

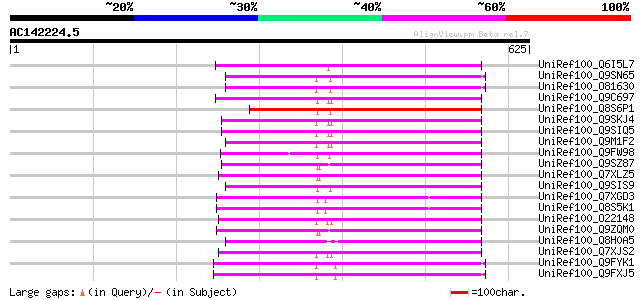
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142224.5 - phase: 0 /pseudo
(625 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6I5L7 Hypothetical protein OSJNBb0088F07.2 [Oryza sat... 258 4e-67
UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis tha... 243 9e-63
UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana] 243 9e-63
UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [A... 239 2e-61
UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa] 238 3e-61
UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcrip... 238 4e-61
UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcrip... 238 4e-61
UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis tha... 237 9e-61
UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcrip... 228 3e-58
UniRef100_Q9SZ87 RNA-directed DNA polymerase-like protein [Arabi... 228 5e-58
UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa] 228 5e-58
UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcrip... 226 2e-57
UniRef100_Q7XGD3 Putative retroelement [Oryza sativa] 224 8e-57
UniRef100_Q8S5K1 Putative retroelement [Oryza sativa] 224 8e-57
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 224 8e-57
UniRef100_Q9ZQM0 Putative non-LTR retroelement reverse transcrip... 220 8e-56
UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa] 217 7e-55
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 216 2e-54
UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana] 213 2e-53
UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana] 213 2e-53
>UniRef100_Q6I5L7 Hypothetical protein OSJNBb0088F07.2 [Oryza sativa]
Length = 1264
Score = 258 bits (659), Expect = 4e-67
Identities = 135/335 (40%), Positives = 201/335 (59%), Gaps = 14/335 (4%)
Query: 248 IVSEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCI 307
+++ G P DT + LI K +P ++K L P+SLCNV YK++SKV+ NRLK +L + I
Sbjct: 822 VLNGGDMPQGWNDTVVVLIPKTKQPDTLKDLRPISLCNVAYKLISKVIVNRLKVVLLEII 881
Query: 308 SDSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVM 367
S SQ+AFVP R I N L +E+ HY+ + KGK G A+K D+SKA+DR++W +L+ +M
Sbjct: 882 SPSQSAFVPRRLITYNVLLAYELTHYLNQRKKGKNGVAAIKLDMSKAYDRVEWDFLRHMM 941
Query: 368 EKK-WDSQMFGLTGSC-------------NV*PIDPHCGIRQGGPFSPYLYIICSEGLNS 413
+ + Q L C + I P G+RQG P SPYL+IIC+EGL++
Sbjct: 942 LRLGFHDQWVNLVMKCVTSVTYRIKINGEHSDQIYPQRGLRQGDPLSPYLFIICAEGLSA 1001
Query: 414 YIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQA 473
++ + G I G ++CR +P I L FADDS + +A ++ L+ +L YE ASGQ
Sbjct: 1002 LLQKAQADGKIEGIKVCRDTPRINHLFFADDSLVLMRAGQNDAQELRRVLNIYEVASGQV 1061
Query: 474 INYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIW 533
IN KSS+ FS NT + L + + + +YLGLP + + ++ F +IK ++W
Sbjct: 1062 INKDKSSVLFSPNTLQSDRMEVRSALCINQEAKNERYLGLPVSIGKSRRKAFEYIKRKVW 1121
Query: 534 KNIQNWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
IQ W LS+AGKE+L+K++AQ IPTY+ + F
Sbjct: 1122 LRIQGWQEKLLSKAGKEILVKAVAQAIPTYAMSCF 1156
>UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis thaliana]
Length = 1294
Score = 243 bits (621), Expect = 9e-63
Identities = 133/327 (40%), Positives = 189/327 (57%), Gaps = 15/327 (4%)
Query: 261 TNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFI 320
TNI +I KI P+++ P++LCNV+YKI+SK L RLK L +SDSQAAF+P R +
Sbjct: 849 TNICMIPKITNPETLSDYRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFIPGRLV 908
Query: 321 LENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVME-----KKWDSQM 375
+N + E++H +K + + + +A+K DVSKA+DR++W++L+ M + W +
Sbjct: 909 NDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSETWIKWI 968
Query: 376 FGLTGSCNV*---------PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHG 426
G S N I P GIRQG P SPYL+I+C++ LN IK+ G I G
Sbjct: 969 MGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEGDIRG 1028
Query: 427 TRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRN 486
RI G P +T L FADDS F ++ V LK++ YE SGQ IN KS I F
Sbjct: 1029 IRIGNGVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQKINMSKSMITFGSR 1088
Query: 487 TDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSR 546
+ + ++LG+ GKYLGLP R K+ +F++I ER+ K +WSA LS
Sbjct: 1089 VHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIERVKKRTSSWSAKYLSP 1148
Query: 547 AGKEVLLKSIAQYIPTYSWAIFCSLPL 573
AGKE++LKS+A +P Y+ + F LPL
Sbjct: 1149 AGKEIMLKSVAMSMPVYAMSCF-KLPL 1174
>UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana]
Length = 1662
Score = 243 bits (621), Expect = 9e-63
Identities = 133/327 (40%), Positives = 189/327 (57%), Gaps = 15/327 (4%)
Query: 261 TNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFI 320
TNI +I KI P+++ P++LCNV+YKI+SK L RLK L +SDSQAAF+P R +
Sbjct: 869 TNICMIPKITNPETLSDYRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFIPGRLV 928
Query: 321 LENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVME-----KKWDSQM 375
+N + E++H +K + + + +A+K DVSKA+DR++W++L+ M + W +
Sbjct: 929 NDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGFSETWIKWI 988
Query: 376 FGLTGSCNV*---------PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHG 426
G S N I P GIRQG P SPYL+I+C++ LN IK+ G I G
Sbjct: 989 MGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHLIKNRVAEGDIRG 1048
Query: 427 TRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRN 486
RI G P +T L FADDS F ++ V LK++ YE SGQ IN KS I F
Sbjct: 1049 IRIGNGVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQKINMSKSMITFGSR 1108
Query: 487 TDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSR 546
+ + ++LG+ GKYLGLP R K+ +F++I ER+ K +WSA LS
Sbjct: 1109 VHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIERVKKRTSSWSAKYLSP 1168
Query: 547 AGKEVLLKSIAQYIPTYSWAIFCSLPL 573
AGKE++LKS+A +P Y+ + F LPL
Sbjct: 1169 AGKEIMLKSVAMSMPVYAMSCF-KLPL 1194
>UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
Length = 1142
Score = 239 bits (610), Expect = 2e-61
Identities = 134/334 (40%), Positives = 196/334 (58%), Gaps = 14/334 (4%)
Query: 249 VSEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCIS 308
+ EG F L TNI LI K +RP M L P+SLCNV YK++SK+L RLK +L IS
Sbjct: 259 LQEGVFDKRLNTTNICLIPKTERPTRMTELRPISLCNVGYKVISKILCQRLKTVLPNLIS 318
Query: 309 DSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVME 368
++Q+AFV R I +N L E+ H ++ + K+ +A+K D+SKA+D+++W++++A++
Sbjct: 319 ETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKDKFMAIKTDMSKAYDQVEWNFIEALLR 378
Query: 369 K-----KWDSQMFGLTGSC------NV*P---IDPHCGIRQGGPFSPYLYIICSEGLNSY 414
K KW S + + N P I P G+RQG P SPYL+I+C+E L +
Sbjct: 379 KMGFCEKWISWIMWCITTVQYKVLINGQPKGLIIPERGLRQGDPLSPYLFILCTEVLIAN 438
Query: 415 IKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAI 474
I+ E LI G ++ SP+++ LLFADDS F KA + I+ IL YE+ SGQ I
Sbjct: 439 IRKAERQNLITGIKVATPSPAVSHLLFADDSLFFCKANKEQCGIILEILKQYESVSGQQI 498
Query: 475 NYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWK 534
N+ KSSI F + +I +LG+ G YLGLP + K +FSF+++R+
Sbjct: 499 NFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMGSYLGLPESLGGSKTKVFSFVRDRLQS 558
Query: 535 NIQNWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
I WSA LS+ GKEV++KS+A +P Y + F
Sbjct: 559 RINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCF 592
>UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa]
Length = 1509
Score = 238 bits (608), Expect = 3e-61
Identities = 126/293 (43%), Positives = 177/293 (60%), Gaps = 14/293 (4%)
Query: 290 ILSKVLANRLKQILHKCISDSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQ 349
++ KVLANRLK+IL IS +Q+AFVP R I +N L +E+ HYM+ K G+ G A K
Sbjct: 717 LIPKVLANRLKKILPDVISPAQSAFVPGRLISDNILIAYEMTHYMRNKRSGQVGYAAFKL 776
Query: 350 DVSKAFDRIKWSYLQAVMEK-----KWDSQMFGLTGSCNV*---------PIDPHCGIRQ 395
D+SKA+DR++WS+L +M K W + + + P G+RQ
Sbjct: 777 DMSKAYDRVEWSFLHDMMLKLGFHTDWVNLIMKCVSTVTYRIRVNGELSESFSPERGLRQ 836
Query: 396 GGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSE 455
G P SPYL+++C+EG ++ + E G +HG RIC+G+PS++ LLFADDS + +A E
Sbjct: 837 GDPLSPYLFLLCAEGFSALLSKTEEEGRLHGIRICQGAPSVSHLLFADDSLILCRANGGE 896
Query: 456 DTILKNILYTYEAASGQAINYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPS 515
L+ IL YE SGQ IN KS++ FS NT S+ + L + + KYLGLP
Sbjct: 897 AQQLQTILQIYEECSGQVINKDKSAVMFSPNTSSLEKGAVMAALNMQRETTNEKYLGLPV 956
Query: 516 MVERDKKSIFSFIKERIWKNIQNWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
V R + IFS++KERIW+ IQ W LSRAGKE+L+K++AQ IPT++ F
Sbjct: 957 FVGRSRTKIFSYLKERIWQRIQGWKEKLLSRAGKEILIKAVAQVIPTFAMGCF 1009
>UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1750
Score = 238 bits (607), Expect = 4e-61
Identities = 132/327 (40%), Positives = 186/327 (56%), Gaps = 14/327 (4%)
Query: 256 PNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFV 315
P++ TNI +I KI P ++ P++LCNV+YK++SK L NRLK L+ +SDSQAAF+
Sbjct: 864 PSINHTNICMIPKITNPTTLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFI 923
Query: 316 PSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVME-----KK 370
P R I +N + EV+H +K + + + +A+K DVSKA+DR++W +L+ M K
Sbjct: 924 PGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNK 983
Query: 371 WDSQMFGLTGSC------NV*P---IDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENT 421
W + S N P I P GIRQG P SPYL+I+C + L+ I ++
Sbjct: 984 WIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASS 1043
Query: 422 GLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSI 481
G + G RI G+P+IT L FADDS F +A V LK++ YE SGQ IN +KS I
Sbjct: 1044 GDLRGVRIGNGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMI 1103
Query: 482 AFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWSA 541
F + +L + GKYLGLP R KK +F +I +R+ K WSA
Sbjct: 1104 TFGSRVYGSTQSRLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSA 1163
Query: 542 WSLSRAGKEVLLKSIAQYIPTYSWAIF 568
LS AGKE++LKS+A +P Y+ + F
Sbjct: 1164 RFLSPAGKEIMLKSVALAMPVYAMSCF 1190
>UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1524
Score = 238 bits (607), Expect = 4e-61
Identities = 132/327 (40%), Positives = 186/327 (56%), Gaps = 14/327 (4%)
Query: 256 PNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFV 315
P++ TNI +I KI P ++ P++LCNV+YK++SK L NRLK L+ +SDSQAAF+
Sbjct: 638 PSINHTNICMIPKITNPTTLSDYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFI 697
Query: 316 PSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVME-----KK 370
P R I +N + EV+H +K + + + +A+K DVSKA+DR++W +L+ M K
Sbjct: 698 PGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNK 757
Query: 371 WDSQMFGLTGSC------NV*P---IDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENT 421
W + S N P I P GIRQG P SPYL+I+C + L+ I ++
Sbjct: 758 WIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHLINGRASS 817
Query: 422 GLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSI 481
G + G RI G+P+IT L FADDS F +A V LK++ YE SGQ IN +KS I
Sbjct: 818 GDLRGVRIGNGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSMI 877
Query: 482 AFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWSA 541
F + +L + GKYLGLP R KK +F +I +R+ K WSA
Sbjct: 878 TFGSRVYGSTQSKLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSA 937
Query: 542 WSLSRAGKEVLLKSIAQYIPTYSWAIF 568
LS AGKE++LKS+A +P Y+ + F
Sbjct: 938 RFLSPAGKEIMLKSVALAMPVYAMSCF 964
>UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis thaliana]
Length = 851
Score = 237 bits (604), Expect = 9e-61
Identities = 132/322 (40%), Positives = 187/322 (57%), Gaps = 14/322 (4%)
Query: 261 TNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFI 320
TNI +I KI P+++ P++LCNV+YK++SK + NRLK L+ +SDSQAAF+P R I
Sbjct: 108 TNICMIPKIQNPQTLSDYRPIALCNVLYKVISKCMVNRLKAHLNSIVSDSQAAFIPGRII 167
Query: 321 LENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVME-----KKWDSQM 375
+N + E++H +K + + + +A+K DVSKA+DR++W +L+ M KW +
Sbjct: 168 NDNVMIAHEIMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGFCDKWIGWI 227
Query: 376 FGLTGSC------NV*P---IDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHG 426
S N P I P GIRQG P SPYL+I+C + L+ IK ++G I G
Sbjct: 228 MAAVKSVHYSVLINGSPHGYISPTRGIRQGDPLSPYLFILCGDILSHLIKVKASSGDIRG 287
Query: 427 TRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRN 486
RI G+P+IT L FADDS F +A V LK++ YE SGQ IN +KS I F
Sbjct: 288 VRIGNGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQKINVQKSLITFGSR 347
Query: 487 TDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSR 546
+ LL + GKYLGLP R KK +F++I +R+ + +WSA LS
Sbjct: 348 VYGSTQTRLKTLLNIPNQGGGGKYLGLPEQFGRKKKEMFNYIIDRVKERTASWSAKFLSP 407
Query: 547 AGKEVLLKSIAQYIPTYSWAIF 568
AGKE+LLKS+A +P Y+ + F
Sbjct: 408 AGKEILLKSVALAMPVYAMSCF 429
>UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1382
Score = 228 bits (582), Expect = 3e-58
Identities = 130/328 (39%), Positives = 180/328 (54%), Gaps = 15/328 (4%)
Query: 255 PPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAF 314
P L D+ + LI K++ + P+SLCNV+YKI SKVLANRLK L +S+ Q+AF
Sbjct: 497 PEGLCDSVVVLIPKVNNASHLSKFRPISLCNVLYKIASKVLANRLKPFLPDIVSEFQSAF 556
Query: 315 VPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEK----- 369
VP R I ++AL +E LH ++ K K ALK D+ KA+DR++W+YL + K
Sbjct: 557 VPGRLITDSALVAYECLHTIR-KQHNKNPFFALKIDMMKAYDRVEWAYLSGCLSKLGFSQ 615
Query: 370 KWDSQMFGLTGSCN---------V*PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHEN 420
W + + S P+ P GIRQG P SPYL+++C+EGL+ + E
Sbjct: 616 DWINTVMRCVSSVRYAVKINGELTKPVVPSRGIRQGDPISPYLFLLCTEGLSCLLHKKEV 675
Query: 421 TGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSS 480
G + G + R P I+ LLFADDS F KA LKN L +Y +ASGQ IN KSS
Sbjct: 676 AGELQGIKNGRHGPPISHLLFADDSIFFAKADSRNVQALKNTLRSYCSASGQKINLHKSS 735
Query: 481 IAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWS 540
I F + ++ L V + YLG+P+ + + F F+ ERIWK + W+
Sbjct: 736 IFFGKRCPDAVKISVKSCLQVDNEVLQDSYLGMPTEIGLATTNFFKFLPERIWKRVNGWT 795
Query: 541 AWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
LSRAG E +LK++AQ IP Y + F
Sbjct: 796 DRPLSRAGMETMLKAVAQAIPNYVMSCF 823
>UniRef100_Q9SZ87 RNA-directed DNA polymerase-like protein [Arabidopsis thaliana]
Length = 1274
Score = 228 bits (580), Expect = 5e-58
Identities = 132/328 (40%), Positives = 189/328 (57%), Gaps = 16/328 (4%)
Query: 256 PNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFV 315
P L +T++ LI KI P+ + P++LCNV YKI++K+L RL+ L + IS Q+AFV
Sbjct: 423 PRLNETHVTLIPKISAPRKVSDYRPIALCNVQYKIVAKILTRRLQPWLSELISLHQSAFV 482
Query: 316 PSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEK-----K 370
P R I +N L E+LH+++ K ++A+K D+SKA+DRIKW++LQ V+ + K
Sbjct: 483 PGRAIADNVLITHEILHFLRVSGAKKYCSMAIKTDMSKAYDRIKWNFLQEVLMRLGFHDK 542
Query: 371 W----------DSQMFGLTGSCNV*PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHEN 420
W S F + GS + P G+RQG P SPYL+I+C+E L+ + +
Sbjct: 543 WIRWVMQCVCTVSYSFLINGSPQG-SVVPSRGLRQGDPLSPYLFILCTEVLSGLCRKAQE 601
Query: 421 TGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSS 480
G++ G R+ RGSP + LLFADD+ F K + L NIL YE ASGQ+IN KS+
Sbjct: 602 KGVMVGIRVARGSPQVNHLLFADDTMFFCKTNPTCCGALSNILKKYELASGQSINLAKSA 661
Query: 481 IAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWS 540
I FS T + L + GKYLGLP R K+ IFS I +RI + +WS
Sbjct: 662 ITFSSKTPQDIKRRVKLSLRIDNEGGIGKYLGLPEHFGRRKRDIFSSIVDRIRQRSHSWS 721
Query: 541 AWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
LS AGK++LLK++ +P+Y+ F
Sbjct: 722 IRFLSSAGKQILLKAVLSSMPSYAMMCF 749
>UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa]
Length = 1205
Score = 228 bits (580), Expect = 5e-58
Identities = 129/331 (38%), Positives = 191/331 (56%), Gaps = 14/331 (4%)
Query: 252 GSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQ 311
G + + DT I +I K + P MK PVSLCNVIYK+++K L NRL+ +L + IS++Q
Sbjct: 466 GEWKEGMNDTVIVMIPKTNAPVEMKDFRPVSLCNVIYKVVAKCLVNRLRPLLQEIISETQ 525
Query: 312 AAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEK-- 369
+AFVP R I +NAL FE H + T+ + ALK D+SKA+DR+ W +L ++K
Sbjct: 526 SAFVPGRMITDNALVAFECFHSIHKCTRESQDFCALKLDLSKAYDRVDWGFLDGALQKLG 585
Query: 370 ------KW-----DSQMFGLTGSCNV*-PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKH 417
KW S + + + N+ P P G+R+G P +PYL++ ++GL++ ++
Sbjct: 586 FGNIWRKWIMSCVTSVRYSVRLNGNMLEPFYPTRGLREGDPLNPYLFLFIADGLSNILQR 645
Query: 418 HENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYR 477
+ I ++CR +P ++ LLFADDS LF+KA V + T +K L YE +GQ IN +
Sbjct: 646 RRDERQIQPLKVCRSAPGVSHLLFADDSLLFFKAEVIQATRIKEALDLYERCTGQLINPK 705
Query: 478 KSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQ 537
+ S+ FS I +L V + + K LGLP+ R K F IKER K +
Sbjct: 706 ECSLLFSALCPQERQDGIKAVLQVERTCFDDKCLGLPTPDGRMKAEQFQPIKERFEKRLT 765
Query: 538 NWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
+WS LS AGKE L+KS+AQ +PTY+ +F
Sbjct: 766 DWSERFLSLAGKEALIKSVAQALPTYTMGVF 796
>UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1715
Score = 226 bits (575), Expect = 2e-57
Identities = 130/322 (40%), Positives = 183/322 (56%), Gaps = 14/322 (4%)
Query: 261 TNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFI 320
T I LI KI PK M P+SLC YKI+SK+L RLKQ L ISDSQAAFVP + I
Sbjct: 849 TQICLIPKIIDPKHMSDYRPISLCTASYKIISKILIKRLKQCLGDVISDSQAAFVPGQNI 908
Query: 321 LENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEK-----KWDSQM 375
+N L E+LH +K + + + G +A+K D+SKA+DR++W++L+ VM + +W +
Sbjct: 909 SDNVLVAHELLHSLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWI 968
Query: 376 FGLTGSCNV*---------PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHG 426
S + I P GIRQG P SPYL++ C+E L++ ++ E IHG
Sbjct: 969 MTCVTSVSYEVLINGSPYGKIFPSRGIRQGDPLSPYLFLFCAEVLSNMLRKAEVNKQIHG 1028
Query: 427 TRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRN 486
+I + +I+ LLFADDS F +A L I YE ASGQ INY KSSI F +
Sbjct: 1029 MKITKDCLAISHLLFADDSLFFCRASNQNIEQLALIFKKYEEASGQKINYAKSSIIFGQK 1088
Query: 487 TDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSR 546
++ + LLG+ GKYLGLP + R K +F +I ++ + + W+ LS
Sbjct: 1089 IPTMRRQRLHRLLGIDNVRGGGKYLGLPEQLGRRKVELFEYIVTKVKERTEGWAYNYLSP 1148
Query: 547 AGKEVLLKSIAQYIPTYSWAIF 568
AGKE+++K+IA +P YS F
Sbjct: 1149 AGKEIVIKAIAMALPVYSMNCF 1170
>UniRef100_Q7XGD3 Putative retroelement [Oryza sativa]
Length = 1694
Score = 224 bits (570), Expect = 8e-57
Identities = 122/333 (36%), Positives = 187/333 (55%), Gaps = 15/333 (4%)
Query: 250 SEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISD 309
S G+ P + +T I LI K ++P+ +K P+SLCNV+YKI+SK L NRL+ IL +S
Sbjct: 1090 SLGTMPSGVNETAIVLIPKTEQPQELKDFRPISLCNVVYKIVSKCLVNRLRPILDDLVSQ 1149
Query: 310 SQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEK 369
+Q+AFVP R I +NAL FE H+++ + A K D+SKA+DR+ W +L+ M K
Sbjct: 1150 NQSAFVPGRLITDNALIAFEYFHHIQRNKNPENAYSAYKLDLSKAYDRVDWEFLEQAMVK 1209
Query: 370 --------KWDSQMFGLT------GSCNV*PIDPHCGIRQGGPFSPYLYIICSEGLNSYI 415
KW L + P G+RQG P SP+L++ ++GL+ +
Sbjct: 1210 LGFAHCWVKWIMACITLVRYAVKLNGTLLNTFAPSRGLRQGDPLSPFLFLFVADGLSFLL 1269
Query: 416 KHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAIN 475
+ + G ++ ICR +P++ LLFADD+ LF+KA + ++K +L TY + +GQ IN
Sbjct: 1270 EEKVDQGAMNPIHICRHAPAVLHLLFADDTPLFFKAANDQAVVMKEVLETYASCTGQLIN 1329
Query: 476 YRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKN 535
K SI F +++ + + I L + S + KYLG P+ R K F ++ER+WK
Sbjct: 1330 PSKCSIMFGQSSPTAVQNQIKQTLQISNS-FEDKYLGFPTPEGRMTKGKFQSLQERLWKR 1388
Query: 536 IQNWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
I W LS GKE+L+K++ Q +P Y +F
Sbjct: 1389 IMLWGEKFLSTGGKEILIKAVLQAMPVYVMGLF 1421
>UniRef100_Q8S5K1 Putative retroelement [Oryza sativa]
Length = 1888
Score = 224 bits (570), Expect = 8e-57
Identities = 122/333 (36%), Positives = 187/333 (55%), Gaps = 15/333 (4%)
Query: 250 SEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISD 309
S G+ P + +T I LI K ++P+ +K P+SLCNV+YKI+SK L NRL+ IL +S
Sbjct: 1047 SLGTMPSGVNETAIVLIPKTEQPQELKDFRPISLCNVVYKIVSKCLVNRLRPILDDLVSQ 1106
Query: 310 SQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEK 369
+Q+AFVP R I +NAL FE H+++ + A K D+SKA+DR+ W +L+ M K
Sbjct: 1107 NQSAFVPGRLITDNALIAFEYFHHIQRNKNPENAYSAYKLDLSKAYDRVDWEFLEQAMVK 1166
Query: 370 --------KWDSQMFGLT------GSCNV*PIDPHCGIRQGGPFSPYLYIICSEGLNSYI 415
KW L + P G+RQG P SP+L++ ++GL+ +
Sbjct: 1167 LGFAHCWVKWIMACITLVRYAVKLNGTLLNTFAPSRGLRQGDPLSPFLFLFVADGLSFLL 1226
Query: 416 KHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAIN 475
+ + G ++ ICR +P++ LLFADD+ LF+KA + ++K +L TY + +GQ IN
Sbjct: 1227 EEKVDQGAMNPIHICRHAPAVLHLLFADDTPLFFKAANDQAVVMKEVLETYASCTGQLIN 1286
Query: 476 YRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKN 535
K SI F +++ + + I L + S + KYLG P+ R K F ++ER+WK
Sbjct: 1287 PSKCSIMFGQSSPTAVQNQIKQTLQISNS-FEDKYLGFPTPEGRMTKGKFQSLQERLWKR 1345
Query: 536 IQNWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
I W LS GKE+L+K++ Q +P Y +F
Sbjct: 1346 IMLWGEKFLSTGGKEILIKAVLQAMPVYVMGLF 1378
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 224 bits (570), Expect = 8e-57
Identities = 129/331 (38%), Positives = 183/331 (54%), Gaps = 14/331 (4%)
Query: 252 GSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQ 311
GS + TNI LI KI + + M P+SLCNVIYK++ K++ANRLK+IL IS++Q
Sbjct: 478 GSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVIYKVIGKLMANRLKKILPSLISETQ 537
Query: 312 AAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVME--- 368
AAFV R I +N L E+LH + K E IA+K D+SKA+DR++W +L+ M
Sbjct: 538 AAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIKTDISKAYDRVEWPFLEKAMRGLG 597
Query: 369 --KKWDSQMFGLTGS------CNV*P---IDPHCGIRQGGPFSPYLYIICSEGLNSYIKH 417
W + S N P I P G+RQG P SPYL++IC+E L ++
Sbjct: 598 FADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICTEMLVKMLQS 657
Query: 418 HENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYR 477
E I G ++ RG+P I+ LLFADDS + K + I+ Y ASGQ +NY
Sbjct: 658 AEQKNQITGLKVARGAPPISHLLFADDSMFYCKVNDEALGQIIRIIEEYSLASGQRVNYL 717
Query: 478 KSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQ 537
KSSI F ++ + LG+ G YLGLP + K + S++K+R+ K +
Sbjct: 718 KSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLPESFQGSKVATLSYLKDRLGKKVL 777
Query: 538 NWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
W + LS GKE+LLK++A +PTY+ + F
Sbjct: 778 GWQSNFLSPGGKEILLKAVAMALPTYTMSCF 808
>UniRef100_Q9ZQM0 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1044
Score = 220 bits (561), Expect = 8e-56
Identities = 131/334 (39%), Positives = 188/334 (56%), Gaps = 16/334 (4%)
Query: 250 SEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISD 309
S + P + T+I LI KI K M P++LC V YKI+SK+L+ RL+ IL + IS+
Sbjct: 302 SSSTLQPTINKTHITLIPKIQSLKRMVDYRPIALCTVFYKIISKLLSRRLQPILQEIISE 361
Query: 310 SQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEK 369
+Q+AFVP R +N L E LHY+K K +A+K ++SKA+DRI+W +++ VM++
Sbjct: 362 NQSAFVPKRASNDNVLITHEALHYLKSLGAEKRCFMAVKTNMSKAYDRIEWDFIKLVMQE 421
Query: 370 --------KW-------DSQMFGLTGSCNV*PIDPHCGIRQGGPFSPYLYIICSEGLNSY 414
W S F L GS + P G+RQG P SP+L+IICSE L+
Sbjct: 422 MGFHQTWISWILQCITTVSYSFLLNGSAQG-AVTPERGLRQGDPLSPFLFIICSEVLSGL 480
Query: 415 IKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAI 474
+ + G + G R+ +G+P + LLFADD+ F ++ + IL YE ASGQ I
Sbjct: 481 CRKAQLDGSLLGLRVSKGNPRVNHLLFADDTIFFCRSDLKSCKTFLCILKKYEEASGQMI 540
Query: 475 NYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWK 534
N KS+I FSR T +LG+ GKYLGLP M R K+ +F+ I +RI +
Sbjct: 541 NKSKSAITFSRKTPDHIKTEAQQILGIQLVGGLGKYLGLPKMFGRKKRDLFNQIVDRIRQ 600
Query: 535 NIQNWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
+WS+ LS AGK +LKS+ +PTY+ + F
Sbjct: 601 RSLSWSSRFLSTAGKTTMLKSVLASMPTYTMSCF 634
>UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa]
Length = 1557
Score = 217 bits (553), Expect = 7e-55
Identities = 121/310 (39%), Positives = 176/310 (56%), Gaps = 7/310 (2%)
Query: 260 DTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRF 319
DT I LI KI++P+ ++ P+SLCNVIYK+LSK L NRL+ L + IS +Q+AFVP R
Sbjct: 874 DTAIVLIPKIEQPRDLRDFRPISLCNVIYKVLSKCLVNRLRPFLDELISKNQSAFVPRRM 933
Query: 320 ILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEKKWDSQMF-GL 378
I +NAL FE H+++ + A K D+SKA+DR+ W +L+ M K S +
Sbjct: 934 ITDNALLAFECFHFIQRNKNPRSAACAYKLDLSKAYDRVDWRFLEQSMYKWGFSHCWVSW 993
Query: 379 TGSCNV*PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQ 438
+C I C +G P+L++ ++GL+ ++ G I R+C +P I+
Sbjct: 994 VMTC----ISTVCD--KGTRCDPFLFLFVADGLSLLLEEKVEQGAISPIRVCHQAPGISH 1047
Query: 439 LLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRNTDSIGHHNITHL 498
LLFADD+ LF+KA +S+ +K + +Y ++GQ IN K SI F + IT+
Sbjct: 1048 LLFADDTLLFFKADLSQSQAIKEVFGSYATSTGQLINPTKCSILFGNSLPIASRDAITNC 1107
Query: 499 LGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSRAGKEVLLKSIAQ 558
L + + + KYLGLP+ R K F ++ERIWK I W LS GKEVL+K++ Q
Sbjct: 1108 LQIASTEFEDKYLGLPTPGGRMHKGRFQSLRERIWKKILQWGENYLSSGGKEVLIKAVIQ 1167
Query: 559 YIPTYSWAIF 568
IP Y IF
Sbjct: 1168 AIPVYVMGIF 1177
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 216 bits (549), Expect = 2e-54
Identities = 130/331 (39%), Positives = 180/331 (54%), Gaps = 14/331 (4%)
Query: 252 GSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQ 311
G P + T+I LI KI P+ M L P+SLC+V+YKI+SK+L RLK+ L +S +Q
Sbjct: 456 GVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSVLYKIISKILTQRLKKHLPAIVSTTQ 515
Query: 312 AAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVM---- 367
+AFVP R I +N L E++H ++ + + ++A K D+SKA+DR++W +L+ +M
Sbjct: 516 SAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMAFKTDMSKAYDRVEWPFLETMMTALG 575
Query: 368 -EKKWDSQMFGLTGS------CNV*P---IDPHCGIRQGGPFSPYLYIICSEGLNSYIKH 417
KW S + S N P I P GIRQG P SP L+++C+E L +
Sbjct: 576 FNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRGIRQGDPLSPALFVLCTEALIHILNK 635
Query: 418 HENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYR 477
E G I G + S+ LLFADD+ L KA E L L Y SGQ IN
Sbjct: 636 AEQAGKITGIQFQDKKVSVNHLLFADDTLLMCKATKQECEELMQCLSQYGQLSGQMINLN 695
Query: 478 KSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKERIWKNIQ 537
KS+I F +N D I G+ GKYLGLP + K+ +F FIKE++ +
Sbjct: 696 KSAITFGKNVDIQIKDWIKSRSGISLEGGTGKYLGLPECLSGSKRDLFGFIKEKLQSRLT 755
Query: 538 NWSAWSLSRAGKEVLLKSIAQYIPTYSWAIF 568
W A +LS+ GKEVLLKSIA +P Y + F
Sbjct: 756 GWYAKTLSQGGKEVLLKSIALALPVYVMSCF 786
>UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana]
Length = 1270
Score = 213 bits (541), Expect = 2e-53
Identities = 120/342 (35%), Positives = 183/342 (53%), Gaps = 15/342 (4%)
Query: 246 KPIVSEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHK 305
K ++G P T++ LI K P M L P+SLC+V+YKI+SK++A RL+ L +
Sbjct: 435 KKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIISKIMAKRLQPWLPE 494
Query: 306 CISDSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQA 365
+SD+Q+AFV R I +N L E++H +K + +A+K D+SKA+DR++WSYL++
Sbjct: 495 IVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMSKAYDRVEWSYLRS 554
Query: 366 VM-----EKKWDSQMFGLTGSCNV*PIDPHC---------GIRQGGPFSPYLYIICSEGL 411
++ KW + + S + C G+RQG P SP+L+++C+EGL
Sbjct: 555 LLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGL 614
Query: 412 NSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASG 471
+ + G + G + P + LLFADDS KA + +L+ IL Y A+G
Sbjct: 615 THLLNKAQWEGALEGIQFSENGPMVHHLLFADDSLFLCKASREQSLVLQKILKVYGNATG 674
Query: 472 QAINYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKER 531
Q IN KSSI F D I LG+ G YLGLP K + ++K+R
Sbjct: 675 QTINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDR 734
Query: 532 IWKNIQNWSAWSLSRAGKEVLLKSIAQYIPTYSWAIFCSLPL 573
+ + + W LS+ GKEVLLKS+A +P ++ + F LP+
Sbjct: 735 LKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCF-KLPI 775
>UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana]
Length = 1254
Score = 213 bits (541), Expect = 2e-53
Identities = 120/342 (35%), Positives = 183/342 (53%), Gaps = 15/342 (4%)
Query: 246 KPIVSEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHK 305
K ++G P T++ LI K P M L P+SLC+V+YKI+SK++A RL+ L +
Sbjct: 438 KKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIISKIMAKRLQPWLPE 497
Query: 306 CISDSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQA 365
+SD+Q+AFV R I +N L E++H +K + +A+K D+SKA+DR++WSYL++
Sbjct: 498 IVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMSKAYDRVEWSYLRS 557
Query: 366 VM-----EKKWDSQMFGLTGSCNV*PIDPHC---------GIRQGGPFSPYLYIICSEGL 411
++ KW + + S + C G+RQG P SP+L+++C+EGL
Sbjct: 558 LLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGL 617
Query: 412 NSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASG 471
+ + G + G + P + LLFADDS KA + +L+ IL Y A+G
Sbjct: 618 THLLNKAQWEGALEGIQFSENGPMVHHLLFADDSLFLCKASREQSLVLQKILKVYGNATG 677
Query: 472 QAINYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLGLPSMVERDKKSIFSFIKER 531
Q IN KSSI F D I LG+ G YLGLP K + ++K+R
Sbjct: 678 QTINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDR 737
Query: 532 IWKNIQNWSAWSLSRAGKEVLLKSIAQYIPTYSWAIFCSLPL 573
+ + + W LS+ GKEVLLKS+A +P ++ + F LP+
Sbjct: 738 LKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCF-KLPI 778
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.342 0.149 0.502
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 958,451,021
Number of Sequences: 2790947
Number of extensions: 37577831
Number of successful extensions: 122885
Number of sequences better than 10.0: 583
Number of HSP's better than 10.0 without gapping: 379
Number of HSP's successfully gapped in prelim test: 204
Number of HSP's that attempted gapping in prelim test: 121492
Number of HSP's gapped (non-prelim): 831
length of query: 625
length of database: 848,049,833
effective HSP length: 134
effective length of query: 491
effective length of database: 474,062,935
effective search space: 232764901085
effective search space used: 232764901085
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 78 (34.7 bits)
Medicago: description of AC142224.5


