

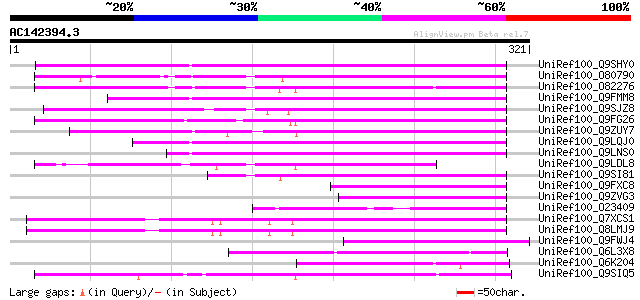
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.3 - phase: 0
(321 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana] 176 1e-42
UniRef100_O80790 Putative non-LTR retroelement reverse transcrip... 153 5e-36
UniRef100_O82276 Putative non-LTR retroelement reverse transcrip... 148 2e-34
UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like... 146 6e-34
UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcrip... 144 3e-33
UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like... 144 3e-33
UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis tha... 134 3e-30
UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana] 126 9e-28
UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana] 118 2e-25
UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis tha... 115 1e-24
UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana] 109 9e-23
UniRef100_Q9FXC8 F12A21.24 [Arabidopsis thaliana] 87 6e-16
UniRef100_Q9ZVG3 T2P11.14 protein [Arabidopsis thaliana] 82 3e-14
UniRef100_O23409 SIMILARITY to probable splicing factor CEPRP21 ... 80 1e-13
UniRef100_Q7XCS1 Putative reverse transcriptase [Oryza sativa] 76 1e-12
UniRef100_Q8LMJ9 Putative reverse transcriptase, 5'-partial [Ory... 76 1e-12
UniRef100_Q9FWJ4 Hypothetical protein F21N10.9 [Arabidopsis thal... 75 2e-12
UniRef100_Q6L3X8 Hypothetical protein [Solanum demissum] 74 7e-12
UniRef100_Q6K204 Hypothetical protein B1469H02.35 [Oryza sativa] 72 2e-11
UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcrip... 72 3e-11
>UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana]
Length = 1055
Score = 176 bits (445), Expect = 1e-42
Identities = 100/293 (34%), Positives = 145/293 (49%), Gaps = 3/293 (1%)
Query: 17 LWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIG-NPMCRFCHDEIESEIHVLRDCPKAT 75
LW +R PER+K F+ ++ + + T +H+ + + +C+ C +ES +HVLRDCP
Sbjct: 400 LWKVRVPERVKTFLWLVGNQAVMTEEERHRRHLSASNVCQVCKGGVESMLHVLRDCPAQL 459
Query: 76 ALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNKETH 135
+W+ VV + FF L W+ NL + + W +A W WR
Sbjct: 460 GIWVRVVPQRRQQGFFSKSLFEWLYDNLGDRSGCED-IPWSTIFAVIIWWGWKWRCGNIF 518
Query: 136 NDNYQRPLHAKNIIMIYVNDYHNAIAKFVIVTSQPRHLEEVGWKEPAIRWVKINTDGACK 195
+N + K + V Y ++ +QPR +GW P + WVK+NTDGA +
Sbjct: 519 GENTKCRDRVKFVKEWAVEVYRAHSGNVLVGITQPRVERMIGWVSPCVGWVKVNTDGASR 578
Query: 196 DG-SIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVD 254
+A GG++R G W GFS +G+C A AELWGV GL A VEL VD
Sbjct: 579 GNPGLASAGGVLRDCTGAWCGGFSLNIGRCSAPQAELWGVYYGLYFAWEKKVPRVELEVD 638
Query: 255 SLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALAN 307
S V+V + + + LV+ LQ +W V++ H YREANR AD LAN
Sbjct: 639 SEVIVGFLKTGISDSHPLSFLVRLCHGFLQKDWLVRIVHVYREANRLADGLAN 691
>UniRef100_O80790 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 970
Score = 153 bits (387), Expect = 5e-36
Identities = 100/301 (33%), Positives = 146/301 (48%), Gaps = 23/301 (7%)
Query: 16 KLWNIRAPERIKHFMSMLYHGRLSTNL---RKHKMGIGNPMCRFCHDEIESEIHVLRDCP 72
++W + APER++ F+ ++ H + TN+ R+H I C C+ ES +HVLRDCP
Sbjct: 646 QIWKLVAPERVRVFIWLVSHMVIMTNVERVRRHLSDIAT--CSVCNGADESILHVLRDCP 703
Query: 73 KATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNK 132
T +W ++ + FF W LFTN+ +W ++ W WR
Sbjct: 704 AMTPIWQRLLPQRRQNEFFSQ--FEW----LFTNLDPAKG-DWPTLFSMGIWWAWKWRCG 756
Query: 133 ETHNDNYQRPLHAKNIIMIYVNDYHNAIAKFVIVT-----SQPRHLEEVGWKEPAIRWVK 187
+ + K ++ D + K + T + R + WK P+ RWVK
Sbjct: 757 DVFGERKLCRDRLK-----FIKDIAEEVRKAHVGTLNNHVKRARVERMIRWKAPSDRWVK 811
Query: 188 INTDGACKDGS-IAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGF 246
+ TDGA + +A G I +GEWL GF+ +G CDA +AELWG GL A GF
Sbjct: 812 LTTDGASRGHQGLAAASGAILNLQGEWLGGFALNIGSCDAPLAELWGAYYGLLIAWDKGF 871
Query: 247 TAVELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALA 306
VELN+DS +VV +++ LV+ + +W V+V H YREANR AD LA
Sbjct: 872 RRVELNLDSELVVGFLSTGISKAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADGLA 931
Query: 307 N 307
N
Sbjct: 932 N 932
>UniRef100_O82276 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 148 bits (373), Expect = 2e-34
Identities = 97/300 (32%), Positives = 147/300 (48%), Gaps = 19/300 (6%)
Query: 16 KLWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIG-NPMCRFCHDEIESEIHVLRDCPKA 74
++W + PER++ F+ ++ + TN+ + + + N +C C+ E+ +HVLRDCP
Sbjct: 905 RIWKLITPERVRVFIWLVSQNVIMTNVERVRRHLSENAICSVCNGAEETILHVLRDCPAM 964
Query: 75 TALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNKET 134
+W ++ FF L W LFTN+ + W + W WR +
Sbjct: 965 EPIWRRLLPLRRHHEFFSQSLLEW----LFTNMDPVKGI-WPTLFGMGIWWAWKWRCCDV 1019
Query: 135 HNDNYQRPLHAKNIIMIYVNDYHNAIAKFVI--VTSQPRHLEE---VGWKEPAIRWVKIN 189
+ K ++ D + + + V ++P + + W+ P+ WVKI
Sbjct: 1020 FGERKICRDRLK-----FIKDMAEEVRRVHVGAVGNRPNGVRVERMIRWQVPSDGWVKIT 1074
Query: 190 TDGACKDG-SIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTA 248
TDGA + +A GG IR +GEWL GF+ +G C A +AELWG GL A GF
Sbjct: 1075 TDGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGAYYGLLIAWDKGFRR 1134
Query: 249 VELNVDSLVVVNIITSERESNASGRS-LVQKIRKLLQMEWKVKVKHSYREANRCADALAN 307
VEL++D +VV + S SNA S LV+ + +W V+V H YREANR AD LAN
Sbjct: 1135 VELDLDCKLVVGFL-STGVSNAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADGLAN 1193
>UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 308
Score = 146 bits (369), Expect = 6e-34
Identities = 84/248 (33%), Positives = 119/248 (47%), Gaps = 2/248 (0%)
Query: 61 IESEIHVLRDCPKATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWA 120
+ES +HV RDCP +W+ V + FF L W+ NL + + W +A
Sbjct: 24 VESVLHVFRDCPAQLGIWVRFVPRRRQQGFFSKSLFEWLYDNLCDRSSCED-IPWSTIFA 82
Query: 121 TACHTIWNWRNKETHNDNYQRPLHAKNIIMIYVNDYHNAIAKFVIVTSQPRHLEEVGWKE 180
W WR +N + K + V Y + ++ ++QPR +GW
Sbjct: 83 VIIWWGWKWRCSNIFGENTKCRDRVKFVKEWVVEVYRAHLGNALVGSTQPRVERLIGWVL 142
Query: 181 PAIRWVKINTDGACKDG-SIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLR 239
P + WVK+NTDGA + +A GG++R EG W GFS +G+C A AELWGV GL
Sbjct: 143 PCVGWVKVNTDGASRGNPGLASAGGVLRDCEGAWCGGFSLNIGRCSAQHAELWGVYYGLY 202
Query: 240 CAKRMGFTAVELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREAN 299
A VEL VDS +V + + + LV+ LQ +W V++ + YREAN
Sbjct: 203 FAWEKKVPRVELEVDSEAIVGFLKTGISDSHPLSFLVRLCHNFLQKDWLVRIVYVYREAN 262
Query: 300 RCADALAN 307
AD LAN
Sbjct: 263 CLADGLAN 270
>UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 321
Score = 144 bits (363), Expect = 3e-33
Identities = 96/293 (32%), Positives = 138/293 (46%), Gaps = 18/293 (6%)
Query: 22 APERIKHFMSMLYHGRLSTNLRKHKMGIGNP-MCRFCHDEIESEIHVLRDCPKATALWLC 80
APER++ F+ ++ + TN+ +++ + + +C+ C E+ +HVLRDCP +W
Sbjct: 2 APERVRVFLWLVVQQVIITNVERYRRHLSDTRVCQICQGGEETILHVLRDCPAMAGIWSR 61
Query: 81 VVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNKETHNDNYQ 140
+V FF L WI NL W +W W WR N +
Sbjct: 62 LVPRDQIRQFFTASLLEWIYKNLRERGSWPTVFVMAVWWG------WKWRCGNIFGGNGK 115
Query: 141 RPLHAKNIIMIYVNDYHN--AIAKFVIVTSQPR--HLEE-VGWKEPAIRWVKINTDGACK 195
K ++ D AIA + ++ R +E V W P WVK+NTDGA +
Sbjct: 116 CRDRVK-----FIKDLAEEVAIANAFVKGNEVRVSRVERLVSWVSPEDGWVKLNTDGASR 170
Query: 196 DG-SIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVD 254
A GG++R G W+ GF+ +G C A +AELWGV GL A G VEL VD
Sbjct: 171 GNPGFATAGGVLRDHNGAWIGGFAVNIGVCSAPLAELWGVYYGLFIAWGRGARRVELEVD 230
Query: 255 SLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALAN 307
S +VV +T+ + L++ L W V++ H YREANR AD LAN
Sbjct: 231 SKMVVGFLTTGIADSHPLSFLLRLCYDFLSKGWIVRISHVYREANRLADGLAN 283
>UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 676
Score = 144 bits (363), Expect = 3e-33
Identities = 95/298 (31%), Positives = 140/298 (46%), Gaps = 11/298 (3%)
Query: 16 KLWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIGNP-MCRFCHDEIESEIHVLRDCPKA 74
++W + PER + F+ ++ + + TN + + + + +C C ES IHVLRDCP
Sbjct: 346 RVWRVMVPERARIFLWLVGNQVVLTNAERVRRHMADSDVCPLCKGASESLIHVLRDCPAM 405
Query: 75 TALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNKET 134
+W+ VV + FFE L W+ NL + W +A W WR
Sbjct: 406 MGIWMRVVPVMEQRRFFETSLLEWMYGNLKERSD-SERRSWPTLFALTVWWGWKWRCGYV 464
Query: 135 HNDNYQRPLHAKNIIMIYVNDYHNAIAKFVIVTSQPRH--LEE--VGWKEPAIRWVKINT 190
++ + ++ + + A + R L E + W++PA WV +NT
Sbjct: 465 FGEDSR----CRDRVKFLKSAVAEVEAAHLAANGDAREDVLVERMIAWRKPAEGWVTMNT 520
Query: 191 DGACKDG-SIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAV 249
DGA A GG+IR G WL GF+ +G C A +AELWGV GL A G+ V
Sbjct: 521 DGASHGNPGQATAGGVIRDEHGSWLVGFALNIGVCSAPLAELWGVYYGLVVAWERGWRRV 580
Query: 250 ELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALAN 307
L VDS +VV + S + LV+ + +W V++ H YREANR AD LAN
Sbjct: 581 RLEVDSALVVGFLQSGIGDSHPLAFLVRLCHGFISKDWIVRITHVYREANRLADGLAN 638
>UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 314
Score = 134 bits (337), Expect = 3e-30
Identities = 86/279 (30%), Positives = 128/279 (45%), Gaps = 16/279 (5%)
Query: 38 LSTNLRKHKMGIGNP-MCRFCHDEIESEIHVLRDCPKATALWLCVVDNAARTSFFEGDLS 96
L TN + + + + +C+ C ++ IH+LRDCP +W+ +V R FF L
Sbjct: 5 LMTNAERRRRHLSDSDICQICKGAEKTIIHILRDCPAMEGIWIRLVPAGKRREFFTQSLL 64
Query: 97 SWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNKE---THNDNYQRPLHAKNIIMIYV 153
W+ NL + W +A + W WR + R K++
Sbjct: 65 EWLFANLGDRRKTCEST-WSTLFALSIWWAWKWRCGNIFGVQDKCRDRVRFLKDLAR--- 120
Query: 154 NDYHNAIAKFVIVTSQPRHLEEV----GWKEPAIRWVKINTDGACKDG-SIAGCGGLIRG 208
++A ++ T H E V W +P W K+NTDGA + +A GG++R
Sbjct: 121 ---ETSMAHVIVRTLSGGHGERVERLIAWSKPEEGWWKLNTDGASRGNPGLASAGGVLRD 177
Query: 209 SEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERES 268
EG W GF+ +G C A +AELWGV GL A T +E+ VDS +VV +
Sbjct: 178 EEGAWRGGFALNIGVCSAPLAELWGVYYGLYIAWERRVTRLEIEVDSEIVVGFLKIGINE 237
Query: 269 NASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALAN 307
LV+ + +W+V++ H YREANR AD LAN
Sbjct: 238 VHPLSFLVRLCHDFISRDWRVRISHVYREANRLADGLAN 276
>UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana]
Length = 272
Score = 126 bits (316), Expect = 9e-28
Identities = 74/232 (31%), Positives = 111/232 (46%), Gaps = 2/232 (0%)
Query: 77 LWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNKETHN 136
+W ++ +SFF L WI NL + N W ++ A W WR
Sbjct: 4 IWTRLLPVRRLSSFFSKSLLEWIYANLGEEIEING-CPWAVTFSQAIWWGWKWRYGNIFG 62
Query: 137 DNYQRPLHAKNIIMIYVNDYHNAIAKFVIVTSQPRHLEEVGWKEPAIRWVKINTDGACKD 196
+N + + I ++ + + K + T R + W P + W K+NTDGA +
Sbjct: 63 ENKKCRDRVRFIKDRALDVWKAHVHKMGVTTRTAREERLIAWSPPRVGWFKLNTDGASRG 122
Query: 197 GS-IAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDS 255
+A GG++R +G W GFS +G C A +AELWG GL A G T +E+ +DS
Sbjct: 123 NPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGAYYGLNIAWERGVTQLEMEIDS 182
Query: 256 LVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALAN 307
+VV + + + + LV+ LL +W V++ H YREANR AD LAN
Sbjct: 183 EMVVGFLRTGIDDSHPLSFLVRLCHGLLSKDWSVRISHVYREANRLADGLAN 234
>UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana]
Length = 253
Score = 118 bits (296), Expect = 2e-25
Identities = 69/211 (32%), Positives = 102/211 (47%), Gaps = 2/211 (0%)
Query: 98 WIKFNLFTNVYWNNNVEWRDFWATACHTIWNWRNKETHNDNYQRPLHAKNIIMIYVNDYH 157
WI NL + N W ++ A W WR +N + + I ++ +
Sbjct: 6 WIYANLGEEIEING-CPWAVTFSQAIWWGWKWRYGNIFGENKKCRDRVRFIKDRALDVWK 64
Query: 158 NAIAKFVIVTSQPRHLEEVGWKEPAIRWVKINTDGACKDGS-IAGCGGLIRGSEGEWLAG 216
+ K + T R + W P + W K+NTDGA + +A GG++R +G W G
Sbjct: 65 AHVHKMGVTTRTAREERLIAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDGNWCYG 124
Query: 217 FSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERESNASGRSLV 276
FS +G C A +AELWG GL A G T +E+ +DS +VV + + + + LV
Sbjct: 125 FSLNIGICSAPLAELWGAYYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHPLSFLV 184
Query: 277 QKIRKLLQMEWKVKVKHSYREANRCADALAN 307
+ LL +W V++ H YREANR AD LAN
Sbjct: 185 RLCHGLLSKDWSVRISHVYREANRLADGLAN 215
>UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis thaliana]
Length = 947
Score = 115 bits (289), Expect = 1e-24
Identities = 85/260 (32%), Positives = 116/260 (43%), Gaps = 37/260 (14%)
Query: 16 KLWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIGNP-MCRFCHDEIESEIHVLRDCPKA 74
+LW + A ER+K F L+H IG+ +C+ C E+ +HVL+DCP
Sbjct: 695 RLWRVVALERVKTF---LWH-------------IGDTSVCQVCKGGDETILHVLKDCPSI 738
Query: 75 TALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTI----WNWR 130
+W +V FF G L W+ NL N E WAT + W WR
Sbjct: 739 AGIWRRLVQVQRSYDFFNGSLFGWLYVNLGMK-----NAETGYAWATLFAIVVWWSWKWR 793
Query: 131 NKETHNDNYQRPLHAKNIIMIYVNDYHNAIAKFVIVTSQPRHLEE-----VGWKEPAIRW 185
+ + K + D ++ + SQ L V WK P W
Sbjct: 794 CGYVFGEVGKCRDRVK-----FFRDLAAEVSHAHAIHSQNGGLRTRVERLVAWKPPDGEW 848
Query: 186 VKINTDGACKDG-SIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRM 244
VK+NTDGA + +A GG++R G W GF+ +G C A +AELWGV GL A
Sbjct: 849 VKLNTDGASRGNLGLATTGGVLRDGIGHWCGGFALDIGVCSAPLAELWGVYYGLYMAWER 908
Query: 245 GFTAVELNVDSLVVVNIITS 264
FT VEL VDS +VV +T+
Sbjct: 909 RFTRVELEVDSELVVGFLTT 928
>UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana]
Length = 233
Score = 109 bits (273), Expect = 9e-23
Identities = 70/191 (36%), Positives = 94/191 (48%), Gaps = 11/191 (5%)
Query: 123 CHTIWNWRNKETHNDNYQRPLHAKNIIMIYVNDYHNAIAKFVIV-----TSQPRHLEEVG 177
C W WR +N + K V D +AK S+ R +V
Sbjct: 10 CWWGWKWRCGNVFGENRKCRDRVK-----LVKDIAQEVAKANNCGSGSNNSRSRMERQVR 64
Query: 178 WKEPAIRWVKINTDGACKDG-SIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLE 236
W +P++ W K+NTDGA +A GG +R GEW GF+ +G+C A +AELWGV
Sbjct: 65 WSKPSLGWCKLNTDGASHGNPGLATAGGALRNEYGEWCFGFALNIGRCLAPLAELWGVYY 124
Query: 237 GLRCAKRMGFTAVELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYR 296
GL A G T +EL VDS +VV + + S+ LV+ L +W V++ H YR
Sbjct: 125 GLFMAWDRGITRLELEVDSEMVVGFLRTGIGSSHPLSFLVRMCHGFLSRDWIVRIGHVYR 184
Query: 297 EANRCADALAN 307
EANR AD LAN
Sbjct: 185 EANRLADGLAN 195
>UniRef100_Q9FXC8 F12A21.24 [Arabidopsis thaliana]
Length = 803
Score = 87.0 bits (214), Expect = 6e-16
Identities = 47/109 (43%), Positives = 60/109 (54%)
Query: 199 IAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVV 258
+A GG+IR G W GF+ +G+C A +AELWGV G A T VEL VDS +V
Sbjct: 657 LATAGGVIRDGAGNWCGGFALNIGRCSAPLAELWGVYYGFYLAWTKALTRVELEVDSELV 716
Query: 259 VNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALAN 307
V + + LV+ LL +W V++ H YREANR AD LAN
Sbjct: 717 VGFLKTGIGDQHPLSFLVRLCHGLLSKDWIVRITHVYREANRLADGLAN 765
>UniRef100_Q9ZVG3 T2P11.14 protein [Arabidopsis thaliana]
Length = 158
Score = 81.6 bits (200), Expect = 3e-14
Identities = 45/104 (43%), Positives = 58/104 (55%)
Query: 204 GLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIIT 263
G +R G+W F+ LG+C A +AELWGV GL A G T +EL VDS +V +T
Sbjct: 17 GALRDEYGDWRGSFALNLGRCTAPLAELWGVYYGLVIAWEKGITRLELEVDSKLVAGFLT 76
Query: 264 SERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALAN 307
+ E + LV+ +W V+V H YREANR AD LAN
Sbjct: 77 TGIEDSHLLSFLVRLCYGFSSKDWIVRVSHVYREANRFADELAN 120
>UniRef100_O23409 SIMILARITY to probable splicing factor CEPRP21 [Arabidopsis
thaliana]
Length = 559
Score = 79.7 bits (195), Expect = 1e-13
Identities = 58/158 (36%), Positives = 74/158 (46%), Gaps = 17/158 (10%)
Query: 151 IYVNDYHNAIAKFVIVTSQPRHLEEVGWKEPAIRWVKINTDGACKDG-SIAGCGGLIRGS 209
I V HN +A S R + W +P WVK++TDGA + A GG+IR
Sbjct: 3 IEVTKAHNLVAGGS--QSVARVERHIAWTKPPEGWVKVSTDGASRGNPGPAAAGGVIRDE 60
Query: 210 EGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERESN 269
+G W+ GF+ L AF+ W G VEL VDS +VV + S
Sbjct: 61 DGLWVGGFALQL----AFVRLRWQSCGG----------RVELEVDSELVVGFLQSGISEA 106
Query: 270 ASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALAN 307
LV+ LL +W V+V H YREANR AD LAN
Sbjct: 107 HPLAFLVRLCHGLLSKDWLVRVSHVYREANRLADGLAN 144
>UniRef100_Q7XCS1 Putative reverse transcriptase [Oryza sativa]
Length = 791
Score = 76.3 bits (186), Expect = 1e-12
Identities = 74/322 (22%), Positives = 137/322 (41%), Gaps = 33/322 (10%)
Query: 11 DEIWLKLWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIG-NPMCRFCHDEIESEIHVLR 69
++ W +W + P+++K F + L+T + K K + + MC+ C E E + H L
Sbjct: 448 NKAWEMIWKCKVPQKVKIFAWRVASNCLATMVNKKKRKLEQSDMCQICDRENEDDAHALC 507
Query: 70 DCPKATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACH--TIW 127
C +A+ LW C+ + G +S IK ++ + + +E + A T+W
Sbjct: 508 RCIQASQLWSCMHKS--------GSVSVDIKASVLGRFWLFDCLEKIPEYEQAMFLMTLW 559
Query: 128 -NW--RNKETHNDNYQRPLHAKNIIMIYVNDYHNA-------------IAKFVIVTSQPR 171
NW RN+ H + ++ I YV+ + + V + P+
Sbjct: 560 RNWYVRNELIHGKSAPPTETSQRFIQSYVDLLFQIRQAPQADLVKGKHVVRTVPLKGGPK 619
Query: 172 HL----EEVGWKEPAIRWVKINTDGACKDGS-IAGCGGLIRGSEGEWLAGFSKFLGKC-D 225
+ + W+ P W+K+N DG+ S G G ++R S G+ + K L +C +
Sbjct: 620 YRVLNNHQPCWERPKDGWMKLNVDGSFDASSGKGGLGMILRNSAGDIIFTSCKPLERCNN 679
Query: 226 AFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQM 285
+EL +EGL+ A +++ D VV ++ + +++ + R LL
Sbjct: 680 PLESELRACVEGLKLAIHWTLLPIQVETDCASVVQLLQGTGRDFSVLANIIHEARHLLVG 739
Query: 286 EWKVKVKHSYREANRCADALAN 307
E + +K R N + LAN
Sbjct: 740 ERVISIKKICRSQNCISHHLAN 761
>UniRef100_Q8LMJ9 Putative reverse transcriptase, 5'-partial [Oryza sativa]
Length = 507
Score = 76.3 bits (186), Expect = 1e-12
Identities = 74/322 (22%), Positives = 137/322 (41%), Gaps = 33/322 (10%)
Query: 11 DEIWLKLWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIG-NPMCRFCHDEIESEIHVLR 69
++ W +W + P+++K F + L+T + K K + + MC+ C E E + H L
Sbjct: 164 NKAWEMIWKCKVPQKVKIFAWRVASNCLATMVNKKKRKLEQSDMCQICDRENEDDAHALC 223
Query: 70 DCPKATALWLCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACH--TIW 127
C +A+ LW C+ + G +S IK ++ + + +E + A T+W
Sbjct: 224 RCIQASQLWSCMHKS--------GSVSVDIKASVLGRFWLFDCLEKIPEYEQAMFLMTLW 275
Query: 128 -NW--RNKETHNDNYQRPLHAKNIIMIYVNDYHNA-------------IAKFVIVTSQPR 171
NW RN+ H + ++ I YV+ + + V + P+
Sbjct: 276 RNWYVRNELIHGKSAPPTETSQRFIQSYVDLLFQIRQAPQADLVKGKHVVRTVPLKGGPK 335
Query: 172 HL----EEVGWKEPAIRWVKINTDGACKDGS-IAGCGGLIRGSEGEWLAGFSKFLGKC-D 225
+ + W+ P W+K+N DG+ S G G ++R S G+ + K L +C +
Sbjct: 336 YRVLNNHQPCWERPKDGWMKLNVDGSFDASSGKGGLGMILRNSAGDIIFTSCKPLERCNN 395
Query: 226 AFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQM 285
+EL +EGL+ A +++ D VV ++ + +++ + R LL
Sbjct: 396 PLESELRACVEGLKLAIHWTLLPIQVETDCASVVQLLQGTGRDFSVLANIIHEARHLLVG 455
Query: 286 EWKVKVKHSYREANRCADALAN 307
E + +K R N + LAN
Sbjct: 456 ERVISIKKICRSQNCISHHLAN 477
>UniRef100_Q9FWJ4 Hypothetical protein F21N10.9 [Arabidopsis thaliana]
Length = 256
Score = 75.5 bits (184), Expect = 2e-12
Identities = 44/115 (38%), Positives = 57/115 (49%)
Query: 207 RGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVDSLVVVNIITSER 266
R +G W GFS +G C A AELWGV GL A T VEL VDS +VV + +
Sbjct: 37 REHQGNWCGGFSVSIGICSAPQAELWGVYYGLYIAWEKWVTRVELEVDSEIVVGFLKTGI 96
Query: 267 ESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALANIGCIMGNEMMFYES 321
+ L + + +W V++ H YREANR AD LAN + F+ S
Sbjct: 97 GDSHPLSFLARLCYGFISKDWIVRISHVYREANRLADGLANYAFTLPLGFHFFHS 151
>UniRef100_Q6L3X8 Hypothetical protein [Solanum demissum]
Length = 1155
Score = 73.6 bits (179), Expect = 7e-12
Identities = 52/174 (29%), Positives = 90/174 (50%), Gaps = 3/174 (1%)
Query: 136 NDNYQRPLH-AKNIIMIYVNDYHNAIAKFVIVTSQPRHLEEVGWKEPAIRWVKINTDGAC 194
N+N L A++I MI V + I + V+ +++ V W +P + K+NTDG+C
Sbjct: 949 NNNIPTTLESAEHIFMIKVENSWENINQIFDVSINHKNVIRVNWIKPPTMFAKLNTDGSC 1008
Query: 195 KDGSIAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLEGLRCAKRMGFTAVELNVD 254
+G G GG++R + G+ + F+ LG+ + AE +L G++ + G + D
Sbjct: 1009 VNGRCGG-GGILRNALGQVIMAFTIKLGEGTSSWAEAMSMLHGMQLCIQRGVNMIIGETD 1067
Query: 255 SLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALANI 308
S+++ IT V+KI+K+++ E + H REAN+ AD LA+I
Sbjct: 1068 SILLAKAITENWSIPWRMYIPVKKIQKMVE-EHGFIINHCLREANQPADKLASI 1120
>UniRef100_Q6K204 Hypothetical protein B1469H02.35 [Oryza sativa]
Length = 212
Score = 72.4 bits (176), Expect = 2e-11
Identities = 44/135 (32%), Positives = 72/135 (52%), Gaps = 4/135 (2%)
Query: 178 WKEPAIRWVKINTDGACKDGS-IAGCGGLIRGSEGEWLAGFSKFLGKCDAFIAELWGVLE 236
W++P W+K+N DG+ K + IA GG+ R EG ++ G+++ +G+ + +AEL +
Sbjct: 51 WRKPEAGWIKLNFDGSSKHATKIASIGGVYRDHEGAFVLGYAERIGRATSSVAELAALRR 110
Query: 237 GLRCAKRMGFTAVELNVDSLVVVNIITSERESNASGRSLVQ--KIRKLLQMEWKVKVKHS 294
GL R G+ V DS VV+++ +R + S L Q +I LL + + V H
Sbjct: 111 GLELVVRNGWRRVWAEGDSKTVVDVV-CDRANVRSEEDLRQCREIAALLPLIDDMAVSHV 169
Query: 295 YREANRCADALANIG 309
YR N+ A A +G
Sbjct: 170 YRSGNKVAHGFARLG 184
>UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1524
Score = 71.6 bits (174), Expect = 3e-11
Identities = 74/304 (24%), Positives = 124/304 (40%), Gaps = 14/304 (4%)
Query: 16 KLWNIRAPERIKHFMSMLYHGRLSTNLRKHKMGIG-NPMCRFCHDEIESEIHVLRDCPKA 74
++WN+ ++KHF+ L+T R G+ +P+C CH E ES H L CP A
Sbjct: 1201 RIWNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPICPRCHRENESINHALFTCPFA 1260
Query: 75 TALW----LCVVDNAARTSFFEGDLSSWIKFNLFTNVYWNNNVEWRDFWATACHTIWNWR 130
T W ++ N ++ FE ++S+ + F T + ++ + W IW R
Sbjct: 1261 TMAWWLSDSSLIRNQLMSNDFEENISNILNFVQDTTM--SDFHKLLPVW--LIWRIWKAR 1316
Query: 131 NKETHNDNYQRPLHAKNIIMIYVNDYHNAIAKFVIVTSQPRHLEE--VGWKEPAIRWVKI 188
N N + P +D+ NA S R + E + W+ P +VK
Sbjct: 1317 NNVVFNKFRESPSKTVLSAKAETHDWLNATQSHKKTPSPTRQIAENKIEWRNPPATYVKC 1376
Query: 189 NTDGACKDGSIAGCGG-LIRGSEGEWLA-GFSKFLGKCDAFIAELWGVLEGLRCAKRMGF 246
N D + GG +IR G ++ G K + AE +L L+ G+
Sbjct: 1377 NFDAGFDVQKLEATGGWIIRNHYGTPISWGSMKLAHTSNPLEAETKALLAALQQTWIRGY 1436
Query: 247 TAVELNVDSLVVVNIITSERESNASGRSLVQKIRKLLQMEWKVKVKHSYREANRCADALA 306
T V + D ++N+I ++S + ++ I ++ R+ N+ A LA
Sbjct: 1437 TQVFMEGDCQTLINLING-ISFHSSLANHLEDISFWANKFASIQFGFIRRKGNKLAHVLA 1495
Query: 307 NIGC 310
GC
Sbjct: 1496 KYGC 1499
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.137 0.450
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 573,136,764
Number of Sequences: 2790947
Number of extensions: 23515417
Number of successful extensions: 47204
Number of sequences better than 10.0: 473
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 427
Number of HSP's that attempted gapping in prelim test: 46766
Number of HSP's gapped (non-prelim): 521
length of query: 321
length of database: 848,049,833
effective HSP length: 127
effective length of query: 194
effective length of database: 493,599,564
effective search space: 95758315416
effective search space used: 95758315416
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 75 (33.5 bits)
Medicago: description of AC142394.3


