

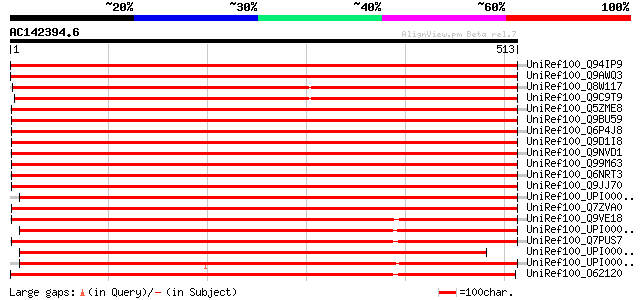
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.6 - phase: 0
(513 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q94IP9 WD-40 repeat protein [Lycopersicon esculentum] 926 0.0
UniRef100_Q9AWQ3 Putative WD-40 repeat protein [Oryza sativa] 920 0.0
UniRef100_Q8W117 At1g73720/F25P22_14 [Arabidopsis thaliana] 908 0.0
UniRef100_Q9C9T9 Hypothetical protein F25P22.14 [Arabidopsis tha... 897 0.0
UniRef100_Q5ZME8 Hypothetical protein [Gallus gallus] 708 0.0
UniRef100_Q9BU59 Smu-1 suppressor of mec-8 and unc-52 homolog [H... 708 0.0
UniRef100_Q6P4J8 Hypothetical protein MGC75979 [Xenopus tropicalis] 706 0.0
UniRef100_Q9D1I8 Mus musculus 18-day embryo whole body cDNA, RIK... 705 0.0
UniRef100_Q9NVD1 Hypothetical protein FLJ10805 [Homo sapiens] 705 0.0
UniRef100_Q99M63 Brain-enriched WD-repeat protein [Rattus norveg... 704 0.0
UniRef100_Q6NRT3 MGC81475 protein [Xenopus laevis] 704 0.0
UniRef100_Q9JJ70 Mus musculus [Mus musculus] 703 0.0
UniRef100_UPI00002DAA1D UPI00002DAA1D UniRef100 entry 699 0.0
UniRef100_Q7ZVA0 Hypothetical protein zgc:56147 [Brachydanio rerio] 699 0.0
UniRef100_Q9VE18 CG5451-PA [Drosophila melanogaster] 671 0.0
UniRef100_UPI0000433250 UPI0000433250 UniRef100 entry 669 0.0
UniRef100_Q7PUS7 ENSANGP00000015224 [Anopheles gambiae str. PEST] 663 0.0
UniRef100_UPI000035F64C UPI000035F64C UniRef100 entry 650 0.0
UniRef100_UPI00003602F7 UPI00003602F7 UniRef100 entry 565 e-159
UniRef100_O62120 SMU-1 [Caenorhabditis elegans] 556 e-157
>UniRef100_Q94IP9 WD-40 repeat protein [Lycopersicon esculentum]
Length = 514
Score = 926 bits (2393), Expect = 0.0
Identities = 444/512 (86%), Positives = 484/512 (93%)
Query: 2 STLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAIL 61
STLEIEARDVIK VLQFCKENSLHQ+FQTLQNECQVSLNTVDS+ETF+ADIN GRWDA+L
Sbjct: 3 STLEIEARDVIKIVLQFCKENSLHQSFQTLQNECQVSLNTVDSLETFIADINSGRWDAVL 62
Query: 62 PQVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHL 121
PQV+QLKLPR LEDLYEQIVLEMIELRE+DTARAILRQT VMGVMKQEQPERYLRLEHL
Sbjct: 63 PQVAQLKLPRKILEDLYEQIVLEMIELREMDTARAILRQTNVMGVMKQEQPERYLRLEHL 122
Query: 122 LVRTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
LVRTYFDP+EAY +STKEKRRAQI+QA++AEV+VVPPSRLMALIGQALKWQQHQGLLPPG
Sbjct: 123 LVRTYFDPHEAYNDSTKEKRRAQISQALSAEVSVVPPSRLMALIGQALKWQQHQGLLPPG 182
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
TQFDLFRGTAAMKQD +DMYPT L HTIKF KSH EC+RFSPDGQ+LVSCS+DGFIEVW
Sbjct: 183 TQFDLFRGTAAMKQDEEDMYPTTLGHTIKFPKKSHPECSRFSPDGQFLVSCSVDGFIEVW 242
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
D+ISGKLKKDLQYQA+ETFMMHD+ VLCVDFSRDSEM+ASGS DGKIKVWRIRT QCLRR
Sbjct: 243 DHISGKLKKDLQYQADETFMMHDDAVLCVDFSRDSEMLASGSQDGKIKVWRIRTGQCLRR 302
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
LE AHSQGVTS+ FSRDG+Q+LSTSFDSTARIHGLKSGK+LKEFRGH+SYVNDA FT DG
Sbjct: 303 LERAHSQGVTSLVFSRDGTQILSTSFDSTARIHGLKSGKLLKEFRGHSSYVNDAIFTFDG 362
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+RVITASSD +VKVWD KTT+CLQTF+PPPPLRGGDASVNSV +FPKN EHI+VCNKTSS
Sbjct: 363 SRVITASSDGSVKVWDAKTTECLQTFRPPPPLRGGDASVNSVHLFPKNNEHIIVCNKTSS 422
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
IY+MTLQGQVVKSFSSGKREGGDFVAAC+SPKGEWIYCVGEDRN+YCFSYQS KLEHLM
Sbjct: 423 IYLMTLQGQVVKSFSSGKREGGDFVAACLSPKGEWIYCVGEDRNLYCFSYQSNKLEHLMK 482
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+THHPH NLVAT+ +D TMKLWKP
Sbjct: 483 VHEKDVIGITHHPHINLVATYGEDCTMKLWKP 514
>UniRef100_Q9AWQ3 Putative WD-40 repeat protein [Oryza sativa]
Length = 517
Score = 920 bits (2377), Expect = 0.0
Identities = 441/512 (86%), Positives = 481/512 (93%)
Query: 2 STLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAIL 61
STLEIEARDV+K VLQFCKENSL QTFQTLQNECQVSLNTVDSI+TF+ADIN GRWDA+L
Sbjct: 6 STLEIEARDVVKIVLQFCKENSLQQTFQTLQNECQVSLNTVDSIDTFIADINAGRWDAVL 65
Query: 62 PQVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHL 121
PQV+QLKLPR LEDLYEQIVLEM ELRELDTARAILRQTQVMGVMKQEQPERYLRLEHL
Sbjct: 66 PQVAQLKLPRKKLEDLYEQIVLEMAELRELDTARAILRQTQVMGVMKQEQPERYLRLEHL 125
Query: 122 LVRTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
LVRTYFDPNEAYQESTKEKRRAQI+QAIA+EV+VVPPSRLMALIGQALKWQQHQGLLPPG
Sbjct: 126 LVRTYFDPNEAYQESTKEKRRAQISQAIASEVSVVPPSRLMALIGQALKWQQHQGLLPPG 185
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
TQFDLFRGTAAMKQD ++ YPT LSH KFG K+H ECARFSPDGQYLVSCS+DG IEVW
Sbjct: 186 TQFDLFRGTAAMKQDEEETYPTTLSHQTKFGKKTHPECARFSPDGQYLVSCSVDGIIEVW 245
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
DYISGKLKKDLQYQA+E+FMMHD+ VL VDFSRDSEM+ASGS DGKIKVWRIRT QCLRR
Sbjct: 246 DYISGKLKKDLQYQADESFMMHDDAVLSVDFSRDSEMLASGSQDGKIKVWRIRTGQCLRR 305
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
LE AH++GVTSV+FSRDG+Q+LS+SFD+TAR+HGLKSGKMLKEFRGH SYVN A F+ DG
Sbjct: 306 LERAHAKGVTSVTFSRDGTQILSSSFDTTARVHGLKSGKMLKEFRGHNSYVNCAIFSTDG 365
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+RVITASSDCTVKVWD KTTDCLQTFKPPPPLRGGDA+VNSV + PKN++HI+VCNKTSS
Sbjct: 366 SRVITASSDCTVKVWDTKTTDCLQTFKPPPPLRGGDATVNSVHLSPKNSDHIIVCNKTSS 425
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
IYIMTLQGQVVKSFSSGKREGGDF+AA +SPKGEWIYCVGED NMYCFSYQSGKLEHLM
Sbjct: 426 IYIMTLQGQVVKSFSSGKREGGDFLAASVSPKGEWIYCVGEDMNMYCFSYQSGKLEHLMK 485
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VHDK++IG+THHPHRNL+AT+++D TMK WKP
Sbjct: 486 VHDKDVIGITHHPHRNLIATYAEDCTMKTWKP 517
>UniRef100_Q8W117 At1g73720/F25P22_14 [Arabidopsis thaliana]
Length = 511
Score = 908 bits (2346), Expect = 0.0
Identities = 433/510 (84%), Positives = 481/510 (93%), Gaps = 1/510 (0%)
Query: 4 LEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQ 63
LEIEARDVIK +LQFCKENSL+QTFQTLQ+ECQVSLNTVDS+ETF++DIN GRWD++LPQ
Sbjct: 3 LEIEARDVIKIMLQFCKENSLNQTFQTLQSECQVSLNTVDSVETFISDINSGRWDSVLPQ 62
Query: 64 VSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLV 123
VSQLKLPRN LEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQ ERYLR+EHLLV
Sbjct: 63 VSQLKLPRNKLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQAERYLRMEHLLV 122
Query: 124 RTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQ 183
R+YFDP+EAY +STKE++RAQIAQA+AAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQ
Sbjct: 123 RSYFDPHEAYGDSTKERKRAQIAQAVAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQ 182
Query: 184 FDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDY 243
FDLFRGTAAMKQDV+D +P L+HTIKFG KSHAECARFSPDGQ+L S S+DGFIEVWDY
Sbjct: 183 FDLFRGTAAMKQDVEDTHPNVLTHTIKFGKKSHAECARFSPDGQFLASSSVDGFIEVWDY 242
Query: 244 ISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLE 303
ISGKLKKDLQYQA+E+FMMHD+PVLC+DFSRDSEM+ASGS DGKIK+WRIRT C+RR +
Sbjct: 243 ISGKLKKDLQYQADESFMMHDDPVLCIDFSRDSEMLASGSQDGKIKIWRIRTGVCIRRFD 302
Query: 304 HAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTR 363
AHSQGVTS+SFSRDGSQLLSTSFD TARIHGLKSGK+LKEFRGHTSYVN A FT+DG+R
Sbjct: 303 -AHSQGVTSLSFSRDGSQLLSTSFDQTARIHGLKSGKLLKEFRGHTSYVNHAIFTSDGSR 361
Query: 364 VITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIY 423
+ITASSDCTVKVWD KTTDCLQTFKPPPPLRG DASVNS+ +FPKNTEHIVVCNKTSSIY
Sbjct: 362 IITASSDCTVKVWDSKTTDCLQTFKPPPPLRGTDASVNSIHLFPKNTEHIVVCNKTSSIY 421
Query: 424 IMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVH 483
IMTLQGQVVKSFSSG REGGDFVAAC+S KG+WIYC+GED+ +YCF+YQSG LEH M VH
Sbjct: 422 IMTLQGQVVKSFSSGNREGGDFVAACVSTKGDWIYCIGEDKKLYCFNYQSGGLEHFMMVH 481
Query: 484 DKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
+K++IG+THHPHRNL+AT+S+D TMKLWKP
Sbjct: 482 EKDVIGITHHPHRNLLATYSEDCTMKLWKP 511
>UniRef100_Q9C9T9 Hypothetical protein F25P22.14 [Arabidopsis thaliana]
Length = 522
Score = 897 bits (2319), Expect = 0.0
Identities = 427/508 (84%), Positives = 476/508 (93%), Gaps = 1/508 (0%)
Query: 6 IEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQVS 65
+E VIK +LQFCKENSL+QTFQTLQ+ECQVSLNTVDS+ETF++DIN GRWD++LPQVS
Sbjct: 16 VECLSVIKIMLQFCKENSLNQTFQTLQSECQVSLNTVDSVETFISDINSGRWDSVLPQVS 75
Query: 66 QLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLVRT 125
QLKLPRN LEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQ ERYLR+EHLLVR+
Sbjct: 76 QLKLPRNKLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQAERYLRMEHLLVRS 135
Query: 126 YFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFD 185
YFDP+EAY +STKE++RAQIAQA+AAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFD
Sbjct: 136 YFDPHEAYGDSTKERKRAQIAQAVAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFD 195
Query: 186 LFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYIS 245
LFRGTAAMKQDV+D +P L+HTIKFG KSHAECARFSPDGQ+L S S+DGFIEVWDYIS
Sbjct: 196 LFRGTAAMKQDVEDTHPNVLTHTIKFGKKSHAECARFSPDGQFLASSSVDGFIEVWDYIS 255
Query: 246 GKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHA 305
GKLKKDLQYQA+E+FMMHD+PVLC+DFSRDSEM+ASGS DGKIK+WRIRT C+RR + A
Sbjct: 256 GKLKKDLQYQADESFMMHDDPVLCIDFSRDSEMLASGSQDGKIKIWRIRTGVCIRRFD-A 314
Query: 306 HSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVI 365
HSQGVTS+SFSRDGSQLLSTSFD TARIHGLKSGK+LKEFRGHTSYVN A FT+DG+R+I
Sbjct: 315 HSQGVTSLSFSRDGSQLLSTSFDQTARIHGLKSGKLLKEFRGHTSYVNHAIFTSDGSRII 374
Query: 366 TASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIM 425
TASSDCTVKVWD KTTDCLQTFKPPPPLRG DASVNS+ +FPKNTEHIVVCNKTSSIYIM
Sbjct: 375 TASSDCTVKVWDSKTTDCLQTFKPPPPLRGTDASVNSIHLFPKNTEHIVVCNKTSSIYIM 434
Query: 426 TLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDK 485
TLQGQVVKSFSSG REGGDFVAAC+S KG+WIYC+GED+ +YCF+YQSG LEH M VH+K
Sbjct: 435 TLQGQVVKSFSSGNREGGDFVAACVSTKGDWIYCIGEDKKLYCFNYQSGGLEHFMMVHEK 494
Query: 486 EIIGVTHHPHRNLVATFSDDHTMKLWKP 513
++IG+THHPHRNL+AT+S+D TMKLWKP
Sbjct: 495 DVIGITHHPHRNLLATYSEDCTMKLWKP 522
>UniRef100_Q5ZME8 Hypothetical protein [Gallus gallus]
Length = 513
Score = 708 bits (1827), Expect = 0.0
Identities = 335/512 (65%), Positives = 419/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVKVW++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKVWNVKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>UniRef100_Q9BU59 Smu-1 suppressor of mec-8 and unc-52 homolog [Homo sapiens]
Length = 513
Score = 708 bits (1827), Expect = 0.0
Identities = 334/512 (65%), Positives = 419/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>UniRef100_Q6P4J8 Hypothetical protein MGC75979 [Xenopus tropicalis]
Length = 513
Score = 706 bits (1822), Expect = 0.0
Identities = 333/512 (65%), Positives = 419/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+T TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRTLATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQNQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MIIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T R+HGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDCSQILSASFDQTIRVHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+ T+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIGTYSEDGLLKLWKP 513
>UniRef100_Q9D1I8 Mus musculus 18-day embryo whole body cDNA, RIKEN full-length
enriched library, clone:1110006E22 product:CDNA FLJ10805
FIS, CLONE NT2RP4000839, WEAKLY SIMILAR TO VEGETATIBLE
INCOMPATIBILITY PROTEIN HET-E-1 homolog [Mus musculus]
Length = 513
Score = 705 bits (1820), Expect = 0.0
Identities = 333/512 (65%), Positives = 418/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEF GH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFLGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>UniRef100_Q9NVD1 Hypothetical protein FLJ10805 [Homo sapiens]
Length = 513
Score = 705 bits (1820), Expect = 0.0
Identities = 333/512 (65%), Positives = 418/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+N +AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNPIATYSEDGLLKLWKP 513
>UniRef100_Q99M63 Brain-enriched WD-repeat protein [Rattus norvegicus]
Length = 513
Score = 704 bits (1817), Expect = 0.0
Identities = 332/512 (64%), Positives = 417/512 (80%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ T E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATFTEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>UniRef100_Q6NRT3 MGC81475 protein [Xenopus laevis]
Length = 513
Score = 704 bits (1816), Expect = 0.0
Identities = 332/512 (64%), Positives = 417/512 (80%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+T TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRTLATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ Q ERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQNQSERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRR IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRTAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDFVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+ T+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIGTYSEDGLLKLWKP 513
>UniRef100_Q9JJ70 Mus musculus [Mus musculus]
Length = 513
Score = 703 bits (1815), Expect = 0.0
Identities = 333/512 (65%), Positives = 417/512 (81%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+ TLQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESSDVIRLIMQYLKENSLHRALATLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQALKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEEKFPTQLSRHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D SQ+LS SFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG
Sbjct: 302 FERAHSKGVTCLSFSKDSSQILSASFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTQDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+ASSD TVK+W++KTT+C TFK G D +VNSV + PKN EH VVCN++++
Sbjct: 362 HYIISASSDGTVKIWNMKTTECSNTFKSLGSTAGTDITVNSVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +S +GEWIYCVGED YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCALSLRGEWIYCVGEDFVFYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+AT+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIATYSEDGLLKLWKP 513
>UniRef100_UPI00002DAA1D UPI00002DAA1D UniRef100 entry
Length = 504
Score = 699 bits (1804), Expect = 0.0
Identities = 328/504 (65%), Positives = 414/504 (82%), Gaps = 1/504 (0%)
Query: 11 VIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQVSQLKLP 70
V++ ++Q+ KEN+L +T TLQ E VSLNTVDSIE+FVADIN G WD +L + LKLP
Sbjct: 1 VVRLIMQYLKENNLQRTLATLQEETTVSLNTVDSIESFVADINSGHWDTVLQAIQSLKLP 60
Query: 71 RNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLVRTYFDPN 130
TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL R+YFDP
Sbjct: 61 DKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLLARSYFDPR 120
Query: 131 EAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFDLFRG 189
EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQ+LKWQQHQGLLPPGT DLFRG
Sbjct: 121 EAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQSLKWQQHQGLLPPGTTIDLFRG 180
Query: 190 TAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLK 249
AA+K ++ +PT LS IKFG KSH EC+RFSPDGQYLV+ S+DGFIEVW++ +GK++
Sbjct: 181 KAAVKDVEEERFPTQLSRQIKFGQKSHVECSRFSPDGQYLVTGSVDGFIEVWNFTTGKIR 240
Query: 250 KDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQG 309
KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR E AHS+G
Sbjct: 241 KDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRRFERAHSKG 300
Query: 310 VTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASS 369
VT VSF +D SQLLSTSFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG +I+ASS
Sbjct: 301 VTCVSFCKDSSQLLSTSFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTPDGHHIISASS 360
Query: 370 DCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQG 429
D TVKVW++KTT+C TFKP G D +VN++ + PKN EH VVCN+++++ IM +QG
Sbjct: 361 DGTVKVWNMKTTECTNTFKPLGTSAGTDITVNNILLLPKNPEHFVVCNRSNTVVIMNMQG 420
Query: 430 QVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIG 489
Q+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +TVH+K++IG
Sbjct: 421 QIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDFVLYCFSTVTGKLERTLTVHEKDVIG 480
Query: 490 VTHHPHRNLVATFSDDHTMKLWKP 513
+ HHPH+NL++T+S+D +KLWKP
Sbjct: 481 IAHHPHQNLISTYSEDGLLKLWKP 504
>UniRef100_Q7ZVA0 Hypothetical protein zgc:56147 [Brachydanio rerio]
Length = 513
Score = 699 bits (1804), Expect = 0.0
Identities = 329/512 (64%), Positives = 416/512 (80%), Gaps = 1/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ ++Q+ KENSLH+T LQ E VSLNTVDSIE+FVADIN G WD +L
Sbjct: 2 SIEIESADVIRLIMQYLKENSLHRTLAMLQEETTVSLNTVDSIESFVADINSGHWDTVLQ 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL
Sbjct: 62 AIQSLKLPDKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLL 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R+YFDP EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQ+LKWQQHQGLLPPG
Sbjct: 122 ARSYFDPREAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQSLKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
DLFRG AA+K ++ +PT L+ IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW
Sbjct: 182 MTIDLFRGKAAVKDVEEERFPTQLARHIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMDDAVLCMSFSRDTEMLATGAQDGKIKVWKIQSGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT +SFS+D +Q+LS SFD T RIHGLKSGK LKEFRGH+S+VN+AT T DG
Sbjct: 302 YERAHSKGVTCLSFSKDSTQILSASFDQTIRIHGLKSGKCLKEFRGHSSFVNEATLTPDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
I+ASSD TVKVW++KTT+C TFK G D +VN+V + PKN EH VVCN++++
Sbjct: 362 HHAISASSDGTVKVWNMKTTECTSTFKSLGTSAGTDITVNNVILLPKNPEHFVVCNRSNT 421
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +T
Sbjct: 422 VVIMNMQGQIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDYVLYCFSTVTGKLERTLT 481
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+ HHPH+NL+ T+S+D +KLWKP
Sbjct: 482 VHEKDVIGIAHHPHQNLIGTYSEDGLLKLWKP 513
>UniRef100_Q9VE18 CG5451-PA [Drosophila melanogaster]
Length = 509
Score = 671 bits (1730), Expect = 0.0
Identities = 316/512 (61%), Positives = 413/512 (79%), Gaps = 5/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ + Q+ KE++L +T QTLQ E VSLNTVDS++ FV DI+ G WD +L
Sbjct: 2 SIEIESADVIRLIQQYLKESNLMKTLQTLQEETGVSLNTVDSVDGFVQDISNGHWDTVLK 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
LKLP L +LYEQIVLE+IELREL AR++LRQT M ++KQ++PERY+ LE++L
Sbjct: 62 VTQSLKLPDKKLLNLYEQIVLELIELRELGAARSLLRQTDPMTMLKQQEPERYIHLENML 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R YFDP EAY E S+KEKRR IAQ ++ EV VVP SRL+AL+GQALKWQQHQGLLPPG
Sbjct: 122 QRAYFDPREAYAEGSSKEKRRTVIAQELSGEVHVVPSSRLLALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
T DLFRG AAMK ++MYPT L IKFG KSH ECA+FSPDGQYL++ S+DGF+EVW
Sbjct: 182 TTIDLFRGKAAMKDQEEEMYPTQLFRQIKFGQKSHVECAQFSPDGQYLITGSVDGFLEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM ++ VL ++FSRDSEM+ASG+ DG+IKVWRI T QCLR+
Sbjct: 242 NFTTGKVRKDLKYQAQDQFMMMEQAVLALNFSRDSEMVASGAQDGQIKVWRIITGQCLRK 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AH++G+T + FSRD SQ+LS SFD T R+HGLKSGKMLKEF+GH+S+VN+ATFT DG
Sbjct: 302 FEKAHTKGITCLQFSRDNSQVLSASFDYTVRLHGLKSGKMLKEFKGHSSFVNEATFTPDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
V++ASSD TVKVW LKTT+C+ T+KP G + +VN+V I PKN EH +VCN++++
Sbjct: 362 HSVLSASSDGTVKVWSLKTTECVATYKP----LGNELAVNTVLILPKNPEHFIVCNRSNT 417
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGG F++A +SP+GE+IYC GED+ +YCFS SGKLE +
Sbjct: 418 VVIMNMQGQIVRSFSSGKREGGAFISATLSPRGEFIYCAGEDQVLYCFSVSSGKLERTLN 477
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+THHPH+NL+A++S+D +KLWKP
Sbjct: 478 VHEKDVIGLTHHPHQNLLASYSEDGLLKLWKP 509
>UniRef100_UPI0000433250 UPI0000433250 UniRef100 entry
Length = 501
Score = 669 bits (1725), Expect = 0.0
Identities = 318/504 (63%), Positives = 402/504 (79%), Gaps = 4/504 (0%)
Query: 11 VIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQVSQLKLP 70
VI+ + Q+ KE++L +T QTLQ E VSLNTVDS++ FVADIN G WD +L + LKLP
Sbjct: 1 VIRLIQQYLKESNLVKTLQTLQEETGVSLNTVDSVDGFVADINNGHWDTVLKAIQSLKLP 60
Query: 71 RNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLVRTYFDPN 130
L DLYEQ+VLE+IELREL AR++LRQT M +MKQ++PERY+ LE+LL R+YFDP
Sbjct: 61 DKKLIDLYEQVVLELIELRELGAARSLLRQTDPMIMMKQQEPERYIHLENLLARSYFDPR 120
Query: 131 EAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFDLFRG 189
EAY + STKEKRRA IA A+A EV+VVP SRL+AL+GQALKWQQHQGLLPPGT DLFRG
Sbjct: 121 EAYPDGSTKEKRRAYIATALAGEVSVVPSSRLLALLGQALKWQQHQGLLPPGTTIDLFRG 180
Query: 190 TAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLK 249
AA++ D+ YPT LS IKFG KSH ECARFSPDGQYLV+ S+DGFIEVW++ +GK++
Sbjct: 181 KAAVRDQEDEKYPTQLSKQIKFGQKSHVECARFSPDGQYLVTGSVDGFIEVWNFTTGKIR 240
Query: 250 KDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQG 309
KDL+YQA++ FMM ++ VL + FSRDSEM+A G DGKIKVWR+++ QCLRR E AHS+G
Sbjct: 241 KDLKYQAQDNFMMMEQAVLSMAFSRDSEMLAGGGQDGKIKVWRVQSGQCLRRFEKAHSKG 300
Query: 310 VTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASS 369
VT + FSRD SQ+LS SFD+T RIHGLKSGK LKEFRGH S+VN+ F DG +I+ASS
Sbjct: 301 VTCLQFSRDNSQILSASFDTTIRIHGLKSGKTLKEFRGHASFVNEVVFAPDGHNLISASS 360
Query: 370 DCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQG 429
D TVKVW LKTT+C+ T+K L D +VNS+ + P+N EH VVCN+++++ IM +QG
Sbjct: 361 DGTVKVWSLKTTECIGTYK---SLGAADLTVNSIHLLPRNPEHFVVCNRSNTVVIMNMQG 417
Query: 430 QVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTVHDKEIIG 489
Q+V+SFSSGKREGGDFV +SP+GE+IYCVGED +YCFS SGKLE + +H+K++IG
Sbjct: 418 QIVRSFSSGKREGGDFVCTVVSPRGEFIYCVGEDLVLYCFSTSSGKLERTLNIHEKDVIG 477
Query: 490 VTHHPHRNLVATFSDDHTMKLWKP 513
+THHPH+NL+ ++S+D +KLWKP
Sbjct: 478 LTHHPHQNLLCSYSEDGLLKLWKP 501
>UniRef100_Q7PUS7 ENSANGP00000015224 [Anopheles gambiae str. PEST]
Length = 509
Score = 663 bits (1711), Expect = 0.0
Identities = 317/512 (61%), Positives = 407/512 (78%), Gaps = 5/512 (0%)
Query: 3 TLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILP 62
++EIE+ DVI+ + Q+ KE++LH+T QTLQ E VSLNTVDS+E FV+DI+ G WD +L
Sbjct: 2 SIEIESADVIRLIQQYLKESNLHKTLQTLQEETGVSLNTVDSVEGFVSDIHNGHWDTVLK 61
Query: 63 QVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLL 122
+ LKLP L DLYEQIVLE+IELREL AR++LRQT M ++KQ++ ERY+ LE++L
Sbjct: 62 AIQSLKLPDKKLIDLYEQIVLELIELRELGAARSLLRQTDPMIMLKQQEAERYIHLENML 121
Query: 123 VRTYFDPNEAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPG 181
R YFDP EAY E S+KEKRRA IA A+A EV+VVP SRL+AL+GQALKWQQHQGLLPPG
Sbjct: 122 QRAYFDPREAYPEGSSKEKRRAAIATALAGEVSVVPSSRLLALLGQALKWQQHQGLLPPG 181
Query: 182 TQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
T DLFRG AA++ D+ YPT L+ IKFG KSH ECA FSPDGQYLV+ S+DGFIEVW
Sbjct: 182 TTIDLFRGKAAIRDQEDESYPTQLARQIKFGQKSHVECALFSPDGQYLVTGSVDGFIEVW 241
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++ +GK++KDL+YQA++ FMM ++ +L + FSRDSEM+A+GS DG++KVW++ T QCLRR
Sbjct: 242 NFTTGKIRKDLKYQAQDNFMMMEQAILSMAFSRDSEMLATGSQDGQVKVWKLLTGQCLRR 301
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
E AHS+GVT + FSRD SQ+L+ SFD R++GLKSGKMLKEFRGHTS+VN+A F+ DG
Sbjct: 302 FEKAHSKGVTCLHFSRDNSQILTASFDHLIRLYGLKSGKMLKEFRGHTSFVNEAAFSPDG 361
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
V++ASSD TVKVW LKTT+C+ TFK G +VNSV I PKN EH VVCN++++
Sbjct: 362 HHVLSASSDGTVKVWSLKTTECISTFK----ALGTHLAVNSVLILPKNPEHFVVCNRSNT 417
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ IM +QGQ+V+SFSSGKREGG F+ A ISP+GE+IYC GED +YCFS SGKLE +
Sbjct: 418 VVIMNMQGQIVRSFSSGKREGGAFICATISPRGEFIYCCGEDLVLYCFSVTSGKLERTLN 477
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+K++IG+THHPH+NL+A++ D +K+WKP
Sbjct: 478 VHEKDVIGLTHHPHQNLLASYGCDGLLKMWKP 509
>UniRef100_UPI000035F64C UPI000035F64C UniRef100 entry
Length = 473
Score = 650 bits (1676), Expect = 0.0
Identities = 312/473 (65%), Positives = 386/473 (80%), Gaps = 1/473 (0%)
Query: 11 VIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQVSQLKLP 70
VI+ ++Q+ KEN+L +T TLQ E VSLNTVDSIE+FVADIN G WD +L + LKLP
Sbjct: 1 VIRLIMQYLKENNLQRTLATLQEETTVSLNTVDSIESFVADINSGHWDTVLQAIQSLKLP 60
Query: 71 RNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLVRTYFDPN 130
TL DLYEQ+VLE+IELREL AR++LRQT M ++KQ QPERY+ LE+LL R+YFDP
Sbjct: 61 DKTLIDLYEQVVLELIELRELGAARSLLRQTDPMIMLKQTQPERYIHLENLLARSYFDPR 120
Query: 131 EAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFDLFRG 189
EAY + S+KEKRRA IAQA+A EV+VVPPSRLMAL+GQ+LKWQQHQGLLPPGT DLFRG
Sbjct: 121 EAYPDGSSKEKRRAAIAQALAGEVSVVPPSRLMALLGQSLKWQQHQGLLPPGTTIDLFRG 180
Query: 190 TAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVWDYISGKLK 249
AA+K ++ +PT LS IKFG KSH EC+RFSPDGQYLV+ S+DGFIEVW++ +GK++
Sbjct: 181 KAAVKDVEEERFPTQLSRQIKFGQKSHVECSRFSPDGQYLVTGSVDGFIEVWNFTTGKIR 240
Query: 250 KDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRRLEHAHSQG 309
KDL+YQA++ FMM D+ VLC+ FSRD+EM+A+G+ DGKIKVW+I++ QCLRR E AHS+G
Sbjct: 241 KDLKYQAQDNFMMMDDAVLCMCFSRDTEMLATGAQDGKIKVWKIQSGQCLRRFERAHSKG 300
Query: 310 VTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDGTRVITASS 369
VT VSF +D SQLLSTSFD T RIHGLKSGK LKEFRGH+S+VN+ATFT DG VI+ASS
Sbjct: 301 VTCVSFCKDSSQLLSTSFDQTIRIHGLKSGKTLKEFRGHSSFVNEATFTPDGHHVISASS 360
Query: 370 DCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSSIYIMTLQG 429
D TVKVW++KTT+C TFKP G D +VN++ + PKN EH VVCN+++++ IM +QG
Sbjct: 361 DGTVKVWNMKTTECTNTFKPLGTSAGTDITVNNILLLPKNPEHFVVCNRSNTVVIMNMQG 420
Query: 430 QVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMTV 482
Q+V+SFSSGKREGGDFV +SP+GEWIYCVGED +YCFS +GKLE +TV
Sbjct: 421 QIVRSFSSGKREGGDFVCCTLSPRGEWIYCVGEDFVLYCFSTVTGKLERTLTV 473
Score = 42.4 bits (98), Expect = 0.032
Identities = 39/169 (23%), Positives = 69/169 (40%), Gaps = 8/169 (4%)
Query: 347 GHTSYVNDATFTNDGTRVITASSDCTVKVWDLKTTDCLQTFKPPPP---LRGGDASVNSV 403
G S+V + F+ DG ++T S D ++VW+ T + K + DA +
Sbjct: 203 GQKSHVECSRFSPDGQYLVTGSVDGFIEVWNFTTGKIRKDLKYQAQDNFMMMDDAVL--C 260
Query: 404 FIFPKNTEHIVVCNKTSSIYIMTLQ-GQVVKSFSSGKREGGDFVAACISPKGEWIYCVGE 462
F ++TE + + I + +Q GQ ++ F +G V+ C +
Sbjct: 261 MCFSRDTEMLATGAQDGKIKVWKIQSGQCLRRFERAHSKGVTCVSFC--KDSSQLLSTSF 318
Query: 463 DRNMYCFSYQSGKLEHLMTVHDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
D+ + +SGK H + T P + V + S D T+K+W
Sbjct: 319 DQTIRIHGLKSGKTLKEFRGHSSFVNEATFTPDGHHVISASSDGTVKVW 367
>UniRef100_UPI00003602F7 UPI00003602F7 UniRef100 entry
Length = 510
Score = 565 bits (1455), Expect = e-159
Identities = 268/512 (52%), Positives = 383/512 (74%), Gaps = 11/512 (2%)
Query: 11 VIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAILPQVSQLKLP 70
VI+ ++Q+ KEN+L++T TLQ E +SLNTV+S+++F+ADIN G WD++L + LKLP
Sbjct: 1 VIRLIMQYLKENNLYRTLTTLQEETTLSLNTVESLDSFIADINCGHWDSVLMAIESLKLP 60
Query: 71 RNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEHLLVRTYFDPN 130
+TL DLYEQ+V+E+IE+RE+ AR++LRQT M ++KQ QP+RYL LE+LL +YFDP
Sbjct: 61 DSTLIDLYEQVVMELIEMREVGAARSLLRQTDPMIMLKQTQPQRYLHLENLLTCSYFDPL 120
Query: 131 EAYQE-STKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPPGTQFDLFRG 189
EAY + S+KEKRR IA+ + EV+VVPPSRLM L+ Q LPP +
Sbjct: 121 EAYPKGSSKEKRRLAIAETLVREVSVVPPSRLMGLLEQVSMELHACPCLPPPKVIVVTVI 180
Query: 190 TAAMKQD------VDDMYPT--NLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEVW 241
T Q DD + T +SH ++FG +SH ECARFSPDG+YL++ S+DGFIEVW
Sbjct: 181 TDVRLQQQQLTCVFDDSFTTVVAVSHELQFGRRSHVECARFSPDGRYLITGSVDGFIEVW 240
Query: 242 DYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLRR 301
++++GK+ KDL+YQA+E+FMM D+ VLC+ FS+D+E++A+G+ +GKIKVW I++ CLRR
Sbjct: 241 NFVTGKISKDLKYQAQESFMMMDDAVLCMCFSQDAELLATGAQNGKIKVWTIQSGLCLRR 300
Query: 302 LEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTNDG 361
EHAH++GVT +SFS+D +Q+LS SF+ + RIH L+SGK LKE GH S+VNDA FT DG
Sbjct: 301 FEHAHNKGVTCLSFSKDSNQILSASFNQSIRIHELRSGKTLKELTGHLSFVNDAYFTQDG 360
Query: 362 TRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPKNTEHIVVCNKTSS 421
+I+AS+D TVK+W+ K+ +C+ T K R + VN+V P++ EH VVCN +++
Sbjct: 361 RHIISASADGTVKIWNTKSWECIHTHKSVS--RASEVPVNNVIPLPQHPEHFVVCNHSNT 418
Query: 422 IYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHLMT 481
+ ++ ++GQ VK+FSSG++EG +FV + +SP+GEWIYCVGED +YCF+Y++GKL+ +T
Sbjct: 419 VVVLNMRGQTVKTFSSGRQEGAEFVCSTVSPRGEWIYCVGEDLVLYCFNYKTGKLQKTLT 478
Query: 482 VHDKEIIGVTHHPHRNLVATFSDDHTMKLWKP 513
VH+KE+IG+THHPH NL+AT+S+D ++LWKP
Sbjct: 479 VHEKEVIGITHHPHENLIATYSEDGLLRLWKP 510
>UniRef100_O62120 SMU-1 [Caenorhabditis elegans]
Length = 510
Score = 556 bits (1432), Expect = e-157
Identities = 263/512 (51%), Positives = 369/512 (71%), Gaps = 5/512 (0%)
Query: 1 MSTLEIEARDVIKTVLQFCKENSLHQTFQTLQNECQVSLNTVDSIETFVADINGGRWDAI 60
MS++EIE+ DVI+ + QF KE++LH+T LQ E VSLNTVDSI+ F +I G WD +
Sbjct: 1 MSSIEIESSDVIRLIEQFLKESNLHRTLAILQEETNVSLNTVDSIDGFCNEITSGNWDNV 60
Query: 61 LPQVSQLKLPRNTLEDLYEQIVLEMIELRELDTARAILRQTQVMGVMKQEQPERYLRLEH 120
L V LKLP L DLYE +++E++ELREL TAR + RQT M ++KQ P+R+ RLE
Sbjct: 61 LKTVQSLKLPAKKLIDLYEHVIIELVELRELATARLVARQTDPMILLKQIDPDRFARLES 120
Query: 121 LLVRTYFDPNEAYQESTKEKRRAQIAQAIAAEVTVVPPSRLMALIGQALKWQQHQGLLPP 180
L+ R YFD E Y + +KEKRR+ IAQ +++EV VV PSRL++L+GQ+LKWQ HQGLLPP
Sbjct: 121 LINRPYFDGQEVYGDVSKEKRRSVIAQTLSSEVHVVAPSRLLSLLGQSLKWQLHQGLLPP 180
Query: 181 GTQFDLFRGTAAMKQDVDDMYPTNLSHTIKFGAKSHAECARFSPDGQYLVSCSIDGFIEV 240
GT DLFRG AA K+ +++ YPT ++ +IKF KS+ E A FSPD YLVS S DGFIEV
Sbjct: 181 GTAIDLFRGKAAQKEQIEERYPTMMARSIKFSTKSYPESAVFSPDANYLVSGSKDGFIEV 240
Query: 241 WDYISGKLKKDLQYQAEETFMMHDEPVLCVDFSRDSEMIASGSTDGKIKVWRIRTAQCLR 300
W+Y++GKL+KDL+YQA++ MM D V C+ FSRDSEM+A+GS DGKIKVW++ T CLR
Sbjct: 241 WNYMNGKLRKDLKYQAQDNLMMMDAAVRCISFSRDSEMLATGSIDGKIKVWKVETGDCLR 300
Query: 301 RLEHAHSQGVTSVSFSRDGSQLLSTSFDSTARIHGLKSGKMLKEFRGHTSYVNDATFTND 360
R + AH++GV +V FS+D S +LS D R+HG+KSGK LKE RGH+SY+ D ++++
Sbjct: 301 RFDRAHTKGVCAVRFSKDNSHILSGGNDHVVRVHGMKSGKCLKEMRGHSSYITDVRYSDE 360
Query: 361 GTRVITASSDCTVKVWDLKTTDCLQTFKPPPPLRGGDASVNSVFIFPK-NTEHIVVCNKT 419
G +I+ S+D +++VW K+ +CL TF+ + D + +V PK + ++VCN++
Sbjct: 361 GNHIISCSTDGSIRVWHGKSGECLSTFR----VGSEDYPILNVIPIPKSDPPQMIVCNRS 416
Query: 420 SSIYIMTLQGQVVKSFSSGKREGGDFVAACISPKGEWIYCVGEDRNMYCFSYQSGKLEHL 479
+++Y++ + GQVV++ +SGKRE GDF+ +SPKGEW Y + ED MYCF SG LE
Sbjct: 417 NTLYVVNISGQVVRTMTSGKREKGDFINCILSPKGEWAYAIAEDGVMYCFMVLSGTLETT 476
Query: 480 MTVHDKEIIGVTHHPHRNLVATFSDDHTMKLW 511
+ V ++ IG+ HHPH+NL+A++++D +KLW
Sbjct: 477 LPVTERLPIGLAHHPHQNLIASYAEDGLLKLW 508
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.134 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 844,229,614
Number of Sequences: 2790947
Number of extensions: 34506576
Number of successful extensions: 140754
Number of sequences better than 10.0: 5678
Number of HSP's better than 10.0 without gapping: 3000
Number of HSP's successfully gapped in prelim test: 2711
Number of HSP's that attempted gapping in prelim test: 90523
Number of HSP's gapped (non-prelim): 26706
length of query: 513
length of database: 848,049,833
effective HSP length: 132
effective length of query: 381
effective length of database: 479,644,829
effective search space: 182744679849
effective search space used: 182744679849
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC142394.6


