PID SYWG+GS+ M V ++LS + V+ LN+TQLSEYRKDGH +++
Sbjct: 294 DGTCYDELYPIDKRSYWGTGSNQEIMKIVGDVLSRVGENVTFLNITQLSEYRKDGHTTVY 353
Query: 406 -RKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLFHFL 446
+ + L EQ ++P +Y DCIHWCLPGVPDTWNE+L+ +L
Sbjct: 354 GERRGKLLTKEQRADPKNYGDCIHWCLPGVPDTWNEILYAYL 395
>UniRef100_Q9LY46 Hypothetical protein F27K19_170 [Arabidopsis thaliana]
Length = 487
Score = 353 bits (905), Expect = 7e-96
Identities = 169/362 (46%), Positives = 231/362 (63%), Gaps = 5/362 (1%)
Query: 87 RKIARRGGSKSSSNRRVSSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTD 146
+KI ++ + + E CD+F+G+WVFDN ++PLY E C +++ Q+ C ++GR D
Sbjct: 119 KKIELFAATEDEEDVELPPEECDLFTGEWVFDNETHPLYKEDQCEFLTAQVTCMRNGRRD 178
Query: 147 LGYQHWRWQPHDCDLKRWNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKR 206
YQ+WRWQP DC L ++ K + EKLR KR+MFVGDSLNR QW SMVCL+QS +P ++
Sbjct: 179 SLYQNWRWQPRDCSLPKFKAKLLLEKLRNKRMMFVGDSLNRNQWESMVCLVQSVVPPGRK 238
Query: 207 SMSPNAHLTIFRAEEYNATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEH 266
S++ L++FR E+YNATVEF WAP L ESNSDDP H + RII P+S+ KH W+
Sbjct: 239 SLNKTGSLSVFRVEDYNATVEFYWAPFLVESNSDDPNMHSILNRIIMPESIEKHGVNWKG 298
Query: 267 ADILVFNTYLWWRQG-PVKLLWTDEENG--ACEELDGRGAMELAMGAWADWVSSKVDPLK 323
D LVFNTY+WW +K+L + G EE++ A M W DWV +DPL+
Sbjct: 299 VDFLVFNTYIWWMNTFAMKVLRGSFDKGDTEYEEIERPVAYRRVMRTWGDWVERNIDPLR 358
Query: 324 KRVFFVTMSPTHLWSREWNPESEGNCYGEKKPI-DVESYWGSGSDLPTMSSVENILSSLN 382
VFF +MSP H+ S +W C E PI ++ + G+D S EN+ SLN
Sbjct: 359 TTVFFASMSPLHIKSLDWENPDGIKCALETTPILNMSMPFSVGTDYRLFSVAENVTHSLN 418
Query: 383 SKVSVLNVTQLSEYRKDGHPSIFR-KFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNEL 441
V LN+T+LSEYRKD H S+ + + L PEQ ++P++Y+DCIHWCLPG+PDTWNE
Sbjct: 419 VPVYFLNITKLSEYRKDAHTSVHTIRQGKMLTPEQQADPNTYADCIHWCLPGLPDTWNEF 478
Query: 442 LF 443
L+
Sbjct: 479 LY 480
>UniRef100_Q9SSL4 F3N23.34 protein [Arabidopsis thaliana]
Length = 417
Score = 352 bits (902), Expect = 2e-95
Identities = 164/356 (46%), Positives = 231/356 (64%), Gaps = 16/356 (4%)
Query: 106 ESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRW- 164
ESC+VF G+WV+DN SYPLY E CPY+ Q C ++GR D YQ+WRW+P CDL R+
Sbjct: 55 ESCNVFEGQWVWDNVSYPLYTEKSCPYLVKQTTCQRNGRPDSYYQNWRWKPSSCDLPRFK 114
Query: 165 -------------NVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPN 211
N ++ + LR KRLMF+GDS+ R + SMVC++QS IP K+S
Sbjct: 115 LAQCLKPLRCFRFNALKLLDVLRNKRLMFIGDSVQRSTFESMVCMVQSVIPEKKKSFHRI 174
Query: 212 AHLTIFRAEEYNATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILV 271
+ IF+AEEYNA++E+ WAP + ES SD NH + +R+++ D++ KH+ WE D+LV
Sbjct: 175 PPMKIFKAEEYNASIEYYWAPFIVESISDHATNHTVHKRLVKLDAIEKHSKSWEGVDVLV 234
Query: 272 FNTYLWWRQGPVKLLWTDEENGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTM 331
F +Y +G + + + E + A ++A+ WA W +K++ K++VFF +M
Sbjct: 235 FESYKSL-EGKLIRICRYGDTSEVREYNVTTAYKMALETWAKWFKTKINSEKQKVFFTSM 293
Query: 332 SPTHLWSREWNPESEGNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSKVSVLNVT 391
SPTHLWS EWNP S+G CY E PID SYWG+GS+ M V ++LS + V+ LN+T
Sbjct: 294 SPTHLWSWEWNPGSDGTCYDELYPIDKRSYWGTGSNQEIMKIVGDVLSRVGENVTFLNIT 353
Query: 392 QLSEYRKDGHPSIF-RKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLFHFL 446
QLSEYRKDGH +++ + + L EQ ++P +Y DCIHWCLPGVPDTWNE+L+ +L
Sbjct: 354 QLSEYRKDGHTTVYGERRGKLLTKEQRADPKNYGDCIHWCLPGVPDTWNEILYAYL 409
>UniRef100_Q8GWS5 Hypothetical protein At3g55990/F27K19_170 [Arabidopsis thaliana]
Length = 487
Score = 351 bits (901), Expect = 2e-95
Identities = 168/362 (46%), Positives = 231/362 (63%), Gaps = 5/362 (1%)
Query: 87 RKIARRGGSKSSSNRRVSSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTD 146
+KI ++ + + E CD+F+G+WVFDN ++PLY E C +++ Q+ C ++GR D
Sbjct: 119 KKIELFAATEDEEDVELPPEECDLFTGEWVFDNETHPLYKEDQCEFLTAQVTCMRNGRRD 178
Query: 147 LGYQHWRWQPHDCDLKRWNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKR 206
YQ+WRWQP DC L ++ K + EKLR KR+MFVGDSLNR QW SMVCL+QS +P ++
Sbjct: 179 SLYQNWRWQPRDCSLPKFKAKLLLEKLRNKRMMFVGDSLNRNQWESMVCLVQSVVPPGRK 238
Query: 207 SMSPNAHLTIFRAEEYNATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEH 266
S++ L++FR E+YNATVEF WAP L ESNSDDP H + RII P+S+ KH W+
Sbjct: 239 SLNKTGSLSVFRVEDYNATVEFYWAPFLVESNSDDPNMHSILNRIIMPESIEKHGVNWKG 298
Query: 267 ADILVFNTYLWWRQG-PVKLLWTDEENG--ACEELDGRGAMELAMGAWADWVSSKVDPLK 323
D LVFNTY+WW +K+L + G EE++ A + W DWV +DPL+
Sbjct: 299 VDFLVFNTYIWWMNTFAMKVLRGSFDKGDTEYEEIERPVAYRRVIRTWGDWVERNIDPLR 358
Query: 324 KRVFFVTMSPTHLWSREWNPESEGNCYGEKKPI-DVESYWGSGSDLPTMSSVENILSSLN 382
VFF +MSP H+ S +W C E PI ++ + G+D S EN+ SLN
Sbjct: 359 TTVFFASMSPLHIKSLDWENPDGIKCALETTPILNMSMPFSVGTDYRLFSVAENVTHSLN 418
Query: 383 SKVSVLNVTQLSEYRKDGHPSIFR-KFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNEL 441
V LN+T+LSEYRKD H S+ + + L PEQ ++P++Y+DCIHWCLPG+PDTWNE
Sbjct: 419 VPVYFLNITKLSEYRKDAHTSVHTIRQGKMLTPEQQADPNTYADCIHWCLPGLPDTWNEF 478
Query: 442 LF 443
L+
Sbjct: 479 LY 480
>UniRef100_Q8LED3 Hypothetical protein [Arabidopsis thaliana]
Length = 434
Score = 348 bits (893), Expect = 2e-94
Identities = 164/345 (47%), Positives = 224/345 (64%), Gaps = 4/345 (1%)
Query: 106 ESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWN 165
E C+V +GKWV++++ PLY + CPY+ Q +C K+G+ + Y W WQP DC + R++
Sbjct: 90 EECNVAAGKWVYNSSIEPLYTDRSCPYIDRQFSCMKNGQPETDYLRWEWQPDDCTIPRFS 149
Query: 166 VKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNAT 225
K KLRGKRL+FVGDSL R QW S VCL++S IP ++SM + +F+A+EYNAT
Sbjct: 150 PKLAMNKLRGKRLLFVGDSLQRSQWESFVCLVESIIPEGEKSMKRSQKYFVFKAKEYNAT 209
Query: 226 VEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQG-PVK 284
+EF WAP + ESN+D PV +RI++ DSV A WE ADILVFNTY+WW G +K
Sbjct: 210 IEFYWAPYIVESNTDIPVISDPKKRIVKVDSVKDRAKFWEGADILVFNTYVWWMSGLRMK 269
Query: 285 LLWTDEENG--ACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREWN 342
LW NG E LD + A L + WA+WV S VDP K RVFF TMSPTH S +W
Sbjct: 270 ALWGSFGNGESGAEALDTQVAYRLGLKTWANWVDSTVDPNKTRVFFTTMSPTHTRSADWG 329
Query: 343 PESEGNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHP 402
+ C+ E KPI + +WG+GS+ M V +++ + + V+V+N+TQLSEYR D H
Sbjct: 330 KPNGTKCFNETKPIKDKKFWGTGSNKQMMKVVSSVIKHMTTHVTVINITQLSEYRIDAHT 389
Query: 403 SIFRKF-WEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLFHFL 446
S++ + + L EQ ++P ++DCIHWCLPG+PDTWN +L L
Sbjct: 390 SVYTETGGKILTAEQRADPMHHADCIHWCLPGLPDTWNRILLAHL 434
>UniRef100_Q5W6Y3 Hypothetical protein OSJNBa0036C12.14 [Oryza sativa]
Length = 454
Score = 344 bits (882), Expect = 3e-93
Identities = 183/451 (40%), Positives = 255/451 (55%), Gaps = 30/451 (6%)
Query: 9 KKVPIFPFIVLLLLVFIAFSTLHNGSTIHKFHENPLHIHQEQASSTTYVKPNLPNHLKKS 68
K+ + PF+ LL+L + FS L+ +E S +P+L + +
Sbjct: 12 KRARVSPFVFLLVLFLLLFSFLYGEDL------------KELLGSQAQARPSLHFNAAAA 59
Query: 69 QEILDRYSRCNSTVGYSGRKIARRGGSK----SSSNRRVSSESCDVFSGKWVFDNASYPL 124
+ ++ + +T GR RR + ++ + E CDVFSG+WV D A+ PL
Sbjct: 60 GDGIELPAATAATT--EGRTTTRRWRGRLPFAANGDGEEEEEECDVFSGRWVRDEAARPL 117
Query: 125 YNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWNVKEMWEKLRGKRLMFVGDS 184
Y E+DCPY+ QLAC HGR + YQ WRWQP C L ++ M ++LRGKR+MFVGDS
Sbjct: 118 YREADCPYIPAQLACEAHGRPETAYQRWRWQPRGCALPAFDAAAMLDRLRGKRVMFVGDS 177
Query: 185 LNRGQWISMVCLLQSEIP---ADKRSMSPNAHLTIFRAEEYNATVEFLWAPLLAESNSDD 241
L RGQ+ S+VCLL + +P A + SP+ ++F A YNATVEF WAP L +SN+D+
Sbjct: 178 LGRGQFTSLVCLLLAAVPDPAARSFATSPDQQRSVFTAAAYNATVEFYWAPFLLQSNADN 237
Query: 242 PVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQGPVKLLWTD------EENGAC 295
HR+ +R++R S+ H WE AD++VFNTYLWW G + D +
Sbjct: 238 AAVHRISDRMVRRGSIGHHGRHWEGADVIVFNTYLWWCTGLQFRILEDGPFDAGGNSSTT 297
Query: 296 EELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREW-NPESEGNCYGEKK 354
+ A +A W +D RVFF +MSPTH S++W E GNCYGE +
Sbjct: 298 TWVSTEEAYAMAFREMLQWAREHMDFATTRVFFTSMSPTHGKSQDWGGGEPGGNCYGETE 357
Query: 355 PIDVESYWGSGSDLPTMSSVENILSSLNSKVSV--LNVTQLSEYRKDGHPSIFRKFWEPL 412
I +YWGS S M ++ +L + V V LNVTQLS YRKD H S+++K W P
Sbjct: 358 MIGDAAYWGSDSRRGVMRAIGEVLDGDGADVPVTFLNVTQLSLYRKDAHTSVYKKQWTPP 417
Query: 413 RPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
PEQL++P +Y+DC+HWCLPG+ DTWNELL+
Sbjct: 418 TPEQLADPKTYADCVHWCLPGLQDTWNELLY 448
>UniRef100_Q94K00 Hypothetical protein At2g40150; T7M7.4 [Arabidopsis thaliana]
Length = 424
Score = 340 bits (872), Expect = 5e-92
Identities = 169/359 (47%), Positives = 232/359 (64%), Gaps = 13/359 (3%)
Query: 97 SSSNRRVSSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQP 156
SSS + + CD+F+G+WVFDN +YPLY E +C ++++Q+ C ++GR D +Q+WRWQP
Sbjct: 60 SSSFVELPPDECDLFTGQWVFDNKTYPLYKEEECEFLTEQVTCLRNGRKDSLFQNWRWQP 119
Query: 157 HDCDLKRWNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTI 216
DC L ++N + + EKLR KRLMFVGDSLNR QW SMVCL+QS IP ++S++ LT+
Sbjct: 120 RDCSLPKFNARVLLEKLRNKRLMFVGDSLNRNQWESMVCLVQSVIPPGRKSLNQTGSLTV 179
Query: 217 FRAEEYNATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYL 276
F+ ++YNATVEF WAP L ESNSDDP H + +RII P+S+ KH W D LVFN+Y+
Sbjct: 180 FKIQDYNATVEFYWAPFLVESNSDDPEKHSIIDRIIMPESIEKHGVNWIGVDFLVFNSYI 239
Query: 277 WWRQG-PVKLLWTDEENGACE--ELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSP 333
WW +K+L ++G E E+ A E + DWV +DPL VFF++MSP
Sbjct: 240 WWMNTVSIKVLRGSFDDGDTEYDEIKRPIAYERVLRTLGDWVDHNIDPLSTTVFFMSMSP 299
Query: 334 THLWSREW-NPESEGNCYGEKKPIDVESY---WGS----GSDLPTMSSVENILSSLNSKV 385
H+ S +W NPE C E PI S+ +G G+D EN+ SL +
Sbjct: 300 LHIKSSDWANPEGI-RCALETTPILNMSFNVAYGQFSAVGTDYRLFPVAENVTQSLKVPI 358
Query: 386 SVLNVTQLSEYRKDGHPSIFR-KFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
LN+T LSEYRKD H S++ K + L EQ ++P++++DCIHWCLPG+PDTWNE L+
Sbjct: 359 HFLNITALSEYRKDAHTSVYTIKQGKLLTREQQNDPANFADCIHWCLPGLPDTWNEFLY 417
>UniRef100_Q5W782 Hypothetical protein OJ1537_B05.7 [Oryza sativa]
Length = 517
Score = 329 bits (843), Expect = 1e-88
Identities = 157/344 (45%), Positives = 216/344 (62%), Gaps = 19/344 (5%)
Query: 106 ESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWN 165
ESCDV+ G+WV+D A+ PLY ES C ++++Q+ C ++GR D YQ WRWQP CDL R++
Sbjct: 166 ESCDVYKGRWVYDEANAPLYKESACEFLTEQVTCMRNGRRDDDYQKWRWQPDGCDLPRFD 225
Query: 166 VKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNAT 225
K + EKLR KRLMFVGDSLNR QW SMVCL+QSE P +K+S+ N L +FR EEYNAT
Sbjct: 226 AKLLLEKLRNKRLMFVGDSLNRNQWESMVCLVQSEAPWEKKSLVKNDSLNVFRLEEYNAT 285
Query: 226 VEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQGP-VK 284
+EF W+P L ESNSDDP H + +RII+P S+ KHA+ WE D L+FNTY+WW P +K
Sbjct: 286 IEFYWSPFLVESNSDDPNMHSIVDRIIKPTSIAKHAANWEGVDYLIFNTYIWWMNTPEMK 345
Query: 285 LLW---TDEENGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREW 341
+L ++ +E++ A + W+ WV VDP + VFF+++SP H+ S W
Sbjct: 346 ILHGGSFSKKPVKYDEMERVAAYRKVLKTWSRWVEKHVDPKRSTVFFMSVSPVHMQSEGW 405
Query: 342 NPESEGNCYGEKKP-IDVESYWGSGSDLPTMSSVENILSSLNS-KVSVLNVTQLSEYRKD 399
C+ E +P I+ G+D S+ ++ ++ V +N+T LSE RKD
Sbjct: 406 GKPDAIKCFSETQPAINYTKKLEVGTDWDLFSTAHHVTKAMKRVPVHFINITALSEIRKD 465
Query: 400 GHPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
H S +NP ++DCIHWCLPG+PDTWNE ++
Sbjct: 466 AHTS-------------KANPRKFADCIHWCLPGLPDTWNEFIY 496
>UniRef100_Q9XEF9 Hypothetical protein T07M07.22 [Arabidopsis thaliana]
Length = 398
Score = 327 bits (838), Expect = 4e-88
Identities = 160/355 (45%), Positives = 208/355 (58%), Gaps = 34/355 (9%)
Query: 102 RVSSESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDL 161
R ESCDVFSGKWV D S PLY E +CPY+ QL C +HGR D YQ WR+
Sbjct: 59 RTPEESCDVFSGKWVRDEVSRPLYEEWECPYIQPQLTCQEHGRPDKDYQFWRF------- 111
Query: 162 KRWNVKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEE 221
N M E LRGKR+M+VGDSLNRG ++SM+CLL IP D++S+ N LT+F A+E
Sbjct: 112 ---NASLMLETLRGKRMMYVGDSLNRGMFVSMICLLHRLIPEDQKSIKTNGSLTVFTAKE 168
Query: 222 YNATVEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWRQG 281
YNAT+EF WAP L ESNSDD + HR+ +R++R S+ KH W+ DI++FNTYLWW G
Sbjct: 169 YNATIEFYWAPFLLESNSDDAIVHRISDRVVRKGSINKHGRHWKGVDIIIFNTYLWWMTG 228
Query: 282 -PVKLLWT------------DEENGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFF 328
+ +L D++ E+ A + M + WV + +D K RVFF
Sbjct: 229 LKMNILEVSDIGGIYRQGSFDDKEKNIVEVSTEDAYRMGMKSMLRWVKNNMDRKKTRVFF 288
Query: 329 VTMSPTHLWSREWNPESEGNCYGEKKPIDVESYWGSGSDLPTMSSVENILSSLNSKVSVL 388
+MSPTH +W E NCY + I+ SYWGS M + + + ++
Sbjct: 289 TSMSPTHAKGIDWGGEPGQNCYNQTTLIEDPSYWGSDCRKSIMKVIGEVFGRSKTPIT-- 346
Query: 389 NVTQLSEYRKDGHPSIFRKFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
KD H SI++K W PL EQL NP+SY+DC+HWCLPG+ DTWNELLF
Sbjct: 347 ---------KDAHTSIYKKQWSPLTAEQLENPTSYADCVHWCLPGLQDTWNELLF 392
>UniRef100_Q9SEZ9 At2g40160/T7M7.25 [Arabidopsis thaliana]
Length = 427
Score = 315 bits (806), Expect = 2e-84
Identities = 168/443 (37%), Positives = 244/443 (54%), Gaps = 41/443 (9%)
Query: 8 KKKVPIFPFIVLLLLVFIAFSTLHNGSTIHKFHENPLHIHQEQASSTTYVKPNLPNHLKK 67
K + + F V+LL VF+ L+N E L + Q Q +T NL H+
Sbjct: 10 KAYLSLLYFAVILLPVFLLGCYLYN--------EKQLRVGQFQEFNTH----NLQEHITP 57
Query: 68 SQEILDRYSRCNSTVGYSGRKIARRGGSKSSSNRRVSSESCDVFSGKWVFDNASYPLYNE 127
Q+ + K V E CDVF+GKWV DN ++PLY E
Sbjct: 58 LQQSKE---------------------DKDKKTDLVPLEFCDVFTGKWVLDNVTHPLYKE 96
Query: 128 SDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWNVKEMWEKLRGKRLMFVGDSLNR 187
+C ++S+ +AC ++GR D YQ WRWQP DC L R++ K + EKLRGK+LMF+GDS++
Sbjct: 97 DECEFLSEWVACTRNGRPDSKYQKWRWQPQDCSLPRFDSKLLLEKLRGKKLMFIGDSIHY 156
Query: 188 GQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNATVEFLWAPLLAESNSDDP-VNHR 246
QW SMVC++QS IP+ K+++ A ++IF EEYNAT+ F WAP L ESN+D P
Sbjct: 157 NQWQSMVCMVQSVIPSGKKTLKHTAQMSIFNIEEYNATISFYWAPFLVESNADPPDKRDG 216
Query: 247 LDERIIRPDSVLKHASLWEHADILVFNTYLWW-RQGPVKLLWTDEEN-GACEELDGRG-- 302
+ +I P+S+ KH W+ AD L+FNTY+WW R +K+L + N G +E + G
Sbjct: 217 KTDPVIIPNSISKHGENWKDADYLIFNTYIWWTRHSTIKVLKQESFNKGDSKEYNEIGIY 276
Query: 303 -AMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWSREWNPESEGNCYGEKKPI-DVES 360
+ + W W+ ++P + +FF +MSPTH+ S +W C E +PI ++
Sbjct: 277 IVYKQVLSTWTKWLEQNINPSQTSIFFSSMSPTHIRSSDWGFNEGSKCEKETEPILNMSK 336
Query: 361 YWGSGSDLPTMSSVENILSSLNSKVSVLNVTQLSEYRKDGHPSIFRKF-WEPLRPEQLSN 419
G++ N S + LN+T +SEYRKDGH S + + + PEQ +
Sbjct: 337 PINVGTNRRLYEIALNATKSTKVPIHFLNITTMSEYRKDGHTSFYGSINGKLMTPEQKLD 396
Query: 420 PSSYSDCIHWCLPGVPDTWNELL 442
P +++DC HWCLPG+PD+WNELL
Sbjct: 397 PRTFADCYHWCLPGLPDSWNELL 419
>UniRef100_Q8S237 P0446G04.14 protein [Oryza sativa]
Length = 522
Score = 305 bits (781), Expect = 2e-81
Identities = 150/349 (42%), Positives = 206/349 (58%), Gaps = 16/349 (4%)
Query: 106 ESCDVFSGKWVFDNASYPLYNESDCPYMSDQLACNKHGRTDLGYQHWRWQPHDCDLKRWN 165
E+CD+ G+WVFDN SYPLY E C +++ Q+ C ++GR D YQ WRWQP DC + R++
Sbjct: 169 ETCDLSKGEWVFDNTSYPLYREEQCEFLTSQVTCMRNGRRDDTYQKWRWQPKDCSMPRFD 228
Query: 166 VKEMWEKLRGKRLMFVGDSLNRGQWISMVCLLQSEIPADKRSMSPNAHLTIFRAEEYNAT 225
K E+LRGKR MFVGDSLNR QW SMVCL+QS + K+ ++ +F A EYNAT
Sbjct: 229 AKLFMERLRGKRFMFVGDSLNRNQWESMVCLVQSAMSPGKKYVTWEDQRVVFHAVEYNAT 288
Query: 226 VEFLWAPLLAESNSDDPVNHRLDERIIRPDSVLKHASLWEHADILVFNTYLWWR------ 279
VEF WAP L ESNSDDP H + RII+ D++ HA W D LVFNTY+WW
Sbjct: 289 VEFYWAPFLVESNSDDPKIHSIQHRIIKADAIAAHAQNWRGVDYLVFNTYIWWMNTLNMK 348
Query: 280 -QGPVKLLWTDEENGACEELDGRGAMELAMGAWADWVSSKVDPLKKRVFFVTMSPTHLWS 338
P W + +E+ A + WA WV+ +DP + VFF+++SP H+
Sbjct: 349 IMRPGGQSWEEH-----DEVVRIEAYRKVLTTWASWVNDNIDPARTSVFFMSISPLHISP 403
Query: 339 REWNPESEGNCYGEKKP-IDVESYWGSGSDLPTMSSVENILSSLNSKVSV--LNVTQLSE 395
W C E P ++ G+D + N+ + ++V + ++VT +SE
Sbjct: 404 EVWGNPGGIRCAKETMPLLNWHGPIWLGTDWDMFHAAANVSRTAATRVPITFVDVTTMSE 463
Query: 396 YRKDGHPSIFR-KFWEPLRPEQLSNPSSYSDCIHWCLPGVPDTWNELLF 443
RKDGH S+ + + L PEQ ++P +Y+DCIHWCLPGVPD WN +L+
Sbjct: 464 RRKDGHTSVHTIRQGKVLTPEQQADPGTYADCIHWCLPGVPDIWNLILY 512
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.134 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 824,139,222
Number of Sequences: 2790947
Number of extensions: 35214309
Number of successful extensions: 79116
Number of sequences better than 10.0: 150
Number of HSP's better than 10.0 without gapping: 145
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 78487
Number of HSP's gapped (non-prelim): 192
length of query: 446
length of database: 848,049,833
effective HSP length: 131
effective length of query: 315
effective length of database: 482,435,776
effective search space: 151967269440
effective search space used: 151967269440
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC143338.3


