

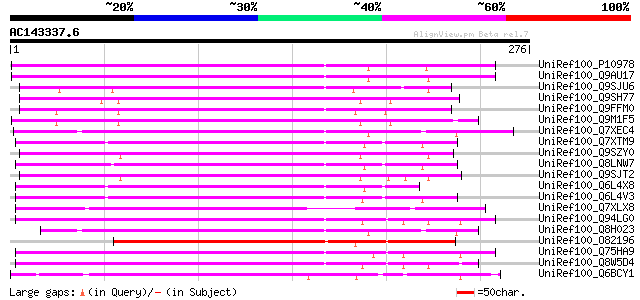
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.6 + phase: 0
(276 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P10978 Retrovirus-related Pol polyprotein from transpo... 176 6e-43
UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense] 164 2e-39
UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidop... 159 8e-38
UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidop... 159 1e-37
UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidops... 157 3e-37
UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana] 154 2e-36
UniRef100_Q7XEC4 Putative polyprotein [Oryza sativa] 154 3e-36
UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa] 151 2e-35
UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana] 150 3e-35
UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa] 148 2e-34
UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidop... 146 5e-34
UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa] 146 7e-34
UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa] 145 9e-34
UniRef100_Q7XLX8 OSJNBa0042I15.12 protein [Oryza sativa] 143 4e-33
UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sa... 143 4e-33
UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Or... 142 9e-33
UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabid... 142 1e-32
UniRef100_Q75HA9 Putative polyprotein [Oryza sativa] 137 2e-31
UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza... 137 2e-31
UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas] 135 1e-30
>UniRef100_P10978 Retrovirus-related Pol polyprotein from transposon TNT 1-94
[Contains: Protease (EC 3.4.23.-); Reverse transcriptase
(EC 2.7.7.49); Endonuclease] [Nicotiana tabacum]
Length = 1328
Score = 176 bits (446), Expect = 6e-43
Identities = 97/267 (36%), Positives = 150/267 (55%), Gaps = 11/267 (4%)
Query: 2 GSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSA 61
G K+++ KF GDN F W+ +M +LIQQ K L + P TM + + ++A SA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 62 VVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQL 121
+ L L D V+ + E TA +W +LESLYM+K+L ++ +LK+QLY+ M E + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 122 TEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRT 181
FN ++ L N+ V++E+EDKAILLL +LP S+++ T+L+GK T+ L++V +AL
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKT-TIELKDVTSALLL 181
Query: 182 KDLTKSKD-------LTHEHGEGLSVTRGNGGGRGNRRKSGNKSRF---ECFNCHKMGHF 231
+ + K +T G + N G G R KS N+S+ C+NC++ GHF
Sbjct: 182 NEKMRKKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRNCYNCNQPGHF 241
Query: 232 KKDCPKINGNSAQIVSEGYEDAGALMV 258
K+DCP + + +D A MV
Sbjct: 242 KRDCPNPRKGKGETSGQKNDDNTAAMV 268
>UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense]
Length = 1328
Score = 164 bits (416), Expect = 2e-39
Identities = 92/268 (34%), Positives = 152/268 (56%), Gaps = 12/268 (4%)
Query: 2 GSKWDIEKFTGDND-FGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARS 60
G K+++ KF GD F +W+ +M+ +LIQQ KAL G+ P +M + ++ +KA S
Sbjct: 3 GVKYEVAKFNGDKPVFSMWQRRMKDLLIQQGLHKALGGKSKKPESMKLEDWEELDEKAAS 62
Query: 61 AVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQ 120
A+ L L D V+ + E +A +W KLE+LYM+K+L ++ +LK+QLY+ M E +
Sbjct: 63 AIRLHLTDDVVNNIVDEESACGIWTKLENLYMSKTLTNKLYLKKQLYTLHMDEGTNFLSH 122
Query: 121 LTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALR 180
L N ++ L N+ V++E+EDK I+LL +LP S+++ T+L+GK+ ++ L++V +AL
Sbjct: 123 LNVLNGLITQLANLGVKIEEEDKRIVLLNSLPSSYDTLSTTILHGKD-SIQLKDVTSALL 181
Query: 181 TKDLTKSKD-------LTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFE---CFNCHKMGH 230
+ + K +T G + N G G R KS +S+ + C+NC + GH
Sbjct: 182 LNEKMRKKPENHGQVFITESRGRSYQRSSSNYGRSGARGKSKVRSKSKARNCYNCDQPGH 241
Query: 231 FKKDCPKINGNSAQIVSEGYEDAGALMV 258
FK+DCP + + +D A MV
Sbjct: 242 FKRDCPNPKRGKGESSGQKNDDNTAAMV 269
>UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 838
Score = 159 bits (402), Expect = 8e-38
Identities = 102/253 (40%), Positives = 147/253 (57%), Gaps = 25/253 (9%)
Query: 6 DIEKFTGDNDFGLWKVKMEA------VLIQQKCEKALKGEGSLPVTMSRAEKTK------ 53
++EK G+ D+ LWK K+ A +L + ++A++ E S T S KT+
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKE 66
Query: 54 MVDKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVE 113
KARS V+L LG+ VLR+V KE TAA M L+ L+M KSL +R +LKQ+LY ++M +
Sbjct: 67 KRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMSD 126
Query: 114 SKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLE 173
S I E + +F K++ DLEN++V + DED+AI+LL +LPK F+ KDT+ YGK T+ L+
Sbjct: 127 SMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGKT-TLALD 185
Query: 174 EVQAALRT---------KDLTKSKDLTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFE--C 222
E+ A+R+ K L S D G S R R N+ +S +KSR + C
Sbjct: 186 EITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSER-NKSQSRSKSREKKVC 244
Query: 223 FNCHKMGHFKKDC 235
+ C K GHFKK C
Sbjct: 245 WVCGKEGHFKKQC 257
>UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1356
Score = 159 bits (401), Expect = 1e-37
Identities = 99/259 (38%), Positives = 149/259 (57%), Gaps = 25/259 (9%)
Query: 6 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMS-----------RAEKTKM 54
++EKF G D+ +WK K+ A + ALK S S + EK +
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 55 VD----KARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFR 110
++ KARSA+VL + D+VLR++ KE+TAA+M L+ LYM+K+L +R + KQ+LYSF+
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 111 MVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGK-EGT 169
M E+ ++ + EF +I+ DLEN+ V + DED+AILLL ALPK+F+ KDT+ Y +
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSI 186
Query: 170 VTLEEVQAALRTKDLT--KSKDLTHEHGEGLSVT-------RGNGGGRGNRRKSGNKSRF 220
+TL+EV AA+ +K+L K EGL V +G G+G +K +K +
Sbjct: 187 LTLDEVAAAIYSKELELGSVKKSIKVQAEGLYVKDKNENKGKGEQKGKGKGKKGKSKKKP 246
Query: 221 ECFNCHKMGHFKKDCPKIN 239
C+ C + GHF+ CP N
Sbjct: 247 GCWTCGEEGHFRSSCPNQN 265
>UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1342
Score = 157 bits (397), Expect = 3e-37
Identities = 94/259 (36%), Positives = 145/259 (55%), Gaps = 30/259 (11%)
Query: 6 DIEKFTGDNDFGLWKVKM----------------EAVLIQQKCEKALKGEGSLPVTMSRA 49
++EKF GD D+ LWK K+ E +++ + G P T +
Sbjct: 7 EVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATSK 66
Query: 50 EKTKMVD----KARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQ 105
+ K++ KARS ++L LG+ VLR+V K+ TAA M L+ L+M KSL +R +LKQ+
Sbjct: 67 LEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQR 126
Query: 106 LYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYG 165
LY ++M E+ + E + +F K++ DLEN++V + DED+AI+LL +LP+ F+ K+T+ Y
Sbjct: 127 LYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYC 186
Query: 166 KEGTVTLEEVQAALRTK--DLTKSKDLTHEHGEGL-------SVTRGNGGGRGNRRKSGN 216
K T+ LEE+ +A+R+K +L S L + +GL S TRG G + R
Sbjct: 187 KT-TLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKNKSRSKSK 245
Query: 217 KSRFECFNCHKMGHFKKDC 235
+ C+ C K GHFKK C
Sbjct: 246 GAGKTCWICGKEGHFKKQC 264
>UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana]
Length = 1363
Score = 154 bits (390), Expect = 2e-36
Identities = 100/273 (36%), Positives = 152/273 (55%), Gaps = 27/273 (9%)
Query: 2 GSKWDIEKFTGDNDFGLWKVKM---------EAVLIQQKCEKALKGEGSLPVTMSRAEKT 52
G++ ++EKF G D+ +WK K+ AVL + + + + + E+
Sbjct: 3 GARIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEERE 62
Query: 53 KMVD------KARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQL 106
KM KARS +VL + D+VLR++ KE +AA+M L+ LYM+K+L +R +LKQ+L
Sbjct: 63 KMEAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKL 122
Query: 107 YSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGK 166
YSF+M E+ +I + EF I+ DLEN+ V + DED+AILLL +LPK F+ KDT+ Y
Sbjct: 123 YSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSS 182
Query: 167 EGTV-TLEEVQAALRTKDLT--KSKDLTHEHGEGLSVT-------RGNGGGRGNRRKSGN 216
TV +L+EV AA+ +++L K EGL V R +G ++S +
Sbjct: 183 GKTVLSLDEVAAAIYSRELEFGSVKKSIKGQAEGLYVKDKAENRGRSEQKDKGKGKRSKS 242
Query: 217 KSRFECFNCHKMGHFKKDCPKINGNSAQIVSEG 249
KS+ C+ C + GH K CP N N Q ++G
Sbjct: 243 KSKRGCWICGEDGHLKSTCP--NKNKPQFKNQG 273
>UniRef100_Q7XEC4 Putative polyprotein [Oryza sativa]
Length = 382
Score = 154 bits (388), Expect = 3e-36
Identities = 90/273 (32%), Positives = 157/273 (56%), Gaps = 12/273 (4%)
Query: 3 SKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAV 62
SK+++ KF G +F LW+++++ +L QQ KAL + ++P + + +M +A + +
Sbjct: 7 SKFEVVKFDGTGNFVLWQMRLKDLLAQQGISKAL--QETMPEKIDADKWNEMKAQAAATI 64
Query: 63 VLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLT 122
L L D V+ +V E + +W KL SL+M+KSL + +LKQQLY ++ E + + +
Sbjct: 65 RLSLSDSVMYQVMDEKSPKEIWDKLASLHMSKSLTSKLYLKQQLYGLQVQEESDLRKHVD 124
Query: 123 EFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTK 182
FN+++ DL ++V+L+DEDKAI+LLC+LP S+E T+ +GK+ TV EE+ ++L +
Sbjct: 125 VFNQLVVDLSKLDVKLDDEDKAIILLCSLPLSYEHVVTTLTHGKD-TVKTEEIISSLLAR 183
Query: 183 DLTKSKD---LTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCP--- 236
DL +SK G+ L V + G + +R C+ CH+ GH +++CP
Sbjct: 184 DLRRSKKNEATKASQGKSLLVKDKHDHEAGVSKSKEKGAR--CYKCHEFGHIRRNCPLLK 241
Query: 237 KINGNSAQIVSEGYE-DAGALMVWCCLEEEKGD 268
K G A + + G + D+G+ + +E G+
Sbjct: 242 KRKGGIASLAARGDDSDSGSHEILTVSDEMSGE 274
>UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa]
Length = 1181
Score = 151 bits (381), Expect = 2e-35
Identities = 94/251 (37%), Positives = 145/251 (57%), Gaps = 19/251 (7%)
Query: 4 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVV 63
K+D+ D F LW+VKM AVL QQ+ + AL G S EK K KA S +
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQELDDALSGFDKRTQDWSNDEK-KRDRKAMSYIH 63
Query: 64 LCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTE 123
L L + +L+EV KE TAA +W KLE + MTK L + LKQ+L+ ++ + ++M+ L+
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSA 123
Query: 124 FNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTKD 183
F +I+ DLE++EV+ +++D A++LLC+LP S+ +F+DT+LY ++ T+TL+EV AL K+
Sbjct: 124 FKEIVADLESMEVKYDEKDLALILLCSLPSSYANFRDTILYSRD-TLTLKEVYDALHAKE 182
Query: 184 LTKS---KDLTHEHGEGLSVTRGNGGGRGNRRKSGNKS------------RFE-CFNCHK 227
K + ++ EGL V RG+ + KS +KS R++ C C +
Sbjct: 183 KMKKMVPSEGSNSQAEGL-VVRGSQQEKNTNNKSRDKSSSSYRGRSKSRGRYKSCKYCKR 241
Query: 228 MGHFKKDCPKI 238
GH C K+
Sbjct: 242 DGHDISKCWKL 252
>UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana]
Length = 1230
Score = 150 bits (379), Expect = 3e-35
Identities = 95/256 (37%), Positives = 143/256 (55%), Gaps = 25/256 (9%)
Query: 6 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDK-------- 57
++EKF G D+ LWK K+ A + AL+ S+ + E+ K +K
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 58 -----ARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMV 112
ARS +VL + D+VLR+ KE TA SM L+ LYM+K+L +R +LKQ+LYS++M
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 113 ESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGK-EGTVT 171
E+ ++ + EF +++ DLEN V + DED+AILLL +LPK F+ KDT+ YG T++
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS 186
Query: 172 LEEVQAALRTKDLT--KSKDLTHEHGEGLSV-TRGNGGGRGNRRKSGNKSRFE------- 221
++EV AA+ +K+L +K EGL V + G +++ GNK R
Sbjct: 187 VDEVVAAIYSKELELGSNKKSIRGQAEGLYVKDKPETRGMSEQKEKGNKGRSRSRSKGWK 246
Query: 222 -CFNCHKMGHFKKDCP 236
C+ C + GHFK CP
Sbjct: 247 GCWICGEEGHFKTSCP 262
>UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa]
Length = 1280
Score = 148 bits (373), Expect = 2e-34
Identities = 93/251 (37%), Positives = 142/251 (56%), Gaps = 19/251 (7%)
Query: 4 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVV 63
K+D+ D F LW+VKM AVL QQ + AL G S EK K KA S +
Sbjct: 40 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKKKD-RKAMSYIH 98
Query: 64 LCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTE 123
L L + +L+EV KE TAA +W KLE + MTK L + LKQ+L+ ++ + ++M+ L+
Sbjct: 99 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLST 158
Query: 124 FNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTKD 183
F +I+ DLE+IEV+ ++ED ++LLC+LP S+ +F+DT+LY + T+ L+EV AL K+
Sbjct: 159 FKEIVADLESIEVKYDEEDLGLILLCSLPSSYANFRDTILYSHD-TLILKEVYDALHAKE 217
Query: 184 LTKS---KDLTHEHGEGLSVTRGNGGGRGNRRKSGNKS------------RFE-CFNCHK 227
K + ++ EGL V RG + + +S +KS R++ C C +
Sbjct: 218 KMKKMVPSEGSNSQAEGL-VVRGRQQEKNTKNQSRDKSSSSYRGRSKSRGRYKSCKYCKR 276
Query: 228 MGHFKKDCPKI 238
GH +C K+
Sbjct: 277 DGHDISECWKL 287
>UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1333
Score = 146 bits (369), Expect = 5e-34
Identities = 100/263 (38%), Positives = 150/263 (57%), Gaps = 28/263 (10%)
Query: 6 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDK-------- 57
++EKF G D+ +WK K+ A L ALK E L ++ + T+ +K
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 58 -------ARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFR 110
ARSA+VL + D+VLR++ KE +AA+M L+ LYM+K+L +R + KQ+LYSF+
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 111 MVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYG-KEGT 169
M E+ +I + EF +I+ DLEN V + DED+AILLL +LPK F+ +DT+ YG T
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVT 186
Query: 170 VTLEEVQAALRTKDLT--KSKDLTHEHGEGLSV-----TRGNGGGRG---NRRKSGNKSR 219
++L+EV AA+ +K+L +K EGL V TRG RG N +KS +KSR
Sbjct: 187 LSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKEKTETRGRTEQRGNNNNNKKSRSKSR 246
Query: 220 FE--CFNCHKMGHFKKDCPKING 240
+ C+ C + + + + NG
Sbjct: 247 SKKGCWICGESSNGSSNYSEANG 269
>UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa]
Length = 1211
Score = 146 bits (368), Expect = 7e-34
Identities = 87/218 (39%), Positives = 129/218 (58%), Gaps = 6/218 (2%)
Query: 4 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVV 63
K+D+ D F LW+VKM AVL QQ + AL G S EK K K S +
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEK-KRDRKTMSYIH 63
Query: 64 LCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTE 123
L L + +L+EV KE TAA +W KLE + MTK L + LKQ+L+ ++ + ++M+ L+
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSA 123
Query: 124 FNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTKD 183
F KI+ DLE++EV+ ++ED ++LLC+LP S+ +F+DT+LY + T+TL+EV AL K+
Sbjct: 124 FKKIVADLESMEVKYDEEDLCLILLCSLPSSYANFRDTILYSCD-TLTLKEVYDALHAKE 182
Query: 184 LTKS---KDLTHEHGEGLSVTRGNGGGRGNRRKSGNKS 218
K + ++ EGL V RG + KS +KS
Sbjct: 183 KIKKMVPSEGSNSQAEGL-VVRGRQQEKNTNSKSRDKS 219
>UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa]
Length = 1243
Score = 145 bits (367), Expect = 9e-34
Identities = 90/251 (35%), Positives = 144/251 (56%), Gaps = 19/251 (7%)
Query: 4 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVV 63
K+D+ D F LW+VKM AVL QQ + AL G S EK K KA S +
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEK-KRDRKAISYIH 63
Query: 64 LCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTE 123
L L + +L+EV KE TAA +W KLE + MTK L + LKQ+L+ ++ + +++M+ L+
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDESVMDHLSA 123
Query: 124 FNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTKD 183
F +I+ DLE++EV+ +++D ++LLC+LP S+ +F+ T+LY ++ T+TL+EV A K+
Sbjct: 124 FKEIVADLESMEVKYDEDDLGLILLCSLPSSYANFRGTILYSRD-TLTLKEVYDAFHAKE 182
Query: 184 LTK---SKDLTHEHGEGLSVTRGNGGGRGNRRKSGNKS------------RFE-CFNCHK 227
K + + ++ EGL V RG + + +S +KS R++ C C +
Sbjct: 183 KMKKMVTSEGSNSQAEGL-VVRGRQQKKNTKNQSRDKSSSSYRGRTKSRGRYKSCKYCKR 241
Query: 228 MGHFKKDCPKI 238
GH +C K+
Sbjct: 242 DGHDISECWKL 252
>UniRef100_Q7XLX8 OSJNBa0042I15.12 protein [Oryza sativa]
Length = 432
Score = 143 bits (361), Expect = 4e-33
Identities = 77/250 (30%), Positives = 136/250 (53%), Gaps = 29/250 (11%)
Query: 4 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVV 63
K+D+ KF G +FGLW+ +++ +L QQ+ KALKG P M + +M +A + +
Sbjct: 43 KFDLVKFDGFGNFGLWQTRVKDLLAQQEVSKALKGVK--PAKMEDDDCEEMQLQAAATIR 100
Query: 64 LCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTE 123
CL D+++ V +E + +W KLE+ +M+K+L ++++LKQ+LY +M E + +
Sbjct: 101 RCLSDQIMYHVMEETSPKIIWEKLEAQFMSKTLTNKRYLKQKLYGLKMQEGADLTAHVNV 160
Query: 124 FNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTKD 183
FN+++ DL ++V+++DEDKAI+LLC+LP+S+E
Sbjct: 161 FNQLVTDLVKMDVEVDDEDKAIVLLCSLPESYEHV------------------------- 195
Query: 184 LTKSKDLTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSA 243
+ K+K+ EGL V + G + + K K +CF C + GH +++CP + G +
Sbjct: 196 MRKNKEGETSQAEGLLVKEDHAGHKVKKNK--KKKMVKCFRCDEWGHIRRECPTLKGKAK 253
Query: 244 QIVSEGYEDA 253
V+ D+
Sbjct: 254 ANVATSSNDS 263
>UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1326
Score = 143 bits (361), Expect = 4e-33
Identities = 94/278 (33%), Positives = 152/278 (53%), Gaps = 25/278 (8%)
Query: 4 KWDIEKFTGDNDFGLWKVKMEAVLIQQK-CEKALKGEGSLPVTMSRAEKTKMVDKARSAV 62
K+D+ F LW+VKM A+L Q ++AL+ G T AE+ + KA +
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAILAQTSDLDEALESFGKKKSTEWTAEEKRKDRKALLLI 64
Query: 63 VLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLT 122
L L + +L+EV +E TAA +W KLES+ M+K L + +K +L+S ++ ES +++ ++
Sbjct: 65 QLHLSNDILQEVLQEKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHIS 124
Query: 123 EFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTK 182
F +I+ DL +IEVQ +DED +LLLC+LP S+ +F+DT+L ++ +TL EV AL+ +
Sbjct: 125 VFKEIVVDLVSIEVQFDDEDLGLLLLCSLPSSYANFRDTILLSRD-ELTLAEVYEALQNR 183
Query: 183 DLTK---SKDLTHEHGEGLSVTRGNGGGR-----GNRRKSGNKSRFE------CFNCHKM 228
+ K D + GE L V RG R +R KS ++ R + C C K
Sbjct: 184 EKMKGMVQSDASSSKGEALQV-RGRSEQRTYNDSSDRDKSQSRGRSKSRGKKFCKYCKKK 242
Query: 229 GHFKKDCPKI--------NGNSAQIVSEGYEDAGALMV 258
HF ++C K+ +G ++ + S D+G +V
Sbjct: 243 NHFIEECWKLQNKEKRKSDGKASVVTSAENSDSGDCLV 280
>UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Oryza sativa]
Length = 556
Score = 142 bits (358), Expect = 9e-33
Identities = 85/239 (35%), Positives = 136/239 (56%), Gaps = 11/239 (4%)
Query: 17 GLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVVLCLGDKVLREVAK 76
G +++++ +L QQ KAL E ++P M + +M +A + + L L D V+ +V
Sbjct: 152 GSSQMRLKDLLAQQGISKAL--EETMPEKMDVGKWVEMKAQATAIIRLSLSDFVMYQVMD 209
Query: 77 EATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEV 136
E T +W KL SLYM+KSL + +LKQQLY +M E + + + FN+++ DL ++V
Sbjct: 210 EKTPKEIWDKLASLYMSKSLTSKLYLKQQLYGLQMQEESDLRKHVDVFNQLVVDLSKLDV 269
Query: 137 QLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTKDLTKSKD---LTHE 193
+L+DEDKAI+LLC+LP SFE T+ +GK+ TV EE+ ++L +DL +SK +
Sbjct: 270 KLDDEDKAIILLCSLPPSFEHVVTTLTHGKD-TVKTEEIISSLLARDLRRSKKNEAMEAS 328
Query: 194 HGEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCP---KINGNSAQIVSEG 249
E L V + G + +R C+ CH+ GH +++CP K G A + + G
Sbjct: 329 QAESLLVKAKHDHEAGVSKSKEKGAR--CYKCHEFGHIRRNCPLLKKRKGGIASLAARG 385
>UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1137
Score = 142 bits (357), Expect = 1e-32
Identities = 74/184 (40%), Positives = 117/184 (63%), Gaps = 4/184 (2%)
Query: 56 DKARSAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESK 115
+KA + + +GDKVLR + TAA WA L+ LY+ KSL +R +L+ ++Y++RM +SK
Sbjct: 42 EKAMDMIFINVGDKVLRNIENSKTAAEAWATLDKLYLVKSLPNRVYLQLKVYNYRMQDSK 101
Query: 116 AIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEV 175
+ E + EF K++ DL N+++Q+ DE +AIL+L ALP S++ K+T+ YG+EG + L++V
Sbjct: 102 TLEENVDEFQKMISDLNNLQIQVPDEVQAILILSALPDSYDMLKETLKYGREG-IKLDDV 160
Query: 176 QAALRTK--DLTKSKDLTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKK 233
+A ++K +L S + GEGL V RG RG+ + + C+ C K GHFK+
Sbjct: 161 ISAAKSKELELRDSSGGSRPVGEGLYV-RGKSQARGSDGPKSTEGKKVCWICGKEGHFKR 219
Query: 234 DCPK 237
C K
Sbjct: 220 QCYK 223
>UniRef100_Q75HA9 Putative polyprotein [Oryza sativa]
Length = 1322
Score = 137 bits (346), Expect = 2e-31
Identities = 90/278 (32%), Positives = 151/278 (53%), Gaps = 25/278 (8%)
Query: 4 KWDIEKFTGDNDFGLWKVKMEAVLIQQK-CEKALKGEGSLPVTMSRAEKTKMVDKARSAV 62
K+D+ F LW+VKM AVL Q ++AL+ G T AE+ + KA S +
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 64
Query: 63 VLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLT 122
L L + +L+EV ++ TAA +W KLES+ M+K L + +K +L+S ++ ES +++ ++
Sbjct: 65 QLHLSNDILQEVLQKKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLHESGSVLNHIS 124
Query: 123 EFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTK 182
F +I+ DL ++EVQ +DED +LLLC+LP S+ +F+ T+L ++ +TL EV AL+ +
Sbjct: 125 VFKEIVADLVSMEVQFDDEDLGLLLLCSLPSSYANFRHTILLSRD-ELTLAEVYEALQNR 183
Query: 183 DLTKSKDLTH---EHGEGLSVTRGNGGGR-----------GNRRKSGNKSRFECFNCHKM 228
+ K ++ GE L V RG R +R +S ++ + C C K
Sbjct: 184 EKMKGMVQSYASSSKGEALQV-RGRSEQRTYNDSNDHDKSQSRGRSKSRGKKFCKYCKKK 242
Query: 229 GHFKKDCPKI--------NGNSAQIVSEGYEDAGALMV 258
HF ++C K+ +G ++ + S D+G +V
Sbjct: 243 NHFIEECWKLQNKEKRKSDGKASVVTSAENSDSGDCLV 280
>UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza sativa]
Length = 1229
Score = 137 bits (346), Expect = 2e-31
Identities = 90/261 (34%), Positives = 146/261 (55%), Gaps = 18/261 (6%)
Query: 4 KWDIEKFTGDNDFGLWKVKMEAVLIQQK-CEKALKGEGSLPVTMSRAEKTKMVDKARSAV 62
K+D+ F LW+VKM AVL Q ++AL+ G T AE+ + KA S +
Sbjct: 2 KYDLPLQDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 61
Query: 63 VLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLT 122
L L + +L++V +E TAA +W KLES+ M+K L + +K +L+S ++ ES +++ ++
Sbjct: 62 QLHLSNDILQKVLQEKTAAELWFKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHIS 121
Query: 123 EFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTK 182
F +I+ DL ++EVQ +DED +LLLC+LP + +F+DT+L ++ +TL EV AL+ +
Sbjct: 122 VFKEIIADLVSMEVQFDDEDLGLLLLCSLPSLYANFRDTILLSRD-ELTLAEVYEALQNR 180
Query: 183 DLTK---SKDLTHEHGEGLSVTRGNGGGR-----GNRRKSGNKSRFE------CFNCHKM 228
+ K D + G+ L V RG R +R KS ++ R + C C K
Sbjct: 181 EKMKGMVQSDASSSKGKALQV-RGRSEQRTYNDSNDRDKSQSRGRSKSRGKKFCKYCKKK 239
Query: 229 GHFKKDCPKINGNSAQIVSEG 249
HF ++C K+ N + S+G
Sbjct: 240 NHFIEECWKLQ-NKEKRKSDG 259
>UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas]
Length = 1298
Score = 135 bits (340), Expect = 1e-30
Identities = 91/277 (32%), Positives = 142/277 (50%), Gaps = 27/277 (9%)
Query: 1 MGSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEK-TKMVDKAR 59
M +K++IEKF G N F LWK+K++A+L + C A+ PV + +K ++M + A
Sbjct: 1 MAAKFEIEKFNGKN-FSLWKLKVKAILRKDNCLAAISER---PVDFTDDKKWSEMNEDAM 56
Query: 60 SAVVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIME 119
+ + L + D VL + ++ TA +W L LY KSL ++ FLK++LY+ RM ES ++ E
Sbjct: 57 ADLYLSIADGVLSSIEEKKTANEIWDHLNRLYEAKSLHNKIFLKRKLYTLRMSESTSVTE 116
Query: 120 QLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFES---------FKDTMLYGKEGTV 170
L N + L ++ ++E +++A LLL +LP S++ D +++
Sbjct: 117 HLNTLNTLFSQLTSLSCKIEPQERAELLLQSLPDSYDQLIINLTNNILTDYLVFDDVAAA 176
Query: 171 TLEEVQAALRTKD----LTKSKDLTHEHGEGLSVTRGNGGGRGNRRKSGNKSRFECFNCH 226
LEE +D L +++ LT G S RG GRG R +K C+NC
Sbjct: 177 VLEEESRRKNKEDRQVNLQQAEALTVMRGR--STERGQSSGRG--RSKSSKKNLTCYNCG 232
Query: 227 KMGHFKKDCPKI--NGNSAQIVSEGYEDAGALMVWCC 261
K GH KKDC + N N V+ +D AL CC
Sbjct: 233 KKGHLKKDCWNLAQNSNPQGNVASTSDDGSAL---CC 266
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 445,596,321
Number of Sequences: 2790947
Number of extensions: 17751588
Number of successful extensions: 71459
Number of sequences better than 10.0: 878
Number of HSP's better than 10.0 without gapping: 489
Number of HSP's successfully gapped in prelim test: 401
Number of HSP's that attempted gapping in prelim test: 70118
Number of HSP's gapped (non-prelim): 1432
length of query: 276
length of database: 848,049,833
effective HSP length: 125
effective length of query: 151
effective length of database: 499,181,458
effective search space: 75376400158
effective search space used: 75376400158
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC143337.6


