

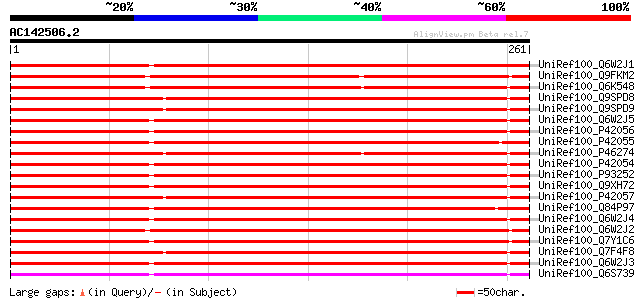
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142506.2 + phase: 1 /partial
(261 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6W2J1 VDAC3.1 [Lotus japonicus] 421 e-116
UniRef100_Q9FKM2 Porin-like protein [Arabidopsis thaliana] 287 2e-76
UniRef100_Q6K548 Outer mitochondrial membrane protein porin [Ory... 230 2e-59
UniRef100_Q9SPD8 Voltage-dependent anion channel protein 1b [Zea... 229 5e-59
UniRef100_Q9SPD9 Voltage-dependent anion channel protein 1a [Zea... 227 2e-58
UniRef100_Q6W2J5 VDAC1.1 [Lotus japonicus] 226 5e-58
UniRef100_P42056 36 kDa outer mitochondrial membrane protein por... 226 5e-58
UniRef100_P42055 34 kDa outer mitochondrial membrane protein por... 225 1e-57
UniRef100_P46274 Outer mitochondrial membrane protein porin [Tri... 223 3e-57
UniRef100_P42054 Outer plastidial membrane protein porin [Pisum ... 222 7e-57
UniRef100_P93252 Porin [Mesembryanthemum crystallinum] 220 2e-56
UniRef100_Q9XH72 Porin [Prunus armeniaca] 220 3e-56
UniRef100_P42057 Outer plastidial membrane protein porin [Zea mays] 219 7e-56
UniRef100_Q84P97 Porin-like protein [Oryza sativa] 218 9e-56
UniRef100_Q6W2J4 VDAC1.2 [Lotus japonicus] 217 3e-55
UniRef100_Q6W2J2 VDAC2.1 [Lotus japonicus] 216 6e-55
UniRef100_Q7Y1C6 PgPOR29 [Pennisetum americanum] 214 1e-54
UniRef100_Q7F4F8 Putative porin [Oryza sativa] 213 3e-54
UniRef100_Q6W2J3 VDAC1.3 [Lotus japonicus] 213 4e-54
UniRef100_Q6S739 Voltage-dependent anion-selective channel prote... 213 5e-54
>UniRef100_Q6W2J1 VDAC3.1 [Lotus japonicus]
Length = 277
Score = 421 bits (1082), Expect = e-116
Identities = 211/261 (80%), Positives = 232/261 (88%), Gaps = 2/261 (0%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDYNFD KF+LSIPSSTGLGL ATGLK+D+ F+GD+NTLYKSGN+TVDVKV+T SNV
Sbjct: 18 LLYKDYNFDQKFNLSIPSSTGLGLIATGLKKDQIFIGDINTLYKSGNLTVDVKVDTYSNV 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
STKVTLND+ KKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNP+P+LELS AI
Sbjct: 78 STKVTLNDIF--RCKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPSPQLELSTAI 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
GSKD+ MGAEVGF+TTSASF+ YNAGI FN+PDFSA LML DKG+SLKASYIH+VDRPDG
Sbjct: 136 GSKDLCMGAEVGFNTTSASFTKYNAGITFNRPDFSAALMLVDKGKSLKASYIHHVDRPDG 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
TVAAE+ H+ SS ENRFT G+SQ IDP TVLKTRFSDDGKAAF CQRAWRP SLITLSA
Sbjct: 196 FTVAAEINHRFSSFENRFTIGSSQLIDPNTVLKTRFSDDGKAAFLCQRAWRPKSLITLSA 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
EYDS KI GS K GLAL+L+
Sbjct: 256 EYDSTKIFGSSTKLGLALALQ 276
>UniRef100_Q9FKM2 Porin-like protein [Arabidopsis thaliana]
Length = 274
Score = 287 bits (735), Expect = 2e-76
Identities = 150/261 (57%), Positives = 189/261 (71%), Gaps = 5/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL KDY FDHKF+L++ S+TG ATGLK+D FF GD++TLYK N VD+K+++ S+V
Sbjct: 18 LLNKDYIFDHKFTLTMLSATGTEFVATGLKKDDFFFGDISTLYKGQNTIVDLKIDSHSSV 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
STKVTL + L S K +SF IPDHKSGKLDVQY+HPHA ++SSIGLNP P L+LSA I
Sbjct: 78 STKVTLKN--LLPSAKAVISFKIPDHKSGKLDVQYVHPHATLNSSIGLNPTPLLDLSATI 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
GS+++ +G EV FDT S+S + YNAGI FN SA L+L DKG+SL+A+Y+H V+
Sbjct: 136 GSQNVCLGGEVSFDTASSSLTKYNAGIGFNNQGVSAALILEDKGESLRATYVHTVN--PT 193
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
+ AEL + S+ N FT G+S S+D TV+KTRFS+ GKA QR WRP S IT SA
Sbjct: 194 TSFGAELIRRFSNYNNSFTVGSSHSVDQFTVVKTRFSNSGKAGMVVQREWRPKSHITFSA 253
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
EYDSK + SP K GLAL+LK
Sbjct: 254 EYDSKAVTSSP-KLGLALALK 273
>UniRef100_Q6K548 Outer mitochondrial membrane protein porin [Oryza sativa]
Length = 274
Score = 230 bits (587), Expect = 2e-59
Identities = 119/261 (45%), Positives = 174/261 (66%), Gaps = 4/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLY+DY DHKF+L+ +S G+ +TAT K+ G++ + K+ N+TVDVK N+DSNV
Sbjct: 17 LLYRDYQTDHKFTLTTYTSNGVAITATSTKKADLIFGEIQSQIKNKNITVDVKANSDSNV 76
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T VT+++ L+ K LSF +PD +SGK ++QY H +A + +SIGL +P + LS+
Sbjct: 77 VTTVTVDE--LTPGLKSILSFAVPDQRSGKFELQYSHDYAGVSASIGLTASPVVNLSSVF 134
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+K +++GA+V DT + + + YNAG++F+ D A+L L +KG SL ASY H V+
Sbjct: 135 GTKALAVGADVSLDTATGNLTKYNAGLSFSNDDLIASLNLNNKGDSLTASYYHIVNH-SA 193
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL H SS+EN TFGT ++DP TV+K RF++ GKA+ Q WRP S+ T+SA
Sbjct: 194 TAVGAELTHSFSSNENSLTFGTQHTLDPLTVVKARFNNSGKASALLQHEWRPKSVWTISA 253
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S +K G+A++LK
Sbjct: 254 EVDTKAIDKS-SKVGIAVALK 273
>UniRef100_Q9SPD8 Voltage-dependent anion channel protein 1b [Zea mays]
Length = 276
Score = 229 bits (584), Expect = 5e-59
Identities = 114/261 (43%), Positives = 172/261 (65%), Gaps = 2/261 (0%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDY DHKF+L+ SS G+ +TAT K+ ++++ K+ N+TVDVK N++SN+
Sbjct: 17 LLYKDYQTDHKFTLTTYSSNGVAVTATSTKKADLIFSEIHSQIKNKNMTVDVKANSESNI 76
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T VT+N+++ K + LSF +PD +SGK+++QYLH +A +++S+ L P LSA
Sbjct: 77 ITTVTVNEIATPGLKTI-LSFAVPDQRSGKVELQYLHDYAGVNASVSLTANPVANLSAGF 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+K +++GAE+ FD+ + +F+ YNAG+ F D A+L + +KG +L +Y H V
Sbjct: 136 GTKALAVGAEISFDSATGNFTKYNAGLMFTHEDLIASLNMNNKGDNLTGAYYHKVSELTN 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL H S++EN TFG ++DP TVLK R ++ GKA+ Q WRP SL+T+SA
Sbjct: 196 TAVGAELTHSFSTNENTLTFGGQHTLDPLTVLKARINNSGKASVLIQHEWRPKSLVTISA 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S +K G+A++LK
Sbjct: 256 EVDTKTIEKS-SKVGIAVALK 275
>UniRef100_Q9SPD9 Voltage-dependent anion channel protein 1a [Zea mays]
Length = 276
Score = 227 bits (579), Expect = 2e-58
Identities = 112/261 (42%), Positives = 173/261 (65%), Gaps = 2/261 (0%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDY DHKF+L+ +S G+ +TA+ K+ +G++ + K+ N+T+DVK N++SN+
Sbjct: 17 LLYKDYQTDHKFTLTTYTSNGVAVTASSTKKADLILGEIQSQIKNKNMTIDVKANSESNI 76
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+++++ K + LSF +PD +SGK+++QYLH +A +++SIGL P + LSAA
Sbjct: 77 ITTITVDEIATPGLKTI-LSFAVPDQRSGKVELQYLHDYAGVNASIGLTANPVVNLSAAF 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+K +++GA++ DT + + + YNA + F D A+L L +KG SL +Y H V
Sbjct: 136 GTKAVALGADISLDTATGNLTKYNAALRFTHEDLVASLNLNNKGDSLTGAYYHKVSELTN 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL H S++EN TFG ++DP TVLK R ++ GKA+ Q WRP SL+T+SA
Sbjct: 196 TAVGAELTHSFSTNENTLTFGGQHALDPLTVLKARINNSGKASALIQHEWRPKSLVTISA 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S +K G+A++LK
Sbjct: 256 EVDTKTIEKS-SKVGIAVALK 275
>UniRef100_Q6W2J5 VDAC1.1 [Lotus japonicus]
Length = 276
Score = 226 bits (576), Expect = 5e-58
Identities = 117/261 (44%), Positives = 166/261 (62%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL+KDY+ D KF+++ S TG+ +T++G K+ + FV D+NT K+ N+T D+KV+TDSN+
Sbjct: 18 LLFKDYHSDQKFTITTFSPTGVAITSSGTKKGELFVADVNTQLKNKNITTDIKVDTDSNL 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T + +ND + K SF +PD +SGK+++QYLH +A I SSIGL P + S I
Sbjct: 78 FTTIAVNDPA--PGLKAIFSFKVPDQRSGKVELQYLHDYAGISSSIGLTANPIVNFSGVI 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +++GA++ FDT + NAG++F K D A L L DKG +L ASY H V
Sbjct: 136 GTNVLALGADLTFDTKIGELTKSNAGLSFTKDDLIAALTLNDKGDALNASYYHVVSPLTN 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AE+ H+ S++EN T GT ++DP T LK R ++ GKA+ Q WRP S T+S
Sbjct: 196 TAVGAEVTHRFSTNENTLTLGTQHALDPLTTLKARVNNFGKASALIQHEWRPKSFFTISG 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S AK GLAL+LK
Sbjct: 256 EVDTKAIEKS-AKVGLALALK 275
>UniRef100_P42056 36 kDa outer mitochondrial membrane protein porin [Solanum
tuberosum]
Length = 275
Score = 226 bits (576), Expect = 5e-58
Identities = 115/261 (44%), Positives = 171/261 (65%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLY+DY DHKF+++ S+TG+ +TA+GLK+ + F+ D++T K+ N+T DVKV+T+SNV
Sbjct: 17 LLYRDYVSDHKFTVTTYSTTGVAITASGLKKGELFLADVSTQLKNKNITTDVKVDTNSNV 76
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+++ + K SF +PD KSGK+++QYLH +A I++SIGL +P + S
Sbjct: 77 YTTITVDEPA--PGLKTIFSFVVPDQKSGKVELQYLHEYAGINTSIGLTASPLVNFSGVA 134
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +++G ++ FDT + +F+ NAG++F+ D A+L L DKG ++ ASY H V
Sbjct: 135 GNNTVALGTDLSFDTATGNFTKCNAGLSFSSSDLIASLALNDKGDTVSASYYHTVKPVTN 194
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL H SS+EN T GT +DP T +K R + GKA+ Q WRP SL T+S
Sbjct: 195 TAVGAELTHSFSSNENTLTIGTQHLLDPLTTVKARVNSYGKASALIQHEWRPKSLFTISG 254
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D++ I S AK GLA++LK
Sbjct: 255 EVDTRAIEKS-AKIGLAVALK 274
>UniRef100_P42055 34 kDa outer mitochondrial membrane protein porin [Solanum
tuberosum]
Length = 275
Score = 225 bits (573), Expect = 1e-57
Identities = 117/261 (44%), Positives = 167/261 (63%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDY DHKFS++ S TG+ +T++G K+ F+ D+NT K+ NVT D+KV+T+SN+
Sbjct: 17 LLYKDYQSDHKFSITTYSPTGVVITSSGSKKGDLFLADVNTQLKNKNVTTDIKVDTNSNL 76
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+++ + K LSF +PD +SGKL+VQYLH +A I +S+GL P + S +
Sbjct: 77 FTTITVDEAA--PGLKTILSFRVPDQRSGKLEVQYLHDYAGICTSVGLTANPIVNFSGVV 134
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ I++G +V FDT + F+ NAG++F D A+L L +KG +L ASY H V
Sbjct: 135 GTNIIALGTDVSFDTKTGDFTKCNAGLSFTNADLVASLNLNNKGDNLTASYYHTVSPLTS 194
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AE+ H S++EN T GT +DP T +K R ++ GKA+ Q WRP SL T+S
Sbjct: 195 TAVGAEVNHSFSTNENIITVGTQHRLDPLTSVKARINNFGKASALLQHEWRPKSLFTVSG 254
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K + AKFGLAL+LK
Sbjct: 255 EVDTKS-VDKGAKFGLALALK 274
>UniRef100_P46274 Outer mitochondrial membrane protein porin [Triticum aestivum]
Length = 275
Score = 223 bits (569), Expect = 3e-57
Identities = 117/261 (44%), Positives = 173/261 (65%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLY+DY DHKF+L+ ++ G +TAT K+ VG++ + K+ N+TVDVK N+ SNV
Sbjct: 17 LLYRDYQTDHKFTLTTYTANGPAITATSTKKADLTVGEIQSQIKNKNITVDVKANSASNV 76
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T +D++ K + LSF +PD KSGK+++QYLH +A I++SIGL P + LS A
Sbjct: 77 ITTITADDLAAPGLKTI-LSFAVPDQKSGKVELQYLHDYAGINASIGLTANPVVNLSGAF 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +++GA+V DT + +F+ YNA +++ D A+L L +KG SL ASY H V++ G
Sbjct: 136 GTSALAVGADVSLDTATKNFAKYNAALSYTNQDLIASLNLNNKGDSLTASYYHIVEK-SG 194
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL H SS+EN TFGT ++DP T++K R ++ GKA+ Q + P SL T+SA
Sbjct: 195 TAVGAELTHSFSSNENSLTFGTQHTLDPLTLVKARINNSGKASALIQHEFMPKSLCTISA 254
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S +K G+A++LK
Sbjct: 255 EVDTKAIEKS-SKVGIAIALK 274
>UniRef100_P42054 Outer plastidial membrane protein porin [Pisum sativum]
Length = 275
Score = 222 bits (566), Expect = 7e-57
Identities = 112/261 (42%), Positives = 169/261 (63%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDY+ D KF++S S TG+ +T++G K+ + F+GD+NT K+ N+T D+KV+T+SN+
Sbjct: 17 LLYKDYHSDKKFTISTYSPTGVAITSSGTKKGELFLGDVNTQLKNKNITTDIKVDTNSNL 76
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+N+ + K LSF +P+ SGK+++QYLH +A I SS+GL P + S+ I
Sbjct: 77 FTTITVNEPA--PGVKAILSFKVPEQTSGKVELQYLHEYAGISSSVGLKANPIVNFSSVI 134
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ ++ GA++ FDT + NA + F K D +L L +KG L ASY H ++
Sbjct: 135 GTNALAFGADISFDTKLGELTKSNAAVNFVKDDLIGSLTLNEKGDLLSASYYHAINPLSN 194
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V +++H+ S+ EN FT GT ++DP T +K R ++ GKA+ Q WRP SLIT+S+
Sbjct: 195 TAVGVDISHRFSTKENTFTLGTQHALDPLTTVKGRVTNSGKASALIQHEWRPKSLITISS 254
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S AK GL+L+LK
Sbjct: 255 EVDTKAIEKS-AKIGLSLALK 274
>UniRef100_P93252 Porin [Mesembryanthemum crystallinum]
Length = 276
Score = 220 bits (561), Expect = 2e-56
Identities = 115/261 (44%), Positives = 167/261 (63%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLY+DY D KFS++ S TG+ +T++G K+ F+ D+NT K+ NVT D+KV+T+SN+
Sbjct: 18 LLYRDYQTDQKFSITTYSPTGVAITSSGTKKGDLFLADVNTQLKNKNVTTDIKVDTNSNL 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+++ + K LSF +PD +SGKL++QYLH +A I +S+GL P + S +
Sbjct: 78 LTTITVDEAA--PGLKTILSFRVPDQRSGKLELQYLHDYAGICTSVGLTANPIVNFSGVL 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ ++ G +V FD+ + +F+ NAG++F D A+L L DKG +L ASY H +
Sbjct: 136 GTNLLAFGTDVSFDSKTGNFTKCNAGLSFANADLIASLTLNDKGDTLNASYYHTISPLTS 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AE+AH S+++N T G +IDP T LK R ++ GKA Q WRP SL+T SA
Sbjct: 196 SAVGAEVAHSFSTNQNTITLGAQHAIDPLTNLKGRVNNFGKATALIQHEWRPKSLVTFSA 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S AK GLAL+LK
Sbjct: 256 EVDTKAIEKS-AKVGLALALK 275
>UniRef100_Q9XH72 Porin [Prunus armeniaca]
Length = 276
Score = 220 bits (560), Expect = 3e-56
Identities = 111/261 (42%), Positives = 169/261 (64%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDY DHKF+++ +STG+ +++TG+++ ++GD++T K+ N+T DVKV+TDSN+
Sbjct: 18 LLYKDYQSDHKFTVTTYTSTGVAISSTGIRKGDLYLGDVSTQLKNKNITTDVKVDTDSNL 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+++ + K SF +PD +SGK+++QY H +A I +SIGL P + S +
Sbjct: 78 RTTITIDEPA--PGLKAIFSFIVPDQRSGKVELQYQHEYAGISTSIGLTANPIVNFSGVV 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +S+G ++ FDT S +F+ NAG+ F D A+L+L DK ++ ASY H V
Sbjct: 136 GNNLLSLGTDLSFDTASGNFTKCNAGLNFTHTDLIASLILNDKADTVTASYYHTVSPLTN 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL+H SS+EN T GT ++DP T +K R ++ G+A+ Q WRP S T+S
Sbjct: 196 TAVGAELSHSFSSNENSLTIGTQHALDPLTTVKGRVNNYGRASALIQHEWRPKSFFTISG 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D++ I S AK GLAL+LK
Sbjct: 256 EVDTRAIEKS-AKIGLALALK 275
>UniRef100_P42057 Outer plastidial membrane protein porin [Zea mays]
Length = 277
Score = 219 bits (557), Expect = 7e-56
Identities = 110/261 (42%), Positives = 167/261 (63%), Gaps = 2/261 (0%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDYN KF L+ S G+ +TA G ++++ G+L+T K+ +TVDVK N++S++
Sbjct: 18 LLYKDYNTHQKFCLTTSSPNGVAITAAGTRKNESIFGELHTQIKNKKLTVDVKANSESDL 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+++ K + ++ +PD +SGKL+ QYLH +A +++S+GLN P + LS A
Sbjct: 78 LTTITVDEFGTPGLKSI-INLVVPDQRSGKLEFQYLHEYAGVNASVGLNSNPMVNLSGAF 136
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
GSK +S+G +V FDT ++ F+ YNA ++ PD A+L L + G +L ASY H V G
Sbjct: 137 GSKALSVGVDVSFDTATSDFTKYNAALSLTSPDLIASLHLNNHGDTLVASYYHLVKNHSG 196
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL+H +S +E+ FG+ S+DP T +KTRF++ G A+ Q WRP S +T+S
Sbjct: 197 TAVGAELSHSMSRNESTLIFGSQHSLDPHTTIKTRFNNYGMASALVQHEWRPKSFVTISG 256
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
+ D+K I S K GL+L LK
Sbjct: 257 DVDTKAIEKS-TKVGLSLVLK 276
>UniRef100_Q84P97 Porin-like protein [Oryza sativa]
Length = 277
Score = 218 bits (556), Expect = 9e-56
Identities = 107/261 (40%), Positives = 174/261 (65%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL KDY +D K ++S SS+G+GLT+T +K+ + D++++YK + VDVKV+T+SN+
Sbjct: 19 LLTKDYTYDQKLTVSTVSSSGVGLTSTAVKKGGLYTLDVSSVYKYKSTLVDVKVDTESNI 78
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
ST +T+ DV S K+ S +PD+ SGK+++QY H +A+ +++G+ P+P +E S
Sbjct: 79 STTLTVFDVL--PSTKLVTSVKLPDYNSGKVEMQYFHENASFATAVGMKPSPVVEFSGTA 136
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G++ ++ GAE GFDT + F+ Y+A I KPD+ A ++LADKG ++K S ++++D
Sbjct: 137 GAQGLAFGAEAGFDTATGKFTKYSAAIGVTKPDYHAAIVLADKGDTVKVSGVYHLDDKQK 196
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
+V AEL +LS++EN T G +DP+T +K R ++ GK A Q +P S++T+S
Sbjct: 197 SSVVAELTRRLSTNENTLTVGGLYKVDPETAVKARLNNTGKLAALLQHEVKPKSVLTISG 256
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E+D+ K + P KFGLAL+L+
Sbjct: 257 EFDT-KALDRPPKFGLALALR 276
>UniRef100_Q6W2J4 VDAC1.2 [Lotus japonicus]
Length = 276
Score = 217 bits (552), Expect = 3e-55
Identities = 109/261 (41%), Positives = 166/261 (62%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL+KDY+ D KF+++ S TG+ +T++G+K+ + FV D+NT K+ N+T D+KV+TDSN+
Sbjct: 18 LLFKDYHSDQKFTITTYSPTGVAITSSGIKKGELFVADVNTQLKNKNITTDLKVDTDSNL 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T VT+++ + K SF +PD +SGK+++QYLH +A I +S+GL P P + S +
Sbjct: 78 FTTVTVDEPA--PGLKAIFSFRVPDQRSGKVELQYLHDYAGISTSVGLTPNPIVNFSGVV 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +++G ++ FDT + +NAG+ F K D A+L + DKG +L ASY H V+
Sbjct: 136 GTNALAVGTDLSFDTKIGELTKFNAGLNFTKADLIASLTVNDKGDALNASYYHIVNPLTN 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
AE+ H+ S++EN T GT ++DP T +K R ++ GKA Q WR S T+S
Sbjct: 196 TAFGAEVTHRFSTNENTLTLGTQHALDPLTSVKARVNNLGKANALIQHEWRAKSFFTISG 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S AK GL L+LK
Sbjct: 256 EVDTKAIEKS-AKIGLGLALK 275
>UniRef100_Q6W2J2 VDAC2.1 [Lotus japonicus]
Length = 276
Score = 216 bits (549), Expect = 6e-55
Identities = 113/261 (43%), Positives = 170/261 (64%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL KDY+ D K ++S S+ G+ LT+T +K+ VGD+ LYK N +DVK++T S +
Sbjct: 18 LLTKDYSSDQKLTVSSYSTAGVALTSTAVKKGGLSVGDVAALYKYKNTLIDVKLDTASII 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
ST +T D L S K SF +PD+ SGKL+VQY H HA + ++ LN +P +++SA
Sbjct: 78 STTLTFTD--LLPSTKAIASFKLPDYNSGKLEVQYFHDHATLTAAAALNQSPIIDVSATF 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ I++GAE G+DT+S F+ Y AGI+ KPD SA+++L DKG S+KASY+H++D+
Sbjct: 136 GTPVIAVGAEAGYDTSSGGFTKYTAGISVTKPDASASIILGDKGDSIKASYLHHLDQLKK 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
AE+ K S++EN FT G S +ID T ++ R ++ GK Q P S++T+S+
Sbjct: 196 SAAVAEITRKFSTNENTFTVGGSFAIDHLTQVRARLNNHGKLGALLQHEIIPKSVLTISS 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K + +P +FGLA++LK
Sbjct: 256 EVDTKALDKNP-RFGLAIALK 275
>UniRef100_Q7Y1C6 PgPOR29 [Pennisetum americanum]
Length = 277
Score = 214 bits (546), Expect = 1e-54
Identities = 105/261 (40%), Positives = 169/261 (64%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL +DY +D K ++S SS+G+GLT+T +K+ + D++++YK N VD+KV+T+SN+
Sbjct: 19 LLTRDYTYDQKLTVSTVSSSGVGLTSTAVKKGGLYTLDVSSVYKYKNTVVDIKVDTESNI 78
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
ST +T+ D S K+ S +PD+ SGK+++ Y H +A++ + +G P+P +ELS +
Sbjct: 79 STTLTVLDAL--PSTKLVTSVKLPDYNSGKVELHYFHENASLATVVGTKPSPVVELSGTV 136
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G++ ++ GAE G+DT S F+ Y A I KPD+ A +LADKG ++K S ++++D
Sbjct: 137 GAQGVTFGAEAGYDTASGKFTKYTAAIGLTKPDYHAAFILADKGDTIKVSGVYHLDEKQK 196
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
+ AEL +LS++ N T G IDP+T +K R ++ G A Q +P SL+T+S
Sbjct: 197 ASAVAELTRRLSTNVNTLTVGGLYKIDPQTAVKARLNNTGTLAALLQHELKPKSLLTISG 256
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E+D+K + +P KFGLAL+LK
Sbjct: 257 EFDTKALDRAP-KFGLALALK 276
>UniRef100_Q7F4F8 Putative porin [Oryza sativa]
Length = 275
Score = 213 bits (543), Expect = 3e-54
Identities = 111/261 (42%), Positives = 166/261 (63%), Gaps = 2/261 (0%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLY+DY HKF+L+ + G+ +TA G ++++ G+L T K+ +TVDVK N++S++
Sbjct: 16 LLYRDYGTHHKFTLTTCTPEGVTITAAGTRKNESVFGELQTQLKNKKLTVDVKANSESDL 75
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T VT+++ K + LS +PD +SGKL++QYLH +A I++S+GLN P + LS
Sbjct: 76 LTTVTVDEFGTPGLKSI-LSLVVPDQRSGKLELQYLHEYAGINASVGLNSNPMVNLSGVF 134
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
GSK++S+G +V FDT +++F+ YNA ++ D A+L L + G +L ASY H V
Sbjct: 135 GSKELSVGVDVAFDTATSNFTKYNAALSLTNSDLIASLHLNNHGDTLIASYYHLVKHHSN 194
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL+H S +E+ FG+ S+DP T +K RF++ G A+ Q WRP SLIT+S
Sbjct: 195 TAVGAELSHSFSRNESTLIFGSQHSLDPHTTVKARFNNYGMASALVQHEWRPKSLITISG 254
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+K I S K GL+L LK
Sbjct: 255 EVDTKAIEKS-TKVGLSLVLK 274
>UniRef100_Q6W2J3 VDAC1.3 [Lotus japonicus]
Length = 276
Score = 213 bits (542), Expect = 4e-54
Identities = 111/261 (42%), Positives = 167/261 (63%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LL+KDY DHKF+++ +S G+ +T+TG+K+ + ++ D++T K+ N+T DVKV+T+SN+
Sbjct: 18 LLFKDYQNDHKFTITTYTSAGVEITSTGVKKGEIYLADVSTKLKNKNITTDVKVDTNSNL 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T +T+++ + K SF PD KSGK+++QY H +A I +SIGL +P + S I
Sbjct: 78 HTTITVDEPA--PGLKTIFSFIFPDQKSGKVELQYQHEYAGISTSIGLTASPIVNFSGII 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +++G ++ FDT S +F+ YNAG+ D A+L L DKG SL ASY H V
Sbjct: 136 GNNLVAVGTDLSFDTASGNFTKYNAGLNVTHADLIASLTLNDKGDSLNASYYHVVSPLTN 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AEL+H SS+EN T GT ++DP T+LK R ++ G+A+ Q W P + ++L
Sbjct: 196 TAVGAELSHSFSSNENILTIGTQHALDPITLLKARVNNYGRASALIQHEWNPRTRVSLVG 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E D+ I S AK GLAL+LK
Sbjct: 256 EVDTGAIEKS-AKVGLALALK 275
>UniRef100_Q6S739 Voltage-dependent anion-selective channel protein [Brassica
campestris]
Length = 276
Score = 213 bits (541), Expect = 5e-54
Identities = 112/261 (42%), Positives = 158/261 (59%), Gaps = 3/261 (1%)
Query: 1 LLYKDYNFDHKFSLSIPSSTGLGLTATGLKRDKFFVGDLNTLYKSGNVTVDVKVNTDSNV 60
LLYKDY D K S++ SSTG+ +T +G + F+GD+ T K+ N T D+KV +DS++
Sbjct: 18 LLYKDYQGDQKLSITAYSSTGVAITTSGTNKGDLFLGDVVTQIKNKNFTADIKVASDSSI 77
Query: 61 STKVTLNDVSLSHSKKVALSFNIPDHKSGKLDVQYLHPHAAIDSSIGLNPAPKLELSAAI 120
T T ++ + K +S +PD KS K+++QY+HPHA I +S+GL P + S I
Sbjct: 78 LTTFTYDEAT--PGLKAIVSAKVPDQKSAKVELQYMHPHAGICTSVGLTANPVVNFSGVI 135
Query: 121 GSKDISMGAEVGFDTTSASFSTYNAGIAFNKPDFSATLMLADKGQSLKASYIHYVDRPDG 180
G+ +++G +V FDT S +F +N G++F K D A+L L DKG+ L ASY H V+
Sbjct: 136 GTSVLALGTDVSFDTESGNFKHFNTGVSFTKDDLIASLTLNDKGEKLNASYYHIVNPLKN 195
Query: 181 LTVAAELAHKLSSSENRFTFGTSQSIDPKTVLKTRFSDDGKAAFQCQRAWRPNSLITLSA 240
V AE++H L S N T GT ++DP T +K R ++ G A Q WRP S IT+S
Sbjct: 196 TVVGAEVSHNLKSQVNSITVGTQHALDPLTTVKARVNNAGIANALIQHEWRPKSFITISG 255
Query: 241 EYDSKKIIGSPAKFGLALSLK 261
E DSK I S AK G AL+LK
Sbjct: 256 EVDSKAIEKS-AKVGFALALK 275
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 414,476,551
Number of Sequences: 2790947
Number of extensions: 16162729
Number of successful extensions: 32226
Number of sequences better than 10.0: 138
Number of HSP's better than 10.0 without gapping: 88
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 31930
Number of HSP's gapped (non-prelim): 183
length of query: 261
length of database: 848,049,833
effective HSP length: 125
effective length of query: 136
effective length of database: 499,181,458
effective search space: 67888678288
effective search space used: 67888678288
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Medicago: description of AC142506.2


