

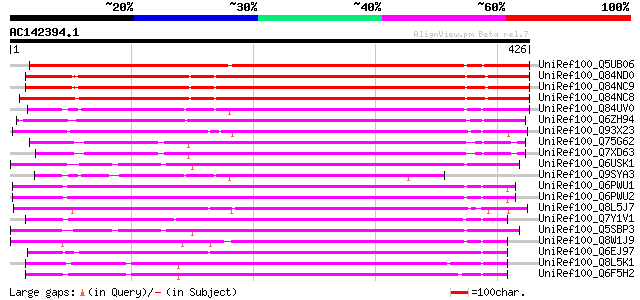
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.1 - phase: 0 /pseudo
(426 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5UB06 Wound-inducible putative chloroplast terpene sy... 390 e-107
UniRef100_Q84ND0 Myrcene synthase Oc15 [Antirrhinum majus] 309 1e-82
UniRef100_Q84NC9 Myrcene synthase 1e20 [Antirrhinum majus] 309 1e-82
UniRef100_Q84NC8 (E)-b-ocimene synthase [Antirrhinum majus] 306 9e-82
UniRef100_Q84UV0 Linalool synthase [Arabidopsis thaliana] 304 4e-81
UniRef100_Q6ZH94 Putative linalool synthase [Oryza sativa] 256 9e-67
UniRef100_Q93X23 Putative chloroplast terpene synthase precursor... 247 4e-64
UniRef100_Q75G62 Putative terpene synthase [Oryza sativa] 237 5e-61
UniRef100_Q7XD63 Putative monoterpene synthase [Oryza sativa] 236 9e-61
UniRef100_Q6USK1 Geraniol synthase [Ocimum basilicum] 231 4e-59
UniRef100_Q9SYA3 T13M11.3 protein [Arabidopsis thaliana] 230 7e-59
UniRef100_Q6PWU1 Terpenoid synthase [Vitis vinifera] 229 1e-58
UniRef100_Q6PWU2 Terpenoid synthase [Vitis vinifera] 228 2e-58
UniRef100_Q8L5J7 3-carene synthase [Salvia stenophylla] 220 7e-56
UniRef100_Q7Y1V1 Putative monoterpene synthase [Melaleuca altern... 219 1e-55
UniRef100_Q5SBP3 R-linalool synthase [Ocimum basilicum] 219 1e-55
UniRef100_Q8W1J9 Linalool synthase [Perilla frutescens var. crispa] 217 4e-55
UniRef100_Q6EJ97 Isoprene synthase [Pueraria lobata] 216 8e-55
UniRef100_Q8L5K1 (+)-limonene synthase 2 [Citrus limon] 213 6e-54
UniRef100_Q6F5H2 D-limonene synthase [Citrus unshiu] 213 6e-54
>UniRef100_Q5UB06 Wound-inducible putative chloroplast terpene synthase 3 [Medicago
truncatula]
Length = 573
Score = 390 bits (1001), Expect = e-107
Identities = 200/410 (48%), Positives = 271/410 (65%), Gaps = 6/410 (1%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E ++D IQRLG+E+ F EEI+ LE++ +L L + A FR+LRQ G
Sbjct: 76 ECLSMVDSIQRLGMEYNFEEEIEATLERKHTMLRFQNFQRNEYQGLSQAAFQFRMLRQEG 135
Query: 77 YYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQAW 136
YY++ ++FD +K L+ + ED+ G+IAL+EASQLSIEGED L+++G + L W
Sbjct: 136 YYISPDIFDKFCDNKGKLKYTFSEDINGMIALFEASQLSIEGEDCLDNVGQFCGQYLNDW 195
Query: 137 LPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSIVKF 196
H QA +V++TL YP H LSRF + + N + + +K+++ +V
Sbjct: 196 SSTFHGHPQAKFVAHTLMYPTHKTLSRFTPTIMQSQNATWTNS---IQQFSKIDTQMVSS 252
Query: 197 MNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPISLVY 256
+ E V KWW+DLGL K+++FA P+KWY W MAC DP FS ER+ELTKP+SL+Y
Sbjct: 253 SHLKEIFAVSKWWKDLGLPKDLEFARDEPIKWYSWSMACLPDPQFSEERIELTKPLSLIY 312
Query: 257 IIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKKHG 316
IIDDIFD +G +D+LTLFT+AV RW++ E LP+ MKV +LY +TN FA Y KHG
Sbjct: 313 IIDDIFDFYGNIDELTLFTDAVKRWDLSAIEQLPDCMKVCFKALYDITNEFALRTYIKHG 372
Query: 317 FNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHA 376
+NP+ +L SWVRLLNAFL+EA W SG +P +EEYL N IVSTGVHV+L+HAFF M
Sbjct: 373 WNPLTSLIKSWVRLLNAFLQEAKWFASGNVPKSEEYLKNAIVSTGVHVILVHAFFCM--G 430
Query: 377 KGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
+GIT++T+S++D +FP II + AKILRL DDLEG K + G DGSY C
Sbjct: 431 QGITEKTVSLMD-DFPTIISTTAKILRLCDDLEGDKDVNCEGNDGSYSKC 479
>UniRef100_Q84ND0 Myrcene synthase Oc15 [Antirrhinum majus]
Length = 581
Score = 309 bits (791), Expect = 1e-82
Identities = 171/415 (41%), Positives = 255/415 (61%), Gaps = 9/415 (2%)
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLR 73
DP E+ +D RLG+ H+F +EI+ L K + + S PI H L+EV+L FRL+R
Sbjct: 95 DPIESLIFVDATLRLGVNHHFQKEIEEILRKSYATMKS-PI-ICEYHTLHEVSLFFRLMR 152
Query: 74 QGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELL 133
Q G YV+A++F+ K + + D +GL+ LYEA+QLS EGE L++ S ++L
Sbjct: 153 QHGRYVSADVFNNFKGESGRFKEELKRDTRGLVELYEAAQLSFEGERILDEAENFSRQIL 212
Query: 134 QAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKW-ICLDELAKMNSS 192
L +D+++ V N L+YP H ++RF ++ + +DL +W L ELA M+
Sbjct: 213 HGNLAGMEDNLRRS-VGNKLRYPFHTSIARFTGRN-YDDDLGGMYEWGKTLRELALMDLQ 270
Query: 193 IVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPC-FSNERVELTKP 251
+ + + + E ++V KWW +LGL K++ A P ++Y W M D S +RVELTK
Sbjct: 271 VERSVYQEELLQVSKWWNELGLYKKLNLARNRPFEFYTWSMVILADYINLSEQRVELTKS 330
Query: 252 ISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMV 311
++ +Y+IDDIFDV+GTLD+L +FTEAVN+W+ + LP MK+ +L N ++ +
Sbjct: 331 VAFIYLIDDIFDVYGTLDELIIFTEAVNKWDYSATDTLPENMKMCCMTLLDTINGTSQKI 390
Query: 312 YKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFF 371
Y+KHG+NPID+LK +W L +AFL EA W SG LP+ EYL N VS+GV+VVL+H F
Sbjct: 391 YEKHGYNPIDSLKTTWKSLCSAFLVEAKWSASGSLPSANEYLENEKVSSGVYVVLVHLFC 450
Query: 372 LMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
LM G ++ ++ + D + ++ S+A I RL +DL K+ QNG DGSYLNC
Sbjct: 451 LMG-LGGTSRGSIELNDTQ--ELMSSIAIIFRLWNDLGSAKNEHQNGKDGSYLNC 502
>UniRef100_Q84NC9 Myrcene synthase 1e20 [Antirrhinum majus]
Length = 584
Score = 309 bits (791), Expect = 1e-82
Identities = 171/415 (41%), Positives = 255/415 (61%), Gaps = 9/415 (2%)
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLR 73
DP E+ +D RLG+ H+F +EI+ L K + + S PI H L+EV+L FRL+R
Sbjct: 98 DPIESLIFVDATLRLGVNHHFQKEIEEILRKSYATMKS-PI-ICEYHTLHEVSLFFRLMR 155
Query: 74 QGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELL 133
Q G YV+A++F+ K + + D +GL+ LYEA+QLS EGE L++ S ++L
Sbjct: 156 QHGRYVSADVFNNFKGESGRFKEELKRDTRGLVELYEAAQLSFEGERILDEAENFSRQIL 215
Query: 134 QAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKW-ICLDELAKMNSS 192
L +D+++ V N L+YP H ++RF ++ + +DL +W L ELA M+
Sbjct: 216 HGNLAGMEDNLRRS-VGNKLRYPFHTSIARFTGRN-YDDDLGGMYEWGKTLRELALMDLQ 273
Query: 193 IVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPC-FSNERVELTKP 251
+ + + + E ++V KWW +LGL K++ A P ++Y W M D S +RVELTK
Sbjct: 274 VERSVYQEELLQVSKWWNELGLYKKLNLARNRPFEFYTWSMVILADYINLSEQRVELTKS 333
Query: 252 ISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMV 311
++ +Y+IDDIFDV+GTLD+L +FTEAVN+W+ + LP MK+ +L N ++ +
Sbjct: 334 VAFIYLIDDIFDVYGTLDELIIFTEAVNKWDYSATDTLPENMKMCCMTLLDTINGTSQKI 393
Query: 312 YKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFF 371
Y+KHG+NPID+LK +W L +AFL EA W SG LP+ EYL N VS+GV+VVL+H F
Sbjct: 394 YEKHGYNPIDSLKTTWKSLCSAFLVEAKWSASGSLPSANEYLENEKVSSGVYVVLVHLFC 453
Query: 372 LMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
LM G ++ ++ + D + ++ S+A I RL +DL K+ QNG DGSYLNC
Sbjct: 454 LMG-LGGTSRGSIELNDTQ--ELMSSIAIIFRLWNDLGSAKNEHQNGKDGSYLNC 505
>UniRef100_Q84NC8 (E)-b-ocimene synthase [Antirrhinum majus]
Length = 579
Score = 306 bits (783), Expect = 9e-82
Identities = 171/420 (40%), Positives = 256/420 (60%), Gaps = 9/420 (2%)
Query: 9 LVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALA 68
L DP E+ +D QRLG+ H+F +EI+ L K + + S I H L++V+L
Sbjct: 88 LKDENDPIESLIFVDATQRLGVNHHFQKEIEEILRKSYATMKSPSI--CKYHTLHDVSLF 145
Query: 69 FRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYL 128
F L+RQ G YV+A++F+ K + + D +GL+ LYEA+QLS EGE L++
Sbjct: 146 FCLMRQHGRYVSADVFNNFKGESGRFKEELKRDTRGLVELYEAAQLSFEGERILDEAENF 205
Query: 129 SCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKW-ICLDELA 187
S ++L L +D+++ V N L+YP H ++RF + + +DL +W L ELA
Sbjct: 206 SRQILHGNLASMEDNLRRS-VGNKLRYPFHKSIARFTGIN-YDDDLGGMYEWGKTLRELA 263
Query: 188 KMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPC-FSNERV 246
M+ + + + + E ++V KWW +LGL K++ A P ++YMW M TD S +RV
Sbjct: 264 LMDLQVERSVYQEELLQVSKWWNELGLYKKLTLARNRPFEFYMWSMVILTDYINLSEQRV 323
Query: 247 ELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNN 306
ELTK ++ +Y+IDDIFDV+GTLD+L +FTEAVN+W+ + LP+ MK+ +L N
Sbjct: 324 ELTKSVAFIYLIDDIFDVYGTLDELIIFTEAVNKWDYSATDTLPDNMKMCYMTLLDTING 383
Query: 307 FAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVL 366
++ +Y+K+G NPID+LK +W L +AFL EA W SG LP+ EYL N VS+GV+VVL
Sbjct: 384 TSQKIYEKYGHNPIDSLKTTWKSLCSAFLVEAKWSASGSLPSANEYLENEKVSSGVYVVL 443
Query: 367 IHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
IH FFLM G + ++ + D ++ S+A I+R+ +DL K+ QNG DGSYL+C
Sbjct: 444 IHLFFLMG-LGGTNRGSIELNDTR--ELMSSIAIIVRIWNDLGCAKNEHQNGKDGSYLDC 500
>UniRef100_Q84UV0 Linalool synthase [Arabidopsis thaliana]
Length = 569
Score = 304 bits (778), Expect = 4e-81
Identities = 167/417 (40%), Positives = 248/417 (59%), Gaps = 16/417 (3%)
Query: 15 PTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQ 74
P+EN +ID+IQ LG + +F + I+ L ++ + F + +L+E+AL FRLLRQ
Sbjct: 90 PSENLEMIDVIQSLGTDLHFRQGIEQTLH----MIYKEGLQF--NGDLHEIALRFRLLRQ 143
Query: 75 GGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQ 134
G+YV +F + K + DVKGL L+EAS+L +EGE+ L+ + L
Sbjct: 144 EGHYVQESIFKNILDKKGGFKDVVKNDVKGLTELFEASELRVEGEETLDGAREFTYSRLN 203
Query: 135 AWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNK----WI-CLDELAKM 189
+ H Q + +L P H + K F + +K + W+ L +A++
Sbjct: 204 ELCSGRESH-QKQEIMKSLAQPRHKTVRGLTSKR-FTSMIKIAGQEDPEWLQSLLRVAEI 261
Query: 190 NSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELT 249
+S +K + + E + FKWW +LGL K+++ A PLKW+ W M DP + +R++LT
Sbjct: 262 DSIRLKSLTQGEMSQTFKWWTELGLEKDVEKARSQPLKWHTWSMKILQDPTLTEQRLDLT 321
Query: 250 KPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAE 309
KPISLVY+IDDIFDV+G L++LT+FT V RW+ G + LP +M+V +L +T +
Sbjct: 322 KPISLVYVIDDIFDVYGELEELTIFTRVVERWDHKGLKTLPKYMRVCFEALDMITTEISM 381
Query: 310 MVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHA 369
+YK HG+NP L+ SW L AFL EA W NSG LPTTEEY+ NG+VS+GVH+V++HA
Sbjct: 382 KIYKSHGWNPTYALRQSWASLCKAFLVEAKWFNSGYLPTTEEYMKNGVVSSGVHLVMLHA 441
Query: 370 FFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLNC 426
+ L+ + +TKE + ++ E P I+ S A ILRL DDL K +Q+G DGSY+ C
Sbjct: 442 YILL--GEELTKEKVELI-ESNPGIVSSAATILRLWDDLGSAKDENQDGTDGSYVEC 495
>UniRef100_Q6ZH94 Putative linalool synthase [Oryza sativa]
Length = 595
Score = 256 bits (654), Expect = 9e-67
Identities = 143/419 (34%), Positives = 239/419 (56%), Gaps = 13/419 (3%)
Query: 6 HKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEV 65
H+K T+ E +D ++RL ++HYF +E+ A++ L + +L +
Sbjct: 101 HQK--TASGGRERIATVDHLRRLCMDHYFQDEVDDAMDACLLE------ELAHGGDLLDA 152
Query: 66 ALAFRLLRQGGYYVNA-ELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLND 124
LAFRL+R+ G++V+A E+ R+ Y +D++GL++L + S ++I E L
Sbjct: 153 TLAFRLMREAGHHVSADEVLGRFTDDNGEFRLDYRKDIRGLLSLQDISHMNIGQEASLCK 212
Query: 125 LGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLD 184
S L++ + + ++ A YV +L +P H L+++ + + ++
Sbjct: 213 AKEFSTRNLESAINYLEPNL-ARYVRQSLDHPYHVSLNQYKARHHLSYLQTLPIRCTAME 271
Query: 185 ELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNE 244
ELA + + K +++ E E+ +WW DLGLA+E+ A KW++W M S
Sbjct: 272 ELALADFQLNKLLHQMEMQEIKRWWMDLGLAQEIPVARDQVQKWFVWMMTAIQGASLSRC 331
Query: 245 RVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVT 304
R+ELTK +S VYI+DDIFD+ GT ++L+ FT+A+ W++ A++LP+ M+ +L+ VT
Sbjct: 332 RIELTKIVSFVYIVDDIFDLVGTREELSCFTQAIRMWDLAAADSLPSCMRSCFRALHTVT 391
Query: 305 NNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHV 364
N+ A+MV ++HG NPI+ LK +W L + F+ E WL++G +P +EEYL NG+V++GV +
Sbjct: 392 NDIADMVEREHGVNPINHLKKAWAMLFDGFMTETKWLSAGQVPDSEEYLRNGVVTSGVPL 451
Query: 365 VLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSY 423
V +H F++ H +++ +D P +I AKILRL DDL K Q GLDGSY
Sbjct: 452 VFVHLLFMLGH--DVSQNAAEFVD-HIPPVISCPAKILRLWDDLGSAKDEAQEGLDGSY 507
>UniRef100_Q93X23 Putative chloroplast terpene synthase precursor [Quercus ilex]
Length = 597
Score = 247 bits (631), Expect = 4e-64
Identities = 150/429 (34%), Positives = 246/429 (56%), Gaps = 13/429 (3%)
Query: 3 KQVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
+QV L +P E LI+I+QRLG+ ++F EEIK L+ + + + D + L
Sbjct: 93 EQVRMMLHKVVNPLEQLELIEILQRLGLSYHFEEEIKRILDGVYN--NDHGGDTWKAENL 150
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL 122
Y AL FRLLRQ GY V+ E+F+ K + S + ED KG+++LYEAS IEGE+ L
Sbjct: 151 YATALKFRLLRQHGYSVSQEVFNSFKDERGSFKACLCEDTKGMLSLYEASFFLIEGENIL 210
Query: 123 NDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWI- 181
+ S + L+ ++ ++++ A V+++L++P+H+ + R ++ FIN + N+ +
Sbjct: 211 EEARDFSTKHLEEYVKQNKEKNLATLVNHSLEFPLHWRMPR-LEARWFIN-IYRHNQDVN 268
Query: 182 -CLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPC 240
L E A+++ +IV+ ++ + +V WW+ GL + + FA P++ + W + P
Sbjct: 269 PILLEFAELDFNIVQAAHQADLKQVSTWWKSTGLVENLSFARDRPVENFFWTVGLIFQPQ 328
Query: 241 FSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSL 300
F R TK +L+ IDD++DV+GTLD+L LFT+ V RW+++ + LP++MK+ +L
Sbjct: 329 FGYCRRMFTKVFALITTIDDVYDVYGTLDELELFTDVVERWDINAMDQLPDYMKICFLTL 388
Query: 301 YKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVST 360
+ N A K+ F+ I LK +WV L +L EA W ++ P+ +EY+ N +S
Sbjct: 389 HNSVNEMALDTMKEQRFHIIKYLKKAWVDLCRYYLVEAKWYSNKYRPSLQEYIENAWISI 448
Query: 361 GVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL----EGVKSGDQ 416
G +L+HA+F + + ITKE L L EE+PNII + I RL+DDL + +K GD
Sbjct: 449 GAPTILVHAYFFVTNP--ITKEALDCL-EEYPNIIRWSSIIARLADDLGTSTDELKRGDV 505
Query: 417 NGLDGSYLN 425
Y+N
Sbjct: 506 PKAIQCYMN 514
>UniRef100_Q75G62 Putative terpene synthase [Oryza sativa]
Length = 505
Score = 237 bits (604), Expect = 5e-61
Identities = 135/411 (32%), Positives = 225/411 (53%), Gaps = 26/411 (6%)
Query: 17 ENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGG 76
E +D ++RL I+HYF +E+ A++ L+ +L + LAFRL+R+ G
Sbjct: 36 ERMATVDHLKRLCIDHYFQDEVDGAMDAHLEELAHGG-------DLLDATLAFRLMREAG 88
Query: 77 YYVNA-ELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQA 135
++V+ E+ + Y +D++GL++L + S ++I E L Y + E
Sbjct: 89 HHVSPDEVLGRFTDGNGDFSMAYSKDIRGLLSLQDISHMNIGAEASL----YKAKEFTSR 144
Query: 136 WLPRHQDHIQ---AIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSS 192
L D+++ A YV +L++P H L ++ + + ++ELA N
Sbjct: 145 NLQSAIDYLEPGLARYVRQSLEHPYHVSLMQYKARHHLSYLQTLPTRCTAMEELALANFK 204
Query: 193 IVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPI 252
+ K +++ E E+ +WW +LGLA+E+ A KWY+W M F FS R+ELTK
Sbjct: 205 LNKLLHQMEMQEIKRWWMNLGLAQEIPVARDQVQKWYVWIMTAFQGASFSRYRIELTKIA 264
Query: 253 SLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVY 312
S VYI+DDIFD+ T ++ + FT+A+ W+ A++LP+ M+ ++Y VTN+ A+MV
Sbjct: 265 SFVYIMDDIFDLVSTQEERSCFTQAIKMWDFAAADSLPSCMRSCYRAIYTVTNDIADMVE 324
Query: 313 KKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFL 372
++HG NPI+ LK +W L + + EA WL +P++E+YL NG++++GV ++ +H F+
Sbjct: 325 REHGVNPINHLKKAWAVLFDGLMTEAKWLTDSHVPSSEDYLRNGVITSGVPLMFLHLLFM 384
Query: 373 MDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSY 423
+ H + ++D P +I AKI RL DD+ K GLDGSY
Sbjct: 385 LGH------DAAELID-NIPPVISCPAKIFRLWDDIGNAK----EGLDGSY 424
>UniRef100_Q7XD63 Putative monoterpene synthase [Oryza sativa]
Length = 468
Score = 236 bits (602), Expect = 9e-61
Identities = 134/406 (33%), Positives = 224/406 (55%), Gaps = 26/406 (6%)
Query: 22 IDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGGYYVNA 81
+D ++RL I+HYF +E+ A++ L+ +L + LAFRL+R+ G++V+
Sbjct: 4 VDHLKRLCIDHYFQDEVDGAMDAHLEELAHGG-------DLLDATLAFRLMREAGHHVSP 56
Query: 82 -ELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQAWLPRH 140
E+ + Y +D++GL++L + S ++I E L Y + E L
Sbjct: 57 DEVLGRFTDGNGDFSMAYSKDIRGLLSLQDISHMNIGAEASL----YKAKEFTSRNLQSA 112
Query: 141 QDHIQ---AIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSIVKFM 197
D+++ A YV +L++P H L ++ + + ++ELA N + K +
Sbjct: 113 IDYLEPGLARYVRQSLEHPYHVSLMQYKARHHLSYLQTLPTRCTAMEELALANFKLNKLL 172
Query: 198 NKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPISLVYI 257
++ E E+ +WW +LGLA+E+ A KWY+W M F FS R+ELTK S VYI
Sbjct: 173 HQMEMQEIKRWWMNLGLAQEIPVARDQVQKWYVWIMTAFQGASFSRYRIELTKIASFVYI 232
Query: 258 IDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKKHGF 317
+DDIFD+ T ++ + FT+A+ W+ A++LP+ M+ ++Y VTN+ A+MV ++HG
Sbjct: 233 MDDIFDLVSTQEERSCFTQAIKMWDFAAADSLPSCMRSCYRAIYTVTNDIADMVEREHGV 292
Query: 318 NPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHAK 377
NPI+ LK +W L + + EA WL +P++E+YL NG++++GV ++ +H F++ H
Sbjct: 293 NPINHLKKAWAVLFDGLMTEAKWLTDSHVPSSEDYLRNGVITSGVPLMFLHLLFMLGH-- 350
Query: 378 GITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSY 423
+ ++D P +I AKI RL DD+ K GLDGSY
Sbjct: 351 ----DAAELID-NIPPVISCPAKIFRLWDDIGNAK----EGLDGSY 387
>UniRef100_Q6USK1 Geraniol synthase [Ocimum basilicum]
Length = 567
Score = 231 bits (588), Expect = 4e-59
Identities = 136/424 (32%), Positives = 223/424 (52%), Gaps = 25/424 (5%)
Query: 1 MCKQVHKKLVTSE-DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSS 59
+ ++ +KL ++ + E LID IQ+LGI +YF + I L F
Sbjct: 77 LLEETTRKLQRNDTESVEKLKLIDNIQQLGIGYYFEDAINAVLRSPFS---------TGE 127
Query: 60 HELYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGE-DVKGLIALYEASQLSIEG 118
+L+ AL FRLLR G ++ E+F K R K+ E D GL++LYEAS L + G
Sbjct: 128 EDLFTAALRFRLLRHNGIEISPEIF----LKFKDERGKFDESDTLGLLSLYEASNLGVAG 183
Query: 119 EDGLNDLGYLSCELLQAWLPRHQDHIQAIY---VSNTLQYPIHYGLSRFMDKSIFINDLK 175
E+ L + + E +A L R A V+ L P H ++R + K
Sbjct: 184 EEILEE----AMEFAEARLRRSLSEPAAPLHGEVAQALDVPRHLRMARLEARRFIEQYGK 239
Query: 176 AKNKWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMAC 235
+ L ELA ++ + V+ +++E E+ +WW++LGL ++ F PL+ ++W +
Sbjct: 240 QSDHDGDLLELAILDYNQVQAQHQSELTEIIRWWKELGLVDKLSFGRDRPLECFLWTVGL 299
Query: 236 FTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKV 295
+P +S+ R+EL K IS++ +IDDIFD +G +D L LFT+A+ RW+++ E LP +MK+
Sbjct: 300 LPEPKYSSVRIELAKAISILLVIDDIFDTYGEMDDLILFTDAIRRWDLEAMEGLPEYMKI 359
Query: 296 SLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNN 355
+LY TN V + G + LK +W+ ++ F++EA W N G P EEY+ N
Sbjct: 360 CYMALYNTTNEVCYKVLRDTGRIVLLNLKSTWIDMIEGFMEEAKWFNGGSAPKLEEYIEN 419
Query: 356 GIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEE-FPNIIYSVAKILRLSDDLEGVKSG 414
G+ + G ++ H FFL+ +G+T + + ++ +P + + +ILRL DDL K
Sbjct: 420 GVSTAGAYMAFAHIFFLI--GEGVTHQNSQLFTQKPYPKVFSAAGRILRLWDDLGTAKEE 477
Query: 415 DQNG 418
+ G
Sbjct: 478 QERG 481
>UniRef100_Q9SYA3 T13M11.3 protein [Arabidopsis thaliana]
Length = 421
Score = 230 bits (586), Expect = 7e-59
Identities = 133/348 (38%), Positives = 196/348 (56%), Gaps = 26/348 (7%)
Query: 21 LIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQGGYYVN 80
+ID+IQ LGI+ +F +EI+ L ++ + F + +L+E+AL FRLLRQ G+YV
Sbjct: 1 MIDVIQSLGIDLHFRQEIEQTLH----MIYKEGLQF--NGDLHEIALRFRLLRQEGHYVQ 54
Query: 81 AELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQAWLPRH 140
K + DVKGL L+EAS+L +EGE+ L+ + L
Sbjct: 55 EN-------KKGGFKDVVKNDVKGLTELFEASELRVEGEETLDGAREFTYSRLNELCSGR 107
Query: 141 QDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNK----WI-CLDELAKMNSSIVK 195
+ H Q + +L P H + K F + +K + W+ L +A+++S +K
Sbjct: 108 ESH-QKQEIMKSLAQPRHKTVRGLTSKR-FTSMIKIAGQEDPEWLQSLLRVAEIDSIRLK 165
Query: 196 FMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPISLV 255
+ + E + FKWW +LGL K+++ A PLKW+ W M DP + +R++LTKPISLV
Sbjct: 166 SLTQGEMSQTFKWWTELGLEKDVEKARSQPLKWHTWSMKILQDPTLTEQRLDLTKPISLV 225
Query: 256 YIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYKKH 315
Y+IDDIFDV+G L++LT+FT V RW+ G + LP +M+V +L +T + +YK H
Sbjct: 226 YVIDDIFDVYGELEELTIFTRVVERWDHKGLKTLPKYMRVCFEALDMITTEISMKIYKSH 285
Query: 316 GFNPIDTLKIS------WVRLLNAFLKEAHWLNSGILPTTEEYLNNGI 357
G+NP L+ S W L AFL EA W NSG LPTTEEY+ NG+
Sbjct: 286 GWNPTYALRQSVIIIQNWASLCKAFLVEAKWFNSGYLPTTEEYMKNGV 333
>UniRef100_Q6PWU1 Terpenoid synthase [Vitis vinifera]
Length = 590
Score = 229 bits (584), Expect = 1e-58
Identities = 141/417 (33%), Positives = 227/417 (53%), Gaps = 9/417 (2%)
Query: 3 KQVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
+ V L + P + LID++QRLGI ++F +EIK L + N + +L
Sbjct: 85 RDVKPMLGKVKKPLDQLELIDVLQRLGIYYHFKDEIKRILNS--IYNQYNRHEEWQKDDL 142
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL 122
Y AL FRLLRQ GY V ++F K S + ED+KG++ LYEAS L ++GE +
Sbjct: 143 YATALEFRLLRQHGYDVPQDVFSRFKDDTGSFKACLCEDMKGMLCLYEASYLCVQGESTM 202
Query: 123 NDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWIC 182
+ L L ++ D AI V + L+ P+H+ + R + K ++
Sbjct: 203 EQARDFAHRHLGKGLEQNIDQNLAIEVKHALELPLHWRMPRLEARWFIDVYEKRQDMNPI 262
Query: 183 LDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFS 242
L E AK++ ++V+ ++ + + WW L +++ FA ++ ++W + +P +
Sbjct: 263 LLEFAKLDFNMVQATHQEDLRHMSSWWSSTRLGEKLNFARDRLMENFLWTVGVIFEPQYG 322
Query: 243 NERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYK 302
R TK +L+ IIDD++DV+GT+D+L LFT+ V+RW+++ + LP +MK+ +LY
Sbjct: 323 YCRRMSTKVNTLITIIDDVYDVYGTMDELELFTDVVDRWDINAMDPLPEYMKLCFLALYN 382
Query: 303 VTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGV 362
TN A K+HG + + L+ +W L ++L EA W S P+ +EY++N +S
Sbjct: 383 STNEMAYDALKEHGLHIVSYLRKAWSDLCKSYLLEAKWYYSRYTPSLQEYISNSWISISG 442
Query: 363 HVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDD----LEGVKSGD 415
V+L+HA+FL+ A ITKE L L E + NII + ILRLSDD L+ +K GD
Sbjct: 443 PVILVHAYFLV--ANPITKEALQSL-ERYHNIIRWSSMILRLSDDLGTSLDELKRGD 496
>UniRef100_Q6PWU2 Terpenoid synthase [Vitis vinifera]
Length = 590
Score = 228 bits (582), Expect = 2e-58
Identities = 141/417 (33%), Positives = 227/417 (53%), Gaps = 9/417 (2%)
Query: 3 KQVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
+ V L + P + LID++QRLGI ++F +EIK L + N + +L
Sbjct: 85 RDVKPMLGKVKKPLDQLELIDVLQRLGIYYHFKDEIKRILNG--IYNQYNRHEEWQKDDL 142
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL 122
Y AL FRLLRQ GY V ++F K S + ED+KG++ LYEAS L ++GE +
Sbjct: 143 YATALEFRLLRQHGYDVPQDVFSRFKDDTGSFKACLCEDMKGMLCLYEASYLCVQGESTM 202
Query: 123 NDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWIC 182
+ L L ++ D AI V + L+ P+H+ + R + K ++
Sbjct: 203 EQARDFAHRHLGKGLEQNIDQNLAIEVKHALELPLHWRMPRLEARWFIDVYEKRQDMNPI 262
Query: 183 LDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFS 242
L E AK++ ++V+ ++ + + WW L +++ FA ++ ++W + +P +
Sbjct: 263 LLEFAKLDFNMVQATHQEDLRHMSSWWSSTRLGEKLNFARDRLMENFLWTVGVIFEPQYG 322
Query: 243 NERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYK 302
R TK +L+ IIDD++DV+GT+D+L LFT+ V+RW+++ + LP +MK+ +LY
Sbjct: 323 YCRRMSTKVNTLITIIDDVYDVYGTMDELELFTDVVDRWDINAMDPLPEYMKLCFLALYN 382
Query: 303 VTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGV 362
TN A K+HG + + L+ +W L ++L EA W S P+ +EY++N +S
Sbjct: 383 STNEMAYDALKEHGLHIVSYLRKAWSDLCKSYLLEAKWYYSRYTPSLQEYISNSWISISG 442
Query: 363 HVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDD----LEGVKSGD 415
V+L+HA+FL+ A ITKE L L E + NII + ILRLSDD L+ +K GD
Sbjct: 443 PVILVHAYFLV--ANPITKEALQSL-ERYHNIIRWSSMILRLSDDLGTSLDELKRGD 496
>UniRef100_Q8L5J7 3-carene synthase [Salvia stenophylla]
Length = 597
Score = 220 bits (560), Expect = 7e-56
Identities = 148/434 (34%), Positives = 235/434 (54%), Gaps = 19/434 (4%)
Query: 4 QVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILS--SNPIDFVSSHE 61
+V K L +PT LID +Q+LG+ +F E K L +L N +
Sbjct: 88 EVKKLLEKEPNPTRQLELIDDLQKLGLSDHFNNEFKEILNSVYLDNKYYRNGAMKEVERD 147
Query: 62 LYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDG 121
LY ALAFRLLRQ G+ V ++ +C K +K +D +GL+ LYEAS L EGE+
Sbjct: 148 LYSTALAFRLLRQHGFQVAQDVLECFKNTKGEFEPSLSDDTRGLLQLYEASFLLTEGENT 207
Query: 122 LNDLGYLSCELLQAWLPRHQ-DHIQAI-YVSNTLQYPIHYGLSRFMDKSIFINDLKAKNK 179
L + ++L+ L + D I + ++ ++L+ PIH+ + R ++ S++I+ K +
Sbjct: 208 LELARDFTTKILEEKLRNDEIDDINLVTWIRHSLEIPIHWRIDR-VNTSVWIDVYKRRPD 266
Query: 180 W--ICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFT 237
I L ELA ++S+IV+ + E +WW + LA+++ FA ++ Y W +
Sbjct: 267 MNPIVL-ELAVLDSNIVQAQYQEELKLDLQWWRNTCLAEKLPFARDRLVESYFWGVGVVQ 325
Query: 238 DPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSL 297
R+ + + I+L+ +IDD++DV+GTL++L FTEA+ RW++ + LP++M++
Sbjct: 326 PRQHGIARMAVDRSIALITVIDDVYDVYGTLEELEQFTEAIRRWDISSIDQLPSYMQLCF 385
Query: 298 SSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGI 357
+L N+ A V K+ GFN I L+ SW ++ FL EA W ++G P EEYL NG
Sbjct: 386 LALDNFINDIAYDVLKEQGFNIIPYLRKSWTDMIEGFLLEAKWYHNGHKPKLEEYLENGW 445
Query: 358 VSTGVHVVLIHAFFLMDHAKGITKETLSILDEEF--PNIIYSVAKILRLSDDL----EGV 411
S G VVL HAFF + H+ +TKE +D+ F I+ + +LRL+DDL + V
Sbjct: 446 RSIGSTVVLTHAFFGVTHS--LTKEN---IDQFFGYHEIVRLSSMLLRLADDLGTSTDEV 500
Query: 412 KSGDQNGLDGSYLN 425
GD Y+N
Sbjct: 501 SRGDVPKAIQCYMN 514
>UniRef100_Q7Y1V1 Putative monoterpene synthase [Melaleuca alternifolia]
Length = 583
Score = 219 bits (558), Expect = 1e-55
Identities = 138/395 (34%), Positives = 210/395 (52%), Gaps = 7/395 (1%)
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLR 73
+P F L+D IQRLG+ YF E+I AL + LS + L+ AL+FR+LR
Sbjct: 92 EPMAIFALVDDIQRLGLGRYFEEDISKALRR---CLSQYAVTGSLQKSLHGTALSFRVLR 148
Query: 74 QGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELL 133
Q G+ V+ ++F S G DV+G+++LYEAS L+ E ED L+ + + L
Sbjct: 149 QHGFEVSQDVFKIFMDESGSFMKTLGGDVQGMLSLYEASHLAFEEEDILHKAKTFAIKHL 208
Query: 134 QAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMNSSI 193
+ L D +V++ L+ P+H + + + N + ELA M +
Sbjct: 209 EN-LNHDIDQDLQDHVNHELELPLHRRMPLLEARRFIEAYSRRSNVNPRILELAVMKFNS 267
Query: 194 VKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPIS 253
+ + + ++ WW ++GLAK + FA ++ + W + +P SN R +TK S
Sbjct: 268 SQLTLQRDLQDMLGWWNNVGLAKRLSFARDRLMECFFWAVGIAREPALSNCRKGVTKAFS 327
Query: 254 LVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYK 313
L+ ++DD++DV GTLD+L LFT+AV RW D ENLP +MK+ +LY N+ A K
Sbjct: 328 LILVLDDVYDVFGTLDELELFTDAVRRWHEDAVENLPGYMKLCFLALYNSVNDMAYETLK 387
Query: 314 KHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFFLM 373
+ G N L W L AFL+EA W + I P EEYLNNG VS+ V+L HA+FL
Sbjct: 388 ETGENVTPYLTKVWYDLCKAFLQEAKWSYNKITPGVEEYLNNGWVSSSGQVMLTHAYFL- 446
Query: 374 DHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
+ + KE L L E + +++ + I RL++DL
Sbjct: 447 -SSPSLRKEELESL-EHYHDLLRLPSLIFRLTNDL 479
>UniRef100_Q5SBP3 R-linalool synthase [Ocimum basilicum]
Length = 574
Score = 219 bits (558), Expect = 1e-55
Identities = 136/424 (32%), Positives = 220/424 (51%), Gaps = 24/424 (5%)
Query: 1 MCKQVHKKLVTSE-DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSS 59
+ ++ +KL ++ + E LID IQRLGI +YF + I L +P
Sbjct: 77 LLEETARKLQRNDTESVEKLKLIDNIQRLGIGYYFEDAIDAVLR--------SPFSAEEE 128
Query: 60 HELYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGE-DVKGLIALYEASQLSIEG 118
+L+ AL FRLLR G V E+F K R ++ E D GL++LYEAS L + G
Sbjct: 129 EDLFTAALRFRLLRHNGIQVTPEIF----LKFKDERGEFDESDTLGLLSLYEASNLGVTG 184
Query: 119 EDGLNDLGYLSCELLQAWLPRHQDHIQAIY---VSNTLQYPIHYGLSRFMDKSIFINDLK 175
E+ L + + E + L R + A V+ L P H ++R + K
Sbjct: 185 EEILEE----AMEFAEPRLRRSLSELAAPLRSEVAQALDVPRHLRMARLEARRFIEQYGK 240
Query: 176 AKNKWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMAC 235
+ L ELA ++ + V+ +++E E+ +WW+ LGL +++ F L+ +MW M
Sbjct: 241 QSDHDGDLLELAILDYNQVQAQHQSELTEITRWWKQLGLVEKLGFGRDRALECFMWTMGI 300
Query: 236 FTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKV 295
P +S+ R+E K +L+Y+IDDIFD +G +D+L LFT+A+ RW+++ E LP +MK+
Sbjct: 301 LPHPKYSSSRIESAKAAALLYVIDDIFDTYGKMDELILFTDAIRRWDLEAMEGLPEYMKI 360
Query: 296 SLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNN 355
+LY TN V K G + LK W+ + A++ E W + G P EEY+ N
Sbjct: 361 CYMALYNTTNEICYRVLKDTGRIALPYLKSVWIETIEAYMVEVKWFSGGSAPKLEEYIEN 420
Query: 356 GIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVA-KILRLSDDLEGVKSG 414
G + G ++VL+H FFL+ +G+T + + ++ + +S A +I RL DDL +
Sbjct: 421 GASTVGAYMVLVHLFFLI--GEGLTHQNVLFFKQKPYHKPFSAAGRIFRLWDDLGTSQEE 478
Query: 415 DQNG 418
++ G
Sbjct: 479 EERG 482
>UniRef100_Q8W1J9 Linalool synthase [Perilla frutescens var. crispa]
Length = 607
Score = 217 bits (553), Expect = 4e-55
Identities = 134/420 (31%), Positives = 223/420 (52%), Gaps = 20/420 (4%)
Query: 1 MCKQVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVAL----EKQFLILSSNPIDF 56
+ +QV L + + L+D ++ LG+ ++F ++IK L + S++ I+
Sbjct: 98 LIEQVKMLLEEEMEAVQQLELVDDLKNLGLSYFFEDQIKQILTFIYNEHKCFHSNSIIEA 157
Query: 57 VSSHELYEVALAFRLLRQGGYYVNAELFDCLKCSKKS-LRVKYGEDVKGLIALYEASQLS 115
+LY AL FRLLRQ G+ V+ E+FDC K + S + + G+D KGL+ LYEAS L
Sbjct: 158 EEIRDLYFTALGFRLLRQHGFQVSQEVFDCFKNEEGSDFKARLGDDTKGLLQLYEASFLL 217
Query: 116 IEGEDGLNDLGYLSCELLQAWLPRH--QDHIQAIYVSNTLQYPIHYGLSR-----FMDKS 168
EGED L + + LQ + D+ ++ ++L+ P+H+ + R F+D+
Sbjct: 218 REGEDTLELARQYATKFLQKKVDHELIDDNNLLSWILHSLEIPLHWRIQRLEARWFLDRY 277
Query: 169 IFINDLKAKNKWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKW 228
D+ + ELAK++ +I++ + E ++ +WW+ LA+++ F ++
Sbjct: 278 ATRRDMNQ-----IILELAKLDFNIIQATQQEELKDLSRWWKSTCLAEKLPFVRDRLVES 332
Query: 229 YMWPMACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAEN 288
Y W +A F + R K I+L+ +DD++D++GTLD+L LFT+A+ RW+ +
Sbjct: 333 YFWAIALFEPHQYGYHRKVAAKIITLITSLDDVYDIYGTLDELQLFTDAIQRWDTESISR 392
Query: 289 LPNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPT 348
LP +M++ LY + A K+ GF I L+ SW L+ A+LKEA W +G +P+
Sbjct: 393 LPYYMQLFYMVLYNFVSELAYDGLKEKGFITIPYLQRSWADLVEAYLKEAKWFYNGYVPS 452
Query: 349 TEEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
EEYLNN +S G V+ FF + A I K + L E+ I+ ++RL DDL
Sbjct: 453 MEEYLNNAYISIGATPVISQVFFTL--ATSIDKPVIDSL-YEYHRILRLSGMLVRLPDDL 509
>UniRef100_Q6EJ97 Isoprene synthase [Pueraria lobata]
Length = 608
Score = 216 bits (551), Expect = 8e-55
Identities = 137/396 (34%), Positives = 219/396 (54%), Gaps = 8/396 (2%)
Query: 15 PTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLRQ 74
P LID +QRLG+ + F ++I ALE ++L N + +L+ AL+FRLLRQ
Sbjct: 110 PLSLLELIDDVQRLGLTYKFEKDIIKALEN-IVLLDENK---KNKSDLHATALSFRLLRQ 165
Query: 75 GGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELLQ 134
G+ V+ ++F+ K + + DV+GL++LYEAS L EGE+ L + S L+
Sbjct: 166 HGFEVSQDVFERFKDKEGGFSGELKGDVQGLLSLYEASYLGFEGENLLEEARTFSITHLK 225
Query: 135 AWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWI-CLDELAKMNSSI 193
L + A VS+ L+ P H L R ++ F++ + K L ELAK++ ++
Sbjct: 226 NNLKEGINTKVAEQVSHALELPYHQRLHR-LEARWFLDKYEPKEPHHQLLLELAKLDFNM 284
Query: 194 VKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPIS 253
V+ +++ E ++ +WW ++GLA ++ F ++ Y W + DP F R +TK
Sbjct: 285 VQTLHQKELQDLSRWWTEMGLASKLDFVRDRLMEVYFWALGMAPDPQFGECRKAVTKMFG 344
Query: 254 LVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNNFAEMVYK 313
LV IIDD++DV+GTLD+L LFT+AV RW+++ LP++MK+ +LY N+ + + K
Sbjct: 345 LVTIIDDVYDVYGTLDELQLFTDAVERWDVNAINTLPDYMKLCFLALYNTVNDTSYSILK 404
Query: 314 KHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIV-STGVHVVLIHAFFL 372
+ G N + L SW L AFL+EA W N+ I+P +YL N V S+GV ++ F +
Sbjct: 405 EKGHNNLSYLTKSWRELCKAFLQEAKWSNNKIIPAFSKYLENASVSSSGVALLAPSYFSV 464
Query: 373 MDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
+ I+ L L +F ++ S I RL +DL
Sbjct: 465 CQQQEDISDHALRSL-TDFHGLVRSSCVIFRLCNDL 499
>UniRef100_Q8L5K1 (+)-limonene synthase 2 [Citrus limon]
Length = 606
Score = 213 bits (543), Expect = 6e-54
Identities = 139/400 (34%), Positives = 222/400 (54%), Gaps = 15/400 (3%)
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHE-LYEVALAFRLL 72
+P + LID +QRLG+ + F EI+ L + +N D+V E LY +L FRLL
Sbjct: 100 EPLDQLELIDNLQRLGLAYRFETEIRNILHNIY----NNNKDYVWRKENLYATSLEFRLL 155
Query: 73 RQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCEL 132
RQ GY V+ E+F+ K + +D KG+++L+EAS S+EGE + + + +
Sbjct: 156 RQHGYPVSQEVFNGFKDDQGGFIF---DDFKGILSLHEASYYSLEGESIMEEAWQFTSKH 212
Query: 133 LQAWL---PRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKM 189
L+ + +D A L+ P+H+ + + K ++K L ELAKM
Sbjct: 213 LKEVMISKSMEEDVFVAEQAKRALELPLHWKVPMLEARWFIHVYEKREDKNHLLLELAKM 272
Query: 190 NSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELT 249
+ ++ + + E E+ WW+D GL +++ FA + ++W M +P F+ R LT
Sbjct: 273 EFNTLQAIYQEELKEISGWWKDTGLGEKLSFARNRLVASFLWSMGIAFEPQFAYCRRVLT 332
Query: 250 KPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGA-ENLPNFMKVSLSSLYKVTNNFA 308
I+L+ +IDDI+DV+GTLD+L +FT+AV RW+++ A ++LP +MK+ +LY N FA
Sbjct: 333 ISIALITVIDDIYDVYGTLDELEIFTDAVARWDINYALKHLPGYMKMCFLALYNFVNEFA 392
Query: 309 EMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIH 368
V K+ F+ + ++K +W+ L+ A+L EA W +S P EEYL NG+VS + LI
Sbjct: 393 YYVLKQQDFDMLLSIKNAWLGLIQAYLVEAKWYHSKYTPKLEEYLENGLVS--ITGPLII 450
Query: 369 AFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
A + I K+ L L E P+I++ +KI RL DDL
Sbjct: 451 AISYLSGTNPIIKKELEFL-ESNPDIVHWSSKIFRLQDDL 489
>UniRef100_Q6F5H2 D-limonene synthase [Citrus unshiu]
Length = 608
Score = 213 bits (543), Expect = 6e-54
Identities = 138/402 (34%), Positives = 223/402 (55%), Gaps = 17/402 (4%)
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLR 73
+P + LID +QRLG+ ++F EI+ L I + N LY +L FRLLR
Sbjct: 100 EPLDQLELIDNLQRLGLAYHFEPEIRNILRN---IHNHNKDYNWRKENLYATSLEFRLLR 156
Query: 74 QGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYLSCELL 133
Q GY V+ E+F K K +D KG+++L+EAS S+EGE + + + + L
Sbjct: 157 QHGYPVSQEVFSGFKDDKVGFIC---DDFKGILSLHEASYYSLEGESIMEEAWQFTSKHL 213
Query: 134 QAWL----PRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDL-KAKNKWICLDELAK 188
+ + + +D A L+ P+H+ ++ FI+ K ++K L ELAK
Sbjct: 214 KEMMITSNSKEEDVFVAEQAKRALELPLHWKKVPMLEARWFIHVYEKREDKNHLLLELAK 273
Query: 189 MNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVEL 248
+ + ++ + + E ++ WW+D GL +++ FA + ++W M +P F+ R L
Sbjct: 274 LEFNTLQAIYQEELKDISGWWKDTGLGEKLSFARNRLVASFLWSMGIAFEPQFAYCRRVL 333
Query: 249 TKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGA-ENLPNFMKVSLSSLYKVTNNF 307
T I+L+ +IDDI+DV+GTLD+L +FT+AV RW+++ A ++LP +MK+ +LY N F
Sbjct: 334 TISIALITVIDDIYDVYGTLDELEIFTDAVARWDINYALKHLPGYMKMCFLALYNFVNEF 393
Query: 308 AEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVS-TGVHVVL 366
A V K+ F+ + ++K +W+ L+ A+L EA W +S P EEYL NG+VS TG ++
Sbjct: 394 AYYVLKQQDFDMLLSIKHAWLGLIQAYLVEAKWYHSKYTPKLEEYLENGLVSITGPLIIT 453
Query: 367 IHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
I + I K+ L L E P+I++ +KI RL DDL
Sbjct: 454 IS---YLSGTNPIIKKELEFL-ESNPDIVHWSSKIFRLQDDL 491
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 731,980,261
Number of Sequences: 2790947
Number of extensions: 30952799
Number of successful extensions: 83206
Number of sequences better than 10.0: 353
Number of HSP's better than 10.0 without gapping: 301
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 81975
Number of HSP's gapped (non-prelim): 393
length of query: 426
length of database: 848,049,833
effective HSP length: 130
effective length of query: 296
effective length of database: 485,226,723
effective search space: 143627110008
effective search space used: 143627110008
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC142394.1


