

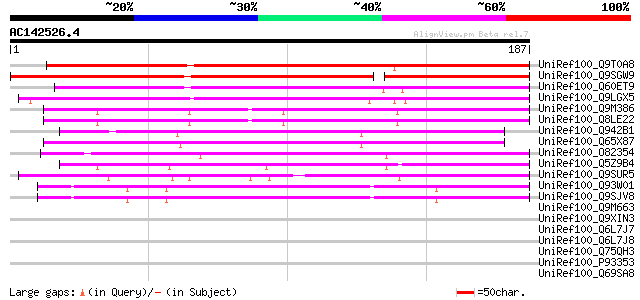
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142526.4 + phase: 0
(187 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9T0A8 Hypothetical protein AT4g23930 [Arabidopsis tha... 192 3e-48
UniRef100_Q9SGW9 F1N19.3 [Arabidopsis thaliana] 171 1e-41
UniRef100_Q60ET9 Hypothetical protein OJ1122_B08.9 [Oryza sativa] 134 2e-30
UniRef100_Q9LGX5 Hypothetical protein [Oryza sativa] 129 4e-29
UniRef100_Q9M386 Hypothetical protein F24B22.160 [Arabidopsis th... 93 3e-18
UniRef100_Q8LE22 Hypothetical protein [Arabidopsis thaliana] 91 1e-17
UniRef100_Q942B1 B1060H01.30 protein [Oryza sativa] 70 4e-11
UniRef100_Q65X87 Hypothetical protein OJ1593_C11.15 [Oryza sativa] 66 5e-10
UniRef100_O82354 Hypothetical protein At2g46150 [Arabidopsis tha... 64 3e-09
UniRef100_Q5Z9B4 Hypothetical protein P0485D10.5 [Oryza sativa] 55 1e-06
UniRef100_Q9SUR5 Hypothetical protein F9D16.80 [Arabidopsis thal... 47 2e-04
UniRef100_Q93W01 At2g01080/F23H14.5 [Arabidopsis thaliana] 45 7e-04
UniRef100_Q9SJV8 Expressed protein [Arabidopsis thaliana] 45 7e-04
UniRef100_Q9M663 Harpin inducing protein [Nicotiana tabacum] 45 0.001
UniRef100_Q9XIN3 Expressed protein [Arabidopsis thaliana] 44 0.002
UniRef100_Q6L7J7 Harpin inducing protein 1-like 18 [Nicotiana ta... 44 0.002
UniRef100_Q6L7J8 Harpin inducing protein 1-like 9 [Nicotiana tab... 44 0.003
UniRef100_Q75QH3 Hin1 like protein [Capsicum chinense] 42 0.006
UniRef100_P93353 Hin1 protein [Nicotiana tabacum] 42 0.008
UniRef100_Q69SA8 Hypothetical protein OSJNBb0052O11.109 [Oryza s... 42 0.010
>UniRef100_Q9T0A8 Hypothetical protein AT4g23930 [Arabidopsis thaliana]
Length = 187
Score = 192 bits (489), Expect = 3e-48
Identities = 95/184 (51%), Positives = 134/184 (72%), Gaps = 12/184 (6%)
Query: 14 TNLASCLVATVFLIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNNSTTVNFTFSQ 73
+NLASC VAT+F++F++I T+Y T+F+P+DP+ISVT+V++PSF++ N+S V+FTFSQ
Sbjct: 6 SNLASCAVATLFIVFLIIAALTVYLTVFRPRDPEISVTSVKVPSFSVANSS--VSFTFSQ 63
Query: 74 YTSVKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQFMAATFTVQSLPLNLE 133
+++V+NPNR FSHY++ QL YG +IG+ FVPAG+I + RT+ M ATF+VQS PL
Sbjct: 64 FSAVRNPNRAAFSHYNNVIQLFYYGNRIGYTFVPAGEIESGRTKRMLATFSVQSFPLAAA 123
Query: 134 PEGL----------QMGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAIAVSDGSVL 183
+ G TVEIES +EM GRVRVL LF+H + AK +CR+AI+ SDGS++
Sbjct: 124 SSSQISAAQFQNSDRSGSTVEIESKLEMAGRVRVLGLFTHRIAAKCNCRIAISSSDGSIV 183
Query: 184 GFHC 187
C
Sbjct: 184 AVRC 187
>UniRef100_Q9SGW9 F1N19.3 [Arabidopsis thaliana]
Length = 342
Score = 171 bits (432), Expect = 1e-41
Identities = 82/131 (62%), Positives = 102/131 (77%), Gaps = 2/131 (1%)
Query: 1 MTKHEHETPPSNRTNLASCLVATVFLIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNL 60
M K S RTNLASC VATVFL+ +L+++ +YFT+FKP+DPKISV AVQLPSF +
Sbjct: 1 MAKPHDRRRSSGRTNLASCAVATVFLLILLVVLLVVYFTVFKPKDPKISVNAVQLPSFAV 60
Query: 61 TNNSTTVNFTFSQYTSVKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQFMA 120
+NN T NF+FSQY +V+NPNR FSHYDSS QLL G Q+GFMF+PAGKI++ R Q+MA
Sbjct: 61 SNN--TANFSFSQYVAVRNPNRAVFSHYDSSIQLLYSGNQVGFMFIPAGKIDSGRIQYMA 118
Query: 121 ATFTVQSLPLN 131
ATFTV S P++
Sbjct: 119 ATFTVHSFPIS 129
Score = 82.8 bits (203), Expect = 4e-15
Identities = 34/52 (65%), Positives = 47/52 (90%)
Query: 136 GLQMGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAIAVSDGSVLGFHC 187
G ++GPT+EIES +E+ GRV+VLH+F+HHV AK+DCRV ++++DGSVLGFHC
Sbjct: 291 GNRVGPTMEIESKMELAGRVKVLHVFTHHVVAKSDCRVTVSIADGSVLGFHC 342
>UniRef100_Q60ET9 Hypothetical protein OJ1122_B08.9 [Oryza sativa]
Length = 233
Score = 134 bits (336), Expect = 2e-30
Identities = 76/209 (36%), Positives = 112/209 (53%), Gaps = 40/209 (19%)
Query: 17 ASCLVATVFLIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNNSTTVNFTFSQYTS 76
ASC+VA +F++ F L++PQ P I+VTAVQLPSF N TV FTF Q S
Sbjct: 27 ASCVVAALFILLAAGGAAAALFLLYRPQAPAIAVTAVQLPSFASRNG--TVAFTFQQLAS 84
Query: 77 VKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQFMAATFTVQSLPLNLE--- 133
V+NPNR +HYDSS ++ G ++G M++PAG+I+ RTQ+MA +FTV + +
Sbjct: 85 VRNPNRSPLAHYDSSLRVAYAGGEVGSMYIPAGQIDGGRTQYMATSFTVPAFAVTSSATA 144
Query: 134 ---------------------------------PEGLQMG--PTVEIESTIEMVGRVRVL 158
P+ Q+ P +E++S + + G+V +L
Sbjct: 145 AASSSPAQTITVPASGPSPAAVGAVALQQEQPPPQQQQVAALPVMEVDSLLVVKGKVTIL 204
Query: 159 HLFSHHVEAKADCRVAIAVSDGSVLGFHC 187
+F+HHV A CR+ ++ +DG VLGF C
Sbjct: 205 RVFTHHVVAAKVCRIGVSPADGRVLGFRC 233
>UniRef100_Q9LGX5 Hypothetical protein [Oryza sativa]
Length = 299
Score = 129 bits (324), Expect = 4e-29
Identities = 80/203 (39%), Positives = 115/203 (56%), Gaps = 20/203 (9%)
Query: 4 HEH-ETPPSNRTNLASCLVATVFLIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNLTN 62
H H T S+ + LASCLVA FL + F LF+P+ P I+V AV+LP+F
Sbjct: 98 HLHVRTARSSSSALASCLVAAAFLALAVGGAGAALFVLFRPRPPDIAVAAVRLPAFASGP 157
Query: 63 NSTTVNFTFSQYTSVKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQFMAAT 122
N T V FTF Q +V+NPNR +H+DSS ++ G ++G +++PAG I+ RT+ M+A+
Sbjct: 158 NGT-VAFTFEQTAAVRNPNRAPLAHFDSSLRVAYAGGELGSVYIPAGLIDGGRTKDMSAS 216
Query: 123 FTVQSL----PLNLEPEGL---------QMGP-----TVEIESTIEMVGRVRVLHLFSHH 164
F V + P +L E + Q P +E++S + + GRV VL + +HH
Sbjct: 217 FAVPAFAAATPPSLPQEQMAAAAAASAQQQQPAAAAAVMEVDSLLVVKGRVTVLRVLTHH 276
Query: 165 VEAKADCRVAIAVSDGSVLGFHC 187
VEA CRV ++ DG VLGF C
Sbjct: 277 VEAAKVCRVGVSPVDGKVLGFRC 299
>UniRef100_Q9M386 Hypothetical protein F24B22.160 [Arabidopsis thaliana]
Length = 235
Score = 93.2 bits (230), Expect = 3e-18
Identities = 63/183 (34%), Positives = 102/183 (55%), Gaps = 9/183 (4%)
Query: 13 RTNLASCLVATVFLIFIL-IIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNN----STTV 67
+ N C+ T+ LI ++ I+I L FTLFKP+ P ++ +V + + N +
Sbjct: 48 KRNCKICICFTILLILLIAIVIVILAFTLFKPKRPTTTIDSVTVDRLQASVNPLLLKVLL 107
Query: 68 NFTFSQYTSVKNPNRGTFSHYDSSFQLLSY-GKQIGFMFVPAGKINARRTQFMAATFTVQ 126
N T + S+KNPNR FS YDSS LL+Y G+ IG +PA +I AR+T + T T+
Sbjct: 108 NLTLNVDLSLKNPNRIGFS-YDSSSALLNYRGQVIGEAPLPANRIAARKTVPLNITLTLM 166
Query: 127 SLPLNLEPEGLQ--MGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAIAVSDGSVLG 184
+ L E + L M + + + +++ G+V VL +F V++ + C ++I+VSD +V
Sbjct: 167 ADRLLSETQLLSDVMAGVIPLNTFVKVTGKVTVLKIFKIKVQSSSSCDLSISVSDRNVTS 226
Query: 185 FHC 187
HC
Sbjct: 227 QHC 229
>UniRef100_Q8LE22 Hypothetical protein [Arabidopsis thaliana]
Length = 235
Score = 91.3 bits (225), Expect = 1e-17
Identities = 62/183 (33%), Positives = 102/183 (54%), Gaps = 9/183 (4%)
Query: 13 RTNLASCLVATVFLIFIL-IIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNN----STTV 67
+ N C+ T+ LI ++ I+I L FTLFKP+ P ++ +V + + N +
Sbjct: 48 KRNCKICICFTILLILLIAIVIVILAFTLFKPKRPTTTIDSVTVDRLQASVNPLLLKVLL 107
Query: 68 NFTFSQYTSVKNPNRGTFSHYDSSFQLLSY-GKQIGFMFVPAGKINARRTQFMAATFTVQ 126
N T + S+KNPNR FS YDSS LL+Y G+ IG +PA +I AR+T + T T+
Sbjct: 108 NLTLNVDLSLKNPNRIGFS-YDSSSALLNYRGQVIGEAPLPANRIAARKTVPLNITLTLM 166
Query: 127 SLPLNLEPEGLQ--MGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAIAVSDGSVLG 184
+ L E + L M + + + +++ G+V VL +F V++ + C ++I+VS+ +V
Sbjct: 167 ADRLLSETQLLSDVMAGVIPLNTFVKVTGKVTVLKIFKIKVQSSSSCDLSISVSNRNVTS 226
Query: 185 FHC 187
HC
Sbjct: 227 QHC 229
>UniRef100_Q942B1 B1060H01.30 protein [Oryza sativa]
Length = 251
Score = 69.7 bits (169), Expect = 4e-11
Identities = 44/166 (26%), Positives = 81/166 (48%), Gaps = 8/166 (4%)
Query: 19 CLVATVFLIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFN----LTNNSTTVNFTFSQY 74
C V + ++ I+I++ L T+FK +DP++++ V L + + + +VN T +
Sbjct: 71 CCVTSAVVVGIVILVLAL--TVFKVKDPELTMNRVTLEGLDGDLGTSRHPVSVNATLNAD 128
Query: 75 TSVKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQFMAATFTV--QSLPLNL 132
S++NPN +F S G+ +G + P G++ A T M T V + N+
Sbjct: 129 VSLRNPNVASFRFDRSETDFYYAGETVGVAYAPEGEVGADSTVRMNVTLDVLADRISPNV 188
Query: 133 EPEGLQMGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAIAVS 178
L G + S E+ GRV VL ++ +++ K +C + + VS
Sbjct: 189 NATDLIFGQGYNLTSYTEISGRVNVLGIYKRNLDIKMNCSITLEVS 234
>UniRef100_Q65X87 Hypothetical protein OJ1593_C11.15 [Oryza sativa]
Length = 253
Score = 65.9 bits (159), Expect = 5e-10
Identities = 45/175 (25%), Positives = 78/175 (43%), Gaps = 9/175 (5%)
Query: 13 RTNLASCLVATVFLIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNL-------TNNST 65
R L C L+ + ++I L T+F+ +DP+I++ V + + + ++
Sbjct: 51 RRALCCCGCCVTTLVVVGLVILVLALTVFRVKDPRITMNGVWVTAISTGPGTGAGIGSTV 110
Query: 66 TVNFTFSQYTSVKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQFMAATFTV 125
N T + SVKNPN + S + GK + +VPAG + A RT M T +
Sbjct: 111 ATNATLTADVSVKNPNAASLRFSRSETDVYYKGKTVSVAYVPAGSVGADRTVRMNITLDL 170
Query: 126 --QSLPLNLEPEGLQMGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAIAVS 178
L L GL +G ++ + M RV VL + ++ + +C V + V+
Sbjct: 171 LADRLASVLNGTGLILGQEYDLTTYTAMRARVSVLGIIKKSLDVRMNCSVILDVA 225
>UniRef100_O82354 Hypothetical protein At2g46150 [Arabidopsis thaliana]
Length = 221
Score = 63.5 bits (153), Expect = 3e-09
Identities = 43/184 (23%), Positives = 81/184 (43%), Gaps = 10/184 (5%)
Query: 12 NRTNLASCLVATVFLIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNNSTTV---- 67
NR + C+ AT ++ I+ TL FT+F+ +DP I + V + + + V
Sbjct: 34 NRIKCSICVTATSLIL--TTIVLTLVFTVFRVKDPIIKMNGVMVNGLDSVTGTNQVQLLG 91
Query: 68 -NFTFSQYTSVKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQFMAATFTVQ 126
N + SVKNPN +F + +++ + G +G GK RT M T +
Sbjct: 92 TNISMIVDVSVKNPNTASFKYSNTTTDIYYKGTLVGEAHGLPGKARPHRTSRMNVTVDIM 151
Query: 127 SLPLNLEP---EGLQMGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAIAVSDGSVL 183
+ +P + V + S + G+V+++ + HV K +C +A+ ++ ++
Sbjct: 152 LDRILSDPGLGREISRSGLVNVWSYTRVGGKVKIMGIVKKHVTVKMNCTMAVNITGQAIQ 211
Query: 184 GFHC 187
C
Sbjct: 212 DVDC 215
>UniRef100_Q5Z9B4 Hypothetical protein P0485D10.5 [Oryza sativa]
Length = 278
Score = 54.7 bits (130), Expect = 1e-06
Identities = 46/180 (25%), Positives = 79/180 (43%), Gaps = 12/180 (6%)
Query: 19 CLVATVFLIFIL-IIIFTLYFTLFKPQDPKISVTAVQLP--SFNLTNNSTTVNFTFSQYT 75
CLV T+ + +L + + L T+F+ +DP + +V++ S NL S +N T
Sbjct: 94 CLVVTIATLALLGVAVLVLSLTVFRVRDPATRLVSVRVVGVSPNLAPPSPQINVTLLLTV 153
Query: 76 SVKNPNRGTFSHYDSS---FQLLSYGKQIGFMFVPAGKINARRTQFMAATFTVQSLPLNL 132
+V NPN +F++ S L G +G V AG+I +R + TV S
Sbjct: 154 AVHNPNPASFTYSSDSGGHADLTYRGAHVGDAVVEAGRIPSRGDGAVQMEMTVLSSSFTG 213
Query: 133 EP-----EGLQMGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAIAVSDGSVLGFHC 187
+ ++ G V +++ + G+V V + H A +DC V V + + C
Sbjct: 214 DVMAELIRDIEAG-AVPFDASARIPGKVAVFGVLKLHAVAYSDCHVVFGVPEMGIRSQEC 272
>UniRef100_Q9SUR5 Hypothetical protein F9D16.80 [Arabidopsis thaliana]
Length = 228
Score = 47.0 bits (110), Expect = 2e-04
Identities = 49/196 (25%), Positives = 85/196 (43%), Gaps = 16/196 (8%)
Query: 4 HEHETPPSNRTNLASCLVATVFLIFILIIIF-TLYFTLFKPQDPKISVTAVQLPS-FNLT 61
H+ ++T L C L ++ + F L T+F P ++V ++ F+
Sbjct: 36 HDRTKYVHSQTKLILCCGFIASLTMLIAVTFIVLSLTVFHLHSPNLTVDSISFNQRFDFV 95
Query: 62 NN--STTVNFTFSQYTSVKNPNRGTF--SHYDSSF---QLLSYGKQIGFMFVPAGKINAR 114
N +T N T S S+ NPN F + + SF +L+ G+ I + I A+
Sbjct: 96 NGKVNTNQNTTVSVEISLHNPNPALFIVKNVNVSFYHGELVVVGESIR----RSETIPAK 151
Query: 115 RTQFMAATFTVQSLPLNLEPEGLQM---GPTVEIESTIEMVGRVRVLHLFSHHVEAKADC 171
RT M T + L GL G V+++S++E+ GRV+ + +F V + DC
Sbjct: 152 RTVKMNLTAEIVKTKLLASLPGLMEDLNGRGVDLKSSVEVRGRVKKMKIFRKTVHLQTDC 211
Query: 172 RVAIAVSDGSVLGFHC 187
+ + ++ F C
Sbjct: 212 FMKMTTNNFLTPTFQC 227
>UniRef100_Q93W01 At2g01080/F23H14.5 [Arabidopsis thaliana]
Length = 231
Score = 45.4 bits (106), Expect = 7e-04
Identities = 40/195 (20%), Positives = 83/195 (42%), Gaps = 20/195 (10%)
Query: 11 SNRTNLASCLVATVFLIFILIIIFTLYFTLF-----KPQDPKISVTAVQL-------PSF 58
S+ +L C +FL+F + + L L KP+ P+ + V + PS
Sbjct: 32 SSSASLKGCCCC-LFLLFAFLALLVLAVVLIVILAVKPKKPQFDLQQVAVVYMGISNPSA 90
Query: 59 NLTNNSTTVNFTFSQYTSVKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQF 118
L + +++ T + NPN+ + +SSF ++ G +G VP +A T+
Sbjct: 91 VLDPTTASLSLTIRMLFTAVNPNKVGIRYGESSFTVMYKGMPLGRATVPGFYQDAHSTKN 150
Query: 119 MAATFTVQSLPLNLEPEGLQMGPTVEIESTIEMV------GRVRVLHLFSHHVEAKADCR 172
+ AT +V + L ++ + + +E+ ++RV++ S V+ +C
Sbjct: 151 VEATISVDRVNL-MQAHAADLVRDASLNDRVELTVRGDVGAKIRVMNFDSPGVQVSVNCG 209
Query: 173 VAIAVSDGSVLGFHC 187
+ I+ +++ C
Sbjct: 210 IGISPRKQALIYKQC 224
>UniRef100_Q9SJV8 Expressed protein [Arabidopsis thaliana]
Length = 231
Score = 45.4 bits (106), Expect = 7e-04
Identities = 40/195 (20%), Positives = 83/195 (42%), Gaps = 20/195 (10%)
Query: 11 SNRTNLASCLVATVFLIFILIIIFTLYFTLF-----KPQDPKISVTAVQL-------PSF 58
S+ +L C +FL+F + + L L KP+ P+ + V + PS
Sbjct: 32 SSSASLKGCCCC-LFLLFAFLALLVLAVVLIVILAVKPKKPQFDLQQVAVVYMGISNPSA 90
Query: 59 NLTNNSTTVNFTFSQYTSVKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQF 118
L + +++ T + NPN+ + +SSF ++ G +G VP +A T+
Sbjct: 91 VLDPTTASLSLTIRMLFTAVNPNKVGIRYGESSFTVMYKGMPLGRATVPGFYQDAHSTKN 150
Query: 119 MAATFTVQSLPLNLEPEGLQMGPTVEIESTIEMV------GRVRVLHLFSHHVEAKADCR 172
+ AT +V + L ++ + + +E+ ++RV++ S V+ +C
Sbjct: 151 VEATISVDRVNL-MQAHAADLVRDASLNDRVELTVRGDVGAKIRVMNFDSPGVQVSVNCG 209
Query: 173 VAIAVSDGSVLGFHC 187
+ I+ +++ C
Sbjct: 210 IGISPRKQALIYKQC 224
>UniRef100_Q9M663 Harpin inducing protein [Nicotiana tabacum]
Length = 229
Score = 45.1 bits (105), Expect = 0.001
Identities = 38/163 (23%), Positives = 73/163 (44%), Gaps = 7/163 (4%)
Query: 16 LASCLVATVF-LIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNNSTTVNFTFSQY 74
L +C +F L+ IL +I + + + +P K VT L F+L+ + T+ + +
Sbjct: 41 LCNCFFKIIFTLLIILGVIALVLWLVLRPNKVKFYVTDATLTQFDLSTTNNTIFYDLALN 100
Query: 75 TSVKNPNRGTFSHYDS-SFQLLSYGKQIGFMFVPAGKINARRTQFMAATFTVQSLPLNLE 133
+++NPN+ +YDS + L G++ + + T + F QSL L +
Sbjct: 101 MTIRNPNKRIGIYYDSIEARALYQGERFDSTNLEPFYQGHKNTSSLRPVFKGQSLVLLGD 160
Query: 134 PEGLQMG-----PTVEIESTIEMVGRVRVLHLFSHHVEAKADC 171
E E+E + M R++V + +H ++ K +C
Sbjct: 161 REKSNYNNEKNLGVYEMEVKLYMRIRLKVGWIKTHKIKPKIEC 203
>UniRef100_Q9XIN3 Expressed protein [Arabidopsis thaliana]
Length = 243
Score = 44.3 bits (103), Expect = 0.002
Identities = 21/80 (26%), Positives = 47/80 (58%), Gaps = 3/80 (3%)
Query: 20 LVATVFLIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNNSTTVNFTFSQYTSVKN 79
+V T FL+ + +I+F ++F + +PQ P +++ ++ + +FN++NN V+ + +N
Sbjct: 64 IVFTTFLLLLGLILF-IFFLIVRPQLPDVNLNSLSVSNFNVSNNQ--VSGKWDLQLQFRN 120
Query: 80 PNRGTFSHYDSSFQLLSYGK 99
PN HY+++ + Y +
Sbjct: 121 PNSKMSLHYETALCAMYYNR 140
>UniRef100_Q6L7J7 Harpin inducing protein 1-like 18 [Nicotiana tabacum]
Length = 229
Score = 44.3 bits (103), Expect = 0.002
Identities = 37/163 (22%), Positives = 73/163 (44%), Gaps = 7/163 (4%)
Query: 16 LASCLVATVF-LIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNNSTTVNFTFSQY 74
L +C +F L+ IL +I + + + +P K VT L F+L+N + T+ + +
Sbjct: 41 LCNCFFKIIFTLLIILGVIALVLWLVLRPNKVKFYVTDATLTQFDLSNTNNTIFYDLALN 100
Query: 75 TSVKNPNRGTFSHYDS-SFQLLSYGKQIGFMFVPAGKINARRTQFMAATFTVQSLPLNLE 133
+++NPN+ +YDS + + G++ + + T + F QSL L +
Sbjct: 101 MTIRNPNKRIGIYYDSIEARAMYQGERFHSTNLKPFYQGHKNTSSLHPVFKGQSLVLLGD 160
Query: 134 PEGLQMG-----PTVEIESTIEMVGRVRVLHLFSHHVEAKADC 171
E E+E + M R++ + +H ++ K +C
Sbjct: 161 REKSNYNNEKNLGVYEMEVKLYMRIRLKAGWIKTHKIKPKIEC 203
>UniRef100_Q6L7J8 Harpin inducing protein 1-like 9 [Nicotiana tabacum]
Length = 229
Score = 43.5 bits (101), Expect = 0.003
Identities = 22/76 (28%), Positives = 39/76 (50%), Gaps = 1/76 (1%)
Query: 16 LASCLVATVFLIFILIIIFTLYFTL-FKPQDPKISVTAVQLPSFNLTNNSTTVNFTFSQY 74
L +C+ F I ++I + L L +P K VT L F+L+ + T+N+ +
Sbjct: 41 LCNCIFQIFFTILVIIGVAALVLWLVLRPNKVKFYVTDATLTQFDLSTTNNTINYDLALN 100
Query: 75 TSVKNPNRGTFSHYDS 90
+++NPN+ +YDS
Sbjct: 101 MTIRNPNKRIGIYYDS 116
>UniRef100_Q75QH3 Hin1 like protein [Capsicum chinense]
Length = 228
Score = 42.4 bits (98), Expect = 0.006
Identities = 35/167 (20%), Positives = 75/167 (43%), Gaps = 7/167 (4%)
Query: 16 LASCLVATVFLIFILI-IIFTLYFTLFKPQDPKISVTAVQLPSFNLTNNSTTVNFTFSQY 74
L +C+ +F I I++ +I + + + +P K VT L F+L+ + T+ + +
Sbjct: 41 LCNCIFQIIFTILIVLGVIALVLWLVLRPNKVKFYVTDATLTQFDLSTANNTLYYDLALN 100
Query: 75 TSVKNPNRGTFSHYDSSFQLLSY-GKQIGFMFVPAGKINARRTQFMAATFTVQSLPLNLE 133
+++NPN+ +YDS Y G++ + + T + QSL L +
Sbjct: 101 MTIRNPNKRIGIYYDSIEARGMYKGERFASQNLEPFYQGHKNTSSLHPVLKGQSLVLLGD 160
Query: 134 PE-----GLQMGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAI 175
E + +IE + M R+++ + +H ++ K +C + +
Sbjct: 161 REKSNYNNEKNSGVYDIEMKLYMRIRLKIGWIKTHKIKPKIECDIKV 207
>UniRef100_P93353 Hin1 protein [Nicotiana tabacum]
Length = 221
Score = 42.0 bits (97), Expect = 0.008
Identities = 35/152 (23%), Positives = 68/152 (44%), Gaps = 6/152 (3%)
Query: 26 LIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNNSTTVNFTFSQYTSVKNPNRGTF 85
L+ IL +I + + + +P K VT L F+L+ + T+ + + +++NPN+
Sbjct: 44 LLIILGVIALVLWLVLRPNKVKFYVTDATLTQFDLSTTNNTIFYDLALNMTIRNPNKRIG 103
Query: 86 SHYDS-SFQLLSYGKQIGFMFVPAGKINARRTQFMAATFTVQSLPLNLEPEGLQMG---- 140
+YDS + L G++ + + T + F QSL L + E
Sbjct: 104 IYYDSIEARALYQGERFDSTNLEPFYQGHKNTSSLRPVFKGQSLVLLGDREKSNYNNEKN 163
Query: 141 -PTVEIESTIEMVGRVRVLHLFSHHVEAKADC 171
E+E + M R++V + +H ++ K +C
Sbjct: 164 LGVYEMEVKLYMRIRLKVGWIKTHKIKPKIEC 195
>UniRef100_Q69SA8 Hypothetical protein OSJNBb0052O11.109 [Oryza sativa]
Length = 213
Score = 41.6 bits (96), Expect = 0.010
Identities = 37/179 (20%), Positives = 76/179 (41%), Gaps = 13/179 (7%)
Query: 7 ETPPSNRTNLASCLVATVFLIFILIIIFTLYFTLFKPQDPKISVTAVQLPSFNLTNNSTT 66
+T P R A CL+ + +++ + +++P+ P++ T V + +
Sbjct: 8 KTKPHRR---ACCLLLAAVAVLGALVL--ALYLVYRPRPPRVVATPVDVTIELFSLVPPK 62
Query: 67 VNFTFSQYTSVKNPNRGTFSHYDSSFQLLSYGKQIGFMFVPAGKINARRTQFMAATFTVQ 126
+ + V NP+ + + +S + +G+++G VP G++ AR T+ + V
Sbjct: 63 LKAAVGVHVVVTNPSNSAYRYGESLASVTYHGERVGASVVPRGEVEARSTRLIEPATAVD 122
Query: 127 SLPLNLEPE-----GLQMGPTVEIESTIEMVGRVRVLHLFSHHVEAKADCRVAIAVSDG 180
+ + P + P V + +T+E G+ VL F V + C V + V G
Sbjct: 123 VVRVAESPHFAHDAAAGVLPFVAV-TTVE--GKALVLRSFEVSVSVEVVCFVQMYVFHG 178
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 287,385,927
Number of Sequences: 2790947
Number of extensions: 10490814
Number of successful extensions: 43470
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 43413
Number of HSP's gapped (non-prelim): 77
length of query: 187
length of database: 848,049,833
effective HSP length: 120
effective length of query: 67
effective length of database: 513,136,193
effective search space: 34380124931
effective search space used: 34380124931
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC142526.4


