

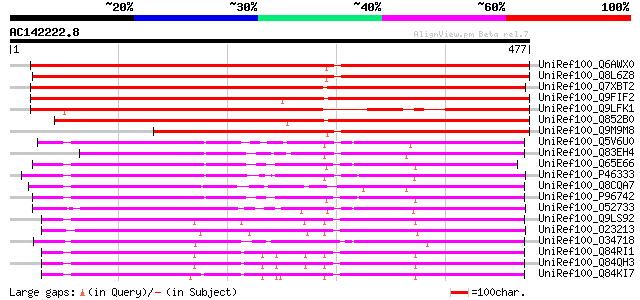
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.8 - phase: 0
(477 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6AWX0 At5g17010 [Arabidopsis thaliana] 649 0.0
UniRef100_Q8L6Z8 Hypothetical protein At3g03090 [Arabidopsis tha... 612 e-174
UniRef100_Q7XBT2 Putative sugar transporter protein [Oryza sativa] 606 e-172
UniRef100_Q9FIF2 Sugar transporter-like protein [Arabidopsis tha... 537 e-151
UniRef100_Q9LFK1 Sugar transporter-like protein [Arabidopsis tha... 491 e-137
UniRef100_Q852B0 Putative sugar transporter protein [Oryza sativa] 484 e-135
UniRef100_Q9M9M8 T17B22.22 protein [Arabidopsis thaliana] 459 e-128
UniRef100_Q5V6U0 Probable metabolite transport protein CsbC [Hal... 263 6e-69
UniRef100_Q83EH4 D-xylose-proton symporter, putative [Coxiella b... 243 9e-63
UniRef100_Q65E66 YwtG [Bacillus licheniformis] 242 2e-62
UniRef100_P46333 Probable metabolite transport protein csbC [Bac... 238 2e-61
UniRef100_Q8CQA7 Bicyclomycin resistance protein TcaB [Staphyloc... 238 3e-61
UniRef100_P96742 YwtG protein [Bacillus subtilis] 238 3e-61
UniRef100_O52733 D-xylose-proton symporter [Lactobacillus brevis] 237 6e-61
UniRef100_Q9LS92 Sugar transporter protein [Arabidopsis thaliana] 231 3e-59
UniRef100_O23213 Sugar transporter like protein [Arabidopsis tha... 229 1e-58
UniRef100_O34718 Metabolite transport protein [Bacillus subtilis] 228 2e-58
UniRef100_Q84RI1 Sorbitol transporter [Malus domestica] 228 3e-58
UniRef100_Q84QH3 Putative sorbitol transporter [Prunus cerasus] 227 5e-58
UniRef100_Q84KI7 Sorbitol transporter [Prunus cerasus] 226 8e-58
>UniRef100_Q6AWX0 At5g17010 [Arabidopsis thaliana]
Length = 503
Score = 649 bits (1675), Expect = 0.0
Identities = 321/461 (69%), Positives = 392/461 (84%), Gaps = 9/461 (1%)
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L FLFPA GGLL+GY+IGATS ATIS+QS SLSGI+WY+L +V++GL+TSGSLYGAL GS
Sbjct: 49 LPFLFPALGGLLYGYEIGATSCATISLQSPSLSGISWYNLSSVDVGLVTSGSLYGALFGS 108
Query: 80 VLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMY 139
++AF IAD +GRR+EL++AAL+YLVGAL+TA AP + +L+IGR+++G+ +GLAMHAAPMY
Sbjct: 109 IVAFTIADVIGRRKELILAALLYLVGALVTALAPTYSVLIIGRVIYGVSVGLAMHAAPMY 168
Query: 140 IAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGM 199
IAETAP+PIRGQLVSLKEFFIV+G+V GYG+GSL V+ +GWRYM+ S P+AVIMG GM
Sbjct: 169 IAETAPSPIRGQLVSLKEFFIVLGMVGGYGIGSLTVNVHSGWRYMYATSVPLAVIMGIGM 228
Query: 200 WWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGE 259
WWLPASPRW+LLR IQ KG+++ ++ AI+SLC L+G F DSA +QV+EI+AE +++GE
Sbjct: 229 WWLPASPRWLLLRVIQGKGNVENQREAAIKSLCCLRGPAFVDSAAEQVNEILAELTFVGE 288
Query: 260 ENDVTLGEMFRGKCRKALVISAGLVLFQQI---QTVTDHGPTKCSLLRCINPTGFSLAAD 316
+ +VT GE+F+GKC KAL+I GLVLFQQI +V + P+ + GFS A D
Sbjct: 289 DKEVTFGELFQGKCLKALIIGGGLVLFQQITGQPSVLYYAPS------ILQTAGFSAAGD 342
Query: 317 ATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLA 376
ATRVSILLG+ KLIMTGVAVVV+DRLGRRPLLLGGV G+V+SLFLLGSYY+F + V+A
Sbjct: 343 ATRVSILLGLLKLIMTGVAVVVIDRLGRRPLLLGGVGGMVVSLFLLGSYYLFFSASPVVA 402
Query: 377 VVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDL 436
VV LLLYVGCYQ+SFGP+GWLMI+EIFPL+LRG+GLS+AVLVNF ANALVTFAFSPLK+L
Sbjct: 403 VVALLLYVGCYQLSFGPIGWLMISEIFPLKLRGRGLSLAVLVNFGANALVTFAFSPLKEL 462
Query: 437 LGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
LGAGILF F I V SLVFI+FIVPETKGLTLEEIEAKCL
Sbjct: 463 LGAGILFCGFGVICVLSLVFIFFIVPETKGLTLEEIEAKCL 503
>UniRef100_Q8L6Z8 Hypothetical protein At3g03090 [Arabidopsis thaliana]
Length = 503
Score = 612 bits (1578), Expect = e-174
Identities = 300/459 (65%), Positives = 377/459 (81%), Gaps = 9/459 (1%)
Query: 22 FLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVL 81
FLFPA G LLFGY+IGATS A +S++S +LSGI+WYDL +V++G++TSGSLYGALIGS++
Sbjct: 51 FLFPALGALLFGYEIGATSCAIMSLKSPTLSGISWYDLSSVDVGIITSGSLYGALIGSIV 110
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIA 141
AF++AD +GRR+EL++AA +YLVGA++T AP F +L+IGR+ +G+GIGL MHAAPMYIA
Sbjct: 111 AFSVADIIGRRKELILAAFLYLVGAIVTVVAPVFSILIIGRVTYGMGIGLTMHAAPMYIA 170
Query: 142 ETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWW 201
ETAP+ IRG+++SLKEF V+G+V GYG+GSL + ++GWRYM+ P VIMG GM W
Sbjct: 171 ETAPSQIRGRMISLKEFSTVLGMVGGYGIGSLWITVISGWRYMYATILPFPVIMGTGMCW 230
Query: 202 LPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEN 261
LPASPRW+LLRA+Q +G+ + L+ AIRSLC+L+G DSA +QV+EI+AE S +GE+
Sbjct: 231 LPASPRWLLLRALQGQGNGENLQQAAIRSLCRLRGSVIADSAAEQVNEILAELSLVGEDK 290
Query: 262 DVTLGEMFRGKCRKALVISAGLVLFQQI---QTVTDHGPTKCSLLRCINPTGFSLAADAT 318
+ T GE+FRGKC KAL I+ GLVLFQQI +V + P+ + GFS AADAT
Sbjct: 291 EATFGELFRGKCLKALTIAGGLVLFQQITGQPSVLYYAPS------ILQTAGFSAAADAT 344
Query: 319 RVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLAVV 378
R+SILLG+ KL+MTGV+V+V+DR+GRRPLLL GVSG+VISLFLLGSYY+F N +AV
Sbjct: 345 RISILLGLLKLVMTGVSVIVIDRVGRRPLLLCGVSGMVISLFLLGSYYMFYKNVPAVAVA 404
Query: 379 GLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLG 438
LLLYVGCYQ+SFGP+GWLMI+EIFPL+LRG+G+S+AVLVNF ANALVTFAFSPLK+LLG
Sbjct: 405 ALLLYVGCYQLSFGPIGWLMISEIFPLKLRGRGISLAVLVNFGANALVTFAFSPLKELLG 464
Query: 439 AGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
AGILF F I V SL FIY+IVPETKGLTLEEIEAKCL
Sbjct: 465 AGILFCAFGVICVVSLFFIYYIVPETKGLTLEEIEAKCL 503
>UniRef100_Q7XBT2 Putative sugar transporter protein [Oryza sativa]
Length = 502
Score = 606 bits (1563), Expect = e-172
Identities = 303/455 (66%), Positives = 369/455 (80%), Gaps = 3/455 (0%)
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L FLFPA GGLL+GYDIGATS ATIS++SS+ SG TWY+L +++ GL+ SGSLYGALIGS
Sbjct: 49 LPFLFPALGGLLYGYDIGATSGATISLKSSTFSGTTWYNLSSLQTGLVVSGSLYGALIGS 108
Query: 80 VLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMY 139
+LAFNIADFLGRRREL+++++ YL+GAL+TA APNFP++V+GR +GIGIGLAMHAAPMY
Sbjct: 109 ILAFNIADFLGRRRELILSSVSYLIGALLTAAAPNFPIMVVGRFFYGIGIGLAMHAAPMY 168
Query: 140 IAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGM 199
IAETAP+ IRG L+SLKEFFIV+G++ GY GSL V+ V+GWRYM+ S+P+ +IMG GM
Sbjct: 169 IAETAPSQIRGMLISLKEFFIVLGMLLGYIAGSLFVEVVSGWRYMYATSTPLCLIMGIGM 228
Query: 200 WWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGE 259
WLPASPRW+LL AIQ K ++ K+ A R LC+L+G+ D +QVD I+ E SY+ +
Sbjct: 229 CWLPASPRWLLLCAIQGKRNIMESKENATRCLCRLRGQASPDLVSEQVDLILDELSYVDQ 288
Query: 260 ENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATR 319
E E+F+GKC KA++I GLV FQQ VT + GFS A+DATR
Sbjct: 289 ERQAGFSEIFQGKCLKAMIIGCGLVFFQQ---VTGQPSVLYYAATILQSAGFSGASDATR 345
Query: 320 VSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLAVVG 379
VS+LLG+ KLIMTGVAV+VVDRLGRRPLL+GGVSGI +SLFLL SYY L +A +AV+
Sbjct: 346 VSVLLGLLKLIMTGVAVLVVDRLGRRPLLIGGVSGIAVSLFLLSSYYTLLKDAPYVAVIA 405
Query: 380 LLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGA 439
LLLYVGCYQ+SFGP+GWLMI+E+FPLRLRG+GLSIAVLVNFA+NALVTFAFSPL+DL+G
Sbjct: 406 LLLYVGCYQLSFGPIGWLMISEVFPLRLRGRGLSIAVLVNFASNALVTFAFSPLEDLIGT 465
Query: 440 GILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
GILF F IAVASLVFI+FIVPETKGLTLEEIEA
Sbjct: 466 GILFSAFGVIAVASLVFIFFIVPETKGLTLEEIEA 500
>UniRef100_Q9FIF2 Sugar transporter-like protein [Arabidopsis thaliana]
Length = 558
Score = 537 bits (1383), Expect = e-151
Identities = 271/461 (58%), Positives = 351/461 (75%), Gaps = 6/461 (1%)
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGS 79
L F+FPA GGLLFGYDIGATS AT+S+QS +LSG TW++ V++GL+ SGSLYGAL+GS
Sbjct: 100 LPFIFPALGGLLFGYDIGATSGATLSLQSPALSGTTWFNFSPVQLGLVVSGSLYGALLGS 159
Query: 80 VLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMY 139
+ + +ADFLGRRREL++AA++YL+G+LIT AP+ +L++GRL++G GIGLAMH AP+Y
Sbjct: 160 ISVYGVADFLGRRRELIIAAVLYLLGSLITGCAPDLNILLVGRLLYGFGIGLAMHGAPLY 219
Query: 140 IAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGM 199
IAET P+ IRG L+SLKE FIV+GI+ G+ +GS +D V GWRYM+G +PVA++MG GM
Sbjct: 220 IAETCPSQIRGTLISLKELFIVLGILLGFSVGSFQIDVVGGWRYMYGFGTPVALLMGLGM 279
Query: 200 WWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQ-VDE--IMAEFSY 256
W LPASPRW+LLRA+Q KG LQ K+ A+ +L +L+GR D ++ VD+ + + +Y
Sbjct: 280 WSLPASPRWLLLRAVQGKGQLQEYKEKAMLALSKLRGRPPGDKISEKLVDDAYLSVKTAY 339
Query: 257 LGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAAD 316
E++ E+F+G KAL I GLVLFQQI T + GFS AAD
Sbjct: 340 EDEKSGGNFLEVFQGPNLKALTIGGGLVLFQQI---TGQPSVLYYAGSILQTAGFSAAAD 396
Query: 317 ATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLA 376
ATRVS+++GVFKL+MT VAV VD LGRRPLL+GGVSGI +SLFLL +YY FL ++A
Sbjct: 397 ATRVSVIIGVFKLLMTWVAVAKVDDLGRRPLLIGGVSGIALSLFLLSAYYKFLGGFPLVA 456
Query: 377 VVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDL 436
V LLLYVGCYQISFGP+ WLM++EIFPLR RG+G+S+AVL NF +NA+VTFAFSPLK+
Sbjct: 457 VGALLLYVGCYQISFGPISWLMVSEIFPLRTRGRGISLAVLTNFGSNAIVTFAFSPLKEF 516
Query: 437 LGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
LGA LF +F IA+ SL+F+ +VPETKGL+LEEIE+K L
Sbjct: 517 LGAENLFLLFGGIALVSLLFVILVVPETKGLSLEEIESKIL 557
>UniRef100_Q9LFK1 Sugar transporter-like protein [Arabidopsis thaliana]
Length = 440
Score = 491 bits (1263), Expect = e-137
Identities = 263/462 (56%), Positives = 326/462 (69%), Gaps = 71/462 (15%)
Query: 20 LRFLFPAFGGLLFGYDIGATSSATISIQSS----SLSGITWYDLDAVEIGLLTSGSLYGA 75
L FLFPA GGLL+GY+IGATS ATIS+Q S + + + ++ +TSGSLYGA
Sbjct: 46 LPFLFPALGGLLYGYEIGATSCATISLQEPMTLLSYYAVPFSAVAFIKWNFMTSGSLYGA 105
Query: 76 LIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHA 135
L GS++AF IAD +GRR+EL++AAL+YLVGAL+TA AP + +L+IGR+++G+ +GLAMHA
Sbjct: 106 LFGSIVAFTIADVIGRRKELILAALLYLVGALVTALAPTYSVLIIGRVIYGVSVGLAMHA 165
Query: 136 APMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIM 195
APMYIAETAP+PIRGQLVSLKEFFIV+G+V GYG+GSL V+ +GWRYM+ S P+AVIM
Sbjct: 166 APMYIAETAPSPIRGQLVSLKEFFIVLGMVGGYGIGSLTVNVHSGWRYMYATSVPLAVIM 225
Query: 196 GFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFS 255
G GMWWLPASPRW+LLR IQ KG+++ ++ AI+SLC L+G F DSA +QV+EI+AE +
Sbjct: 226 GIGMWWLPASPRWLLLRVIQGKGNVENQREAAIKSLCCLRGPAFVDSAAEQVNEILAELT 285
Query: 256 YLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAA 315
++GE+ +VT GE+F+GKC KAL+I GLVLFQQ
Sbjct: 286 FVGEDKEVTFGELFQGKCLKALIIGGGLVLFQQ--------------------------- 318
Query: 316 DATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVL 375
LIMTGVAVVV+DRLGRRPLLLGGV G+ ++ +
Sbjct: 319 -------------LIMTGVAVVVIDRLGRRPLLLGGVGGMRLTSCCC---------SCTA 356
Query: 376 AVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKD 435
A+ GLL EIFPL+LRG+GLS+AVLVNF ANALVTFAFSPLK+
Sbjct: 357 ALCGLL------------------PEIFPLKLRGRGLSLAVLVNFGANALVTFAFSPLKE 398
Query: 436 LLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
LLGAGILF F I V SLVFI+FIVPETKGLTLEEIEAKCL
Sbjct: 399 LLGAGILFCGFGVICVLSLVFIFFIVPETKGLTLEEIEAKCL 440
>UniRef100_Q852B0 Putative sugar transporter protein [Oryza sativa]
Length = 525
Score = 484 bits (1245), Expect = e-135
Identities = 249/440 (56%), Positives = 326/440 (73%), Gaps = 7/440 (1%)
Query: 42 ATISIQ-SSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFLGRRRELLVAAL 100
A +SI S+ LSG TW+ L ++++GL+ SGSLYGAL GS+LA+ +ADFLGRR EL+ AA
Sbjct: 88 APLSISLSAELSGTTWFSLSSIQLGLVASGSLYGALGGSLLAYRVADFLGRRIELVTAAA 147
Query: 101 MYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFI 160
+Y+ GAL+T FAP+F LL+IGRL++GIGIGLAMH AP+YIAET+P+ IRG L+SLKE FI
Sbjct: 148 LYISGALVTGFAPDFVLLIIGRLLYGIGIGLAMHGAPLYIAETSPSRIRGTLISLKELFI 207
Query: 161 VIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDL 220
V+GI+ GY +GSL +D V GWRYMFG +P+AVIM GMW LP SPRW+LLRA+Q K +
Sbjct: 208 VLGILTGYLVGSLEIDVVGGWRYMFGFGAPLAVIMAIGMWNLPPSPRWLLLRAVQGKASV 267
Query: 221 QTLKDTAIRSLCQLQGRTFHDSA-PQQVDEIMAEF--SYLGEENDVTLGEMFRGKCRKAL 277
+ K AI++L L+GR D ++D+ + +Y +E++ + +MF G KAL
Sbjct: 268 EDNKKKAIQALRSLRGRFRSDRVLADEIDDTLLSIKAAYAEQESEGNIWKMFEGASLKAL 327
Query: 278 VISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVV 337
+I GLVLFQQI T + GF+ A+DA +VSIL+G+FKL+MTGVAV
Sbjct: 328 IIGGGLVLFQQI---TGQPSVLYYATSILQTAGFAAASDAAKVSILIGLFKLLMTGVAVF 384
Query: 338 VVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLAVVGLLLYVGCYQISFGPMGWL 397
VD LGRRPLL+GG+ GI +SLFLL +YY L++ +AV LLLYVG YQ+SFGP+ WL
Sbjct: 385 KVDDLGRRPLLIGGIGGIAVSLFLLAAYYKILNSFPFVAVGALLLYVGSYQVSFGPISWL 444
Query: 398 MIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFI 457
M++EIFPLR RG+G+S+AVL NF +NALVTFAFSPL++ LG +F +F AI++ SLVF+
Sbjct: 445 MVSEIFPLRTRGRGISLAVLTNFGSNALVTFAFSPLQEFLGPANIFLLFGAISLLSLVFV 504
Query: 458 YFIVPETKGLTLEEIEAKCL 477
VPETKGLTLEEIE+K L
Sbjct: 505 ILKVPETKGLTLEEIESKLL 524
>UniRef100_Q9M9M8 T17B22.22 protein [Arabidopsis thaliana]
Length = 342
Score = 459 bits (1182), Expect = e-128
Identities = 230/348 (66%), Positives = 280/348 (80%), Gaps = 9/348 (2%)
Query: 133 MHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVA 192
MHAAPMYIAETAP+ IRG+++SLKEF V+G+V GYG+GSL + ++GWRYM+ P
Sbjct: 1 MHAAPMYIAETAPSQIRGRMISLKEFSTVLGMVGGYGIGSLWITVISGWRYMYATILPFP 60
Query: 193 VIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMA 252
VIMG GM WLPASPRW+LLRA+Q +G+ + L+ AIRSLC+L+G DSA +QV+EI+A
Sbjct: 61 VIMGTGMCWLPASPRWLLLRALQGQGNGENLQQAAIRSLCRLRGSVIADSAAEQVNEILA 120
Query: 253 EFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQ---TVTDHGPTKCSLLRCINPT 309
E S +GE+ + T GE+FRGKC KAL I+ GLVLFQQI +V + P+ +
Sbjct: 121 ELSLVGEDKEATFGELFRGKCLKALTIAGGLVLFQQITGQPSVLYYAPS------ILQTA 174
Query: 310 GFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFL 369
GFS AADATR+SILLG+ KL+MTGV+V+V+DR+GRRPLLL GVSG+VISLFLLGSYY+F
Sbjct: 175 GFSAAADATRISILLGLLKLVMTGVSVIVIDRVGRRPLLLCGVSGMVISLFLLGSYYMFY 234
Query: 370 DNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFA 429
N +AV LLLYVGCYQ+SFGP+GWLMI+EIFPL+LRG+G+S+AVLVNF ANALVTFA
Sbjct: 235 KNVPAVAVAALLLYVGCYQLSFGPIGWLMISEIFPLKLRGRGISLAVLVNFGANALVTFA 294
Query: 430 FSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
FSPLK+LLGAGILF F I V SL FIY+IVPETKGLTLEEIEAKCL
Sbjct: 295 FSPLKELLGAGILFCAFGVICVVSLFFIYYIVPETKGLTLEEIEAKCL 342
>UniRef100_Q5V6U0 Probable metabolite transport protein CsbC [Haloarcula marismortui]
Length = 459
Score = 263 bits (673), Expect = 6e-69
Identities = 161/454 (35%), Positives = 247/454 (53%), Gaps = 37/454 (8%)
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNI 85
A GLLFG+D G S A + IQ S + + + G++ SG++ GA G+ + +
Sbjct: 26 ALNGLLFGFDTGIISGAFLFIQDS-------FVMSPLVEGIIVSGAMAGAAAGAAVGGQL 78
Query: 86 ADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAP 145
AD LGRRR +L+AA+++ VG+ A AP P+LV GRL+ G+ IG A P+YI+E AP
Sbjct: 79 ADRLGRRRLILIAAIVFFVGSFTMAVAPTVPVLVAGRLIDGVAIGFASIVGPLYISEIAP 138
Query: 146 TPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPAS 205
IRG L SL + + GI+ Y + D A WR+M G AV++ G+ +P S
Sbjct: 139 PEIRGGLTSLNQLMVTTGILLSYFVNYAFADAGA-WRWMLGAGMVPAVVLAIGILKMPES 197
Query: 206 PRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTL 265
PRW+ +T + A+ RT Q++DEI + +++ +
Sbjct: 198 PRWLFEHG-------RTDEARAVLK------RTRSGGVEQELDEIQ---ETVETQSETGI 241
Query: 266 GEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADATRVSI 322
++ R ALV+ GL +FQQ I V + PT + TG A + ++
Sbjct: 242 WDLLAPWLRPALVVGLGLAVFQQITGINAVIYYAPT------ILESTGLGNVA-SILATV 294
Query: 323 LLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYI---FLDNAAVLAVVG 379
+G ++MT VA+++VDR+GRR LLL GV G+V +L +LG+ + ++A +
Sbjct: 295 GIGTINVVMTVVAIMLVDRVGRRRLLLVGVGGMVATLAVLGTVFYLPGLEGGLGIIATIS 354
Query: 380 LLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGA 439
L+L+V + I GP+ WL+I+EI+PL +RG + + + N+ AN LV+ F L D +G
Sbjct: 355 LMLFVSFFAIGLGPVFWLLISEIYPLSVRGSAMGLVTVANWGANLLVSLTFPVLTDGVGT 414
Query: 440 GILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
F++F ++A LVF+Y VPETKG TLE IE
Sbjct: 415 SATFWLFGLCSLAGLVFVYRYVPETKGRTLEAIE 448
>UniRef100_Q83EH4 D-xylose-proton symporter, putative [Coxiella burnetii]
Length = 409
Score = 243 bits (620), Expect = 9e-63
Identities = 154/415 (37%), Positives = 234/415 (56%), Gaps = 28/415 (6%)
Query: 65 GLLTSGSLYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLV 124
G + S L GA +G++ + ++AD++GR+R L++ AL+++VG I++ + LVIGR++
Sbjct: 3 GFVVSAVLIGAFLGALFSGHLADYIGRKRLLIIDALIFIVGTAISSMTVSISWLVIGRII 62
Query: 125 FGIGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYM 184
GI IG+A ++AP+YI+E +P RG LVSL + + IGI Y + A WR M
Sbjct: 63 VGIAIGIASYSAPLYISEISPPHRRGALVSLNQLAVTIGIFLSYVVDYYFARHDA-WRSM 121
Query: 185 FGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAP 244
F A ++ GM LP SPRWI R ++K A+ L +L+G H A
Sbjct: 122 FAAGVIPAALLLLGMIVLPYSPRWIFSRGHEEK---------ALWILRKLRGHGPH--AE 170
Query: 245 QQVDEIMAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCS 301
Q+++ I A ++ +F R L I+ GL +FQQ I TV + PT
Sbjct: 171 QELEHIRASL----QQQKGDWRTLFSKIIRPTLFIAIGLAVFQQVTGINTVLYYAPT--- 223
Query: 302 LLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFL 361
+ TGF + A ++ +G +I+T +++ ++D LGRRPLL GV + +SL +
Sbjct: 224 ---ILKMTGFQASQTAILATMGIGAVLVIITIISLPLIDSLGRRPLLFIGVGAMTVSLLV 280
Query: 362 LG---SYYIFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLV 418
L + +D +A LL+++ + IS GP+ WLM +EIFPLR+RG G SI
Sbjct: 281 LSWSFKVHGHMDYMRWIAFGSLLVFISGFSISLGPIMWLMFSEIFPLRVRGLGASIGACT 340
Query: 419 NFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
N+A+N LVT F L + LG F+I+ I+V +L+FIY VPETKG+TLE+IE
Sbjct: 341 NWASNWLVTITFLTLIEYLGPSGTFFIYFIISVITLIFIYTSVPETKGVTLEQIE 395
>UniRef100_Q65E66 YwtG [Bacillus licheniformis]
Length = 477
Score = 242 bits (617), Expect = 2e-62
Identities = 150/451 (33%), Positives = 247/451 (54%), Gaps = 37/451 (8%)
Query: 22 FLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVL 81
+ F A GG+L+GYD G S A + ++ L+A GL+ S L GA+ GS L
Sbjct: 11 YFFGALGGVLYGYDTGVISGAILFMKDE-------LGLNAFTEGLVVSAILIGAIFGSGL 63
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIA 141
+ + D GRRR ++ AA++Y +G L TA AP+ +V R+V G+ +G + P+Y++
Sbjct: 64 SGRLTDRFGRRRAIMSAAVLYCIGGLGTALAPSTEYMVAFRIVLGLAVGCSTTIVPLYLS 123
Query: 142 ETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWW 201
E AP RG L SL + I IGI+ Y + D A WR+M G++ ++ + G+++
Sbjct: 124 ELAPKESRGALSSLNQLMITIGILLSYLINYAFSDAGA-WRWMLGLALIPSIGLLIGIFF 182
Query: 202 LPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEN 261
+P SPRW+L + ++K A R L +++G ++VD+ + E +++
Sbjct: 183 MPESPRWLLTKGKEEK---------ARRVLSKMRG-------GERVDQEVKEIKEAEKQD 226
Query: 262 DVTLGEMFRGKCRKALVISAGLVLFQQI---QTVTDHGPTKCSLLRCINPTGFSLAADAT 318
L E+ R AL+ GL QQ T+ + P + GF +A A
Sbjct: 227 QGGLKELLEPWVRPALIAGVGLAFLQQFIGTNTIIYYAP------KTFTNVGFEDSA-AI 279
Query: 319 RVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN---AAVL 375
++ +G ++MT VA+ +DR+GR+PLLL G +G+VISL +L +F N AA
Sbjct: 280 LGTVGIGTVNVLMTLVAIRFIDRIGRKPLLLFGNAGMVISLIVLSFSNLFFGNTSGAAWT 339
Query: 376 AVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKD 435
V+ L +++ + +S+GP+ W+M+ E+FPL +RG G ++ L+ A N +VT +F L +
Sbjct: 340 TVICLGVFIVVFAVSWGPIVWVMLPELFPLHVRGIGTGVSTLMLHAGNLIVTLSFPVLME 399
Query: 436 LLGAGILFYIFSAIAVASLVFIYFIVPETKG 466
+G LF ++AI +A+ +F++F V ETKG
Sbjct: 400 AMGISYLFLCYAAIGIAAFLFVFFKVTETKG 430
>UniRef100_P46333 Probable metabolite transport protein csbC [Bacillus subtilis]
Length = 461
Score = 238 bits (608), Expect = 2e-61
Identities = 153/470 (32%), Positives = 251/470 (52%), Gaps = 38/470 (8%)
Query: 12 LPKTCLKNLRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGS 71
+ K K + + F A GGLL+GYD G S A + I + L + GL+ S
Sbjct: 1 MKKDTRKYMIYFFGALGGLLYGYDTGVISGALLFINND-------IPLTTLTEGLVVSML 53
Query: 72 LYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGL 131
L GA+ GS L+ +D GRR+ + V ++++++GAL AF+ +L+ R++ G+ +G
Sbjct: 54 LLGAIFGSALSGTCSDRWGRRKVVFVLSIIFIIGALACAFSQTIGMLIASRVILGLAVGG 113
Query: 132 AMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPV 191
+ P+Y++E APT IRG L ++ IV GI+ Y + L A WR+M G+++
Sbjct: 114 STALVPVYLSEMAPTKIRGTLGTMNNLMIVTGILLAYIVNYLFTPFEA-WRWMVGLAAVP 172
Query: 192 AVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIM 251
AV++ G+ ++P SPRW++ R +++ R + + HD P+ ++ +
Sbjct: 173 AVLLLIGIAFMPESPRWLVKRGSEEEA----------RRIMNIT----HD--PKDIEMEL 216
Query: 252 AEFSY-LGEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCIN 307
AE E+ + TLG + R L+I GL +FQQ I TV + PT
Sbjct: 217 AEMKQGEAEKKETTLGVLKAKWIRPMLLIGVGLAIFQQAVGINTVIYYAPT------IFT 270
Query: 308 PTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYI 367
G +A A ++ +G+ +IM A++++DR+GR+ LL+ G GI +SL L +
Sbjct: 271 KAGLGTSASALG-TMGIGILNVIMCITAMILIDRVGRKKLLIWGSVGITLSLAALSGVLL 329
Query: 368 FLD---NAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANA 424
L + A + VV L +Y+ YQ ++GP+ W+++ E+FP + RG LV AAN
Sbjct: 330 TLGLSASTAWMTVVFLGVYIVFYQATWGPVVWVLMPELFPSKARGAATGFTTLVLSAANL 389
Query: 425 LVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
+V+ F + +G +F +FS I + S F +++VPETKG +LEEIEA
Sbjct: 390 IVSLVFPLMLSAMGIAWVFMVFSVICLLSFFFAFYMVPETKGKSLEEIEA 439
>UniRef100_Q8CQA7 Bicyclomycin resistance protein TcaB [Staphylococcus epidermidis]
Length = 467
Score = 238 bits (607), Expect = 3e-61
Identities = 150/462 (32%), Positives = 255/462 (54%), Gaps = 38/462 (8%)
Query: 18 KNLRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALI 77
K L F+ A GGLL+GYD G S A + I L++ G++ S L GA++
Sbjct: 26 KYLIFILGALGGLLYGYDNGVISGALLFIHKD-------IPLNSTTEGIVVSSMLIGAIV 78
Query: 78 GSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAP 137
G+ + +AD LGRRR +++ A+++++GALI A + N LL+IGRL+ G+ +G +M P
Sbjct: 79 GAGSSGPLADKLGRRRLVMLIAIVFIIGALILAASTNLALLIIGRLIIGLAVGGSMSTVP 138
Query: 138 MYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGF 197
+Y++E APT RG L SL + I IGI+A Y + D + GWR+M G++ +VI+
Sbjct: 139 VYLSEMAPTEYRGSLGSLNQLMITIGILAAYLVNYAFAD-IEGWRWMLGLAVVPSVILLV 197
Query: 198 GMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYL 257
G++++P SPRW+ L+ + A R + ++ T+ DS + + M E + +
Sbjct: 198 GIYFMPESPRWL----------LENRNEEAARQVMKI---TYDDSEIDKELKEMKEINAI 244
Query: 258 GEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADA 317
E + + G + L++ +FQQ + + + + + F+ A
Sbjct: 245 SESTWTVIKSPWLG---RILIVGCIFAIFQQFIGI--------NAVIFYSSSIFAKAGLG 293
Query: 318 TRVSIL----LGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLG--SYYIFLDN 371
SIL +G +++T VA+ VVD++ R+ LL+GG G++ SL ++ + I + +
Sbjct: 294 EAASILGSVGIGTINVLVTIVAIFVVDKIDRKKLLVGGNIGMIASLLIMAILIWTIGIAS 353
Query: 372 AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFS 431
+A + +V L L++ + IS+GP+ W+M+ E+FP+R RG I+ LV +V+ F
Sbjct: 354 SAWIIIVCLSLFIVFFGISWGPVLWVMLPELFPMRARGAATGISALVLNIGTLIVSLFFP 413
Query: 432 PLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
L D L +F IF+ I V +++F+ +PET+G +LEEIE
Sbjct: 414 ILSDALSTEWVFLIFAFIGVLAMIFVIKFLPETRGRSLEEIE 455
>UniRef100_P96742 YwtG protein [Bacillus subtilis]
Length = 457
Score = 238 bits (607), Expect = 3e-61
Identities = 142/458 (31%), Positives = 250/458 (54%), Gaps = 37/458 (8%)
Query: 22 FLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVL 81
+ F A GG L+GYD G S A + ++ L+A GL+ S L GA++GS
Sbjct: 10 YFFGALGGALYGYDTGVISGAILFMKKE-------LGLNAFTEGLVVSSLLVGAILGSGA 62
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIA 141
A + D GR++ ++ AAL++ +G L A APN ++V+ R++ G+ +G + P+Y++
Sbjct: 63 AGKLTDRFGRKKAIMAAALLFCIGGLGVALAPNTGVMVLFRIILGLAVGTSTTIVPLYLS 122
Query: 142 ETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWW 201
E AP RG L SL + I +GI+ Y + + D A WR+M G+++ ++++ G+ +
Sbjct: 123 ELAPKHKRGALSSLNQLMITVGILLSYIVNYIFADAEA-WRWMLGLAAVPSLLLLIGILF 181
Query: 202 LPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEN 261
+P SPRW+ + K A + L +L+G + +D+ + + +++
Sbjct: 182 MPESPRWLFTNGEESK---------AKKILEKLRG-------TKDIDQEIHDIKEAEKQD 225
Query: 262 DVTLGEMFRGKCRKALVISAGLVLFQQI---QTVTDHGPTKCSLLRCINPTGFSLAADAT 318
+ L E+F R AL+ GL QQ T+ + P + GF +A
Sbjct: 226 EGGLKELFDPWVRPALIAGLGLAFLQQFIGTNTIIYYAP------KTFTNVGFGNSASIL 279
Query: 319 RVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN---AAVL 375
++ +G ++MT VA+ ++D++GR+PLLL G +G+VISL +L +F +N A+
Sbjct: 280 G-TVGIGTVNVLMTLVAIKIIDKIGRKPLLLFGNAGMVISLIVLALVNLFFNNTPAASWT 338
Query: 376 AVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKD 435
V+ L +++ + +S+GP+ W+M+ E+FPL +RG G ++ L+ +V+ + L +
Sbjct: 339 TVICLGVFIVVFAVSWGPVVWVMLPELFPLHVRGIGTGVSTLMLHVGTLIVSLTYPILME 398
Query: 436 LLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+G LF I++AI + + +F+ F V ETKG +LEEIE
Sbjct: 399 AIGISYLFLIYAAIGIMAFLFVRFKVTETKGRSLEEIE 436
>UniRef100_O52733 D-xylose-proton symporter [Lactobacillus brevis]
Length = 457
Score = 237 bits (604), Expect = 6e-61
Identities = 153/462 (33%), Positives = 243/462 (52%), Gaps = 40/462 (8%)
Query: 22 FLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVL 81
+ F A GGLLFGYD G S A + IQ G +W G + S L GA++G+ +
Sbjct: 10 YFFGALGGLLFGYDTGVISGAILFIQKQMNLG-SWQQ------GWVVSAVLLGAILGAAI 62
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIA 141
+D GRR+ LL++A+++ VGAL +AF+P F L+I R++ G+ +G A P Y+A
Sbjct: 63 IGPSSDRFGRRKLLLLSAIIFFVGALGSAFSPEFWTLIISRIILGMAVGAASALIPTYLA 122
Query: 142 ETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWW 201
E AP+ RG + SL + ++ GI+ Y GWR+M G ++ A ++ G
Sbjct: 123 ELAPSDKRGTVSSLFQLMVMTGILLAYITNYSFSGFYTGWRWMLGFAAIPAALLFLGGLI 182
Query: 202 LPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEN 261
LP SPR+++ K G L + L HD + E + + E
Sbjct: 183 LPESPRFLV-----KSGHLDEARHV-------LDTMNKHDQVA-----VNKEINDIQESA 225
Query: 262 DVTLG---EMFRGKCRKALVISAGLVLFQQIQ---TVTDHGPTKCSLLRCINPTGFSLAA 315
+ G E+F R +L+I GL +FQQ+ TV + PT GF ++A
Sbjct: 226 KIVSGGWSELFGKMVRPSLIIGIGLAIFQQVMGCNTVLYYAPT------IFTDVGFGVSA 279
Query: 316 DATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFL---DNA 372
A I +G+F +I+T +AV ++D++ R+ ++ G G+ ISLF++ F A
Sbjct: 280 -ALLAHIGIGIFNVIVTAIAVAIMDKIDRKKIVNIGAVGMGISLFVMSIGMKFSGGSQTA 338
Query: 373 AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSP 432
A+++V+ L +Y+ + ++GP+ W+MI E+FPL +RG G S A ++N+ AN +V+ F
Sbjct: 339 AIISVIALTVYIAFFSATWGPVMWVMIGEVFPLNIRGLGNSFASVINWTANMIVSLTFPS 398
Query: 433 LKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
L D G G LF + + AS+ F+ V ET+ +LE+IEA
Sbjct: 399 LLDFFGTGSLFIGYGILCFASIWFVQKKVFETRNRSLEDIEA 440
>UniRef100_Q9LS92 Sugar transporter protein [Arabidopsis thaliana]
Length = 539
Score = 231 bits (590), Expect = 3e-59
Identities = 146/461 (31%), Positives = 231/461 (49%), Gaps = 30/461 (6%)
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
+L GYDIG S A I I+ ++ ++IG+L +LIGS A +D++
Sbjct: 48 ILLGYDIGVMSGAMIYIKRD-------LKINDLQIGILAGSLNIYSLIGSCAAGRTSDWI 100
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR +++A ++ GA++ +PN+ L+ GR + GIG+G A+ AP+Y AE +P R
Sbjct: 101 GRRYTIVLAGAIFFAGAILMGLSPNYAFLMFGRFIAGIGVGYALMIAPVYTAEVSPASSR 160
Query: 150 GQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
G L S E FI GI+ GY L + GWR M GI + +VI+ G+ +P SPR
Sbjct: 161 GFLNSFPEVFINAGIMLGYVSNLAFSNLPLKVGWRLMLGIGAVPSVILAIGVLAMPESPR 220
Query: 208 WILL--RAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTL 265
W+++ R K L D+ + +L+ P + + + S +
Sbjct: 221 WLVMQGRLGDAKRVLDKTSDSPTEATLRLEDIKHAAGIPADCHDDVVQVSRRNSHGEGVW 280
Query: 266 GEMF---RGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADATR 319
E+ R+ ++ + G+ FQQ I V P R G
Sbjct: 281 RELLIRPTPAVRRVMIAAIGIHFFQQASGIDAVVLFSP------RIFKTAGLKTDHQQLL 334
Query: 320 VSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN-------A 372
++ +GV K VA ++DR+GRRPLLL V G+V+SL LG+ +D A
Sbjct: 335 ATVAVGVVKTSFILVATFLLDRIGRRPLLLTSVGGMVLSLAALGTSLTIIDQSEKKVMWA 394
Query: 373 AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSP 432
V+A+ ++ YV + I GP+ W+ +EIFPLRLR +G S+ V+VN + +++ +F P
Sbjct: 395 VVVAIATVMTYVATFSIGAGPITWVYSSEIFPLRLRSQGSSMGVVVNRVTSGVISISFLP 454
Query: 433 LKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+ + G FY+F IA + VF Y +PET+G LE+++
Sbjct: 455 MSKAMTTGGAFYLFGGIATVAWVFFYTFLPETQGRMLEDMD 495
>UniRef100_O23213 Sugar transporter like protein [Arabidopsis thaliana]
Length = 493
Score = 229 bits (584), Expect = 1e-58
Identities = 146/462 (31%), Positives = 247/462 (52%), Gaps = 30/462 (6%)
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
++FGYD G S A + I+ + + V+I +LT AL+GS+LA +D +
Sbjct: 29 IIFGYDTGVMSGAMVFIEEDLKT-------NDVQIEVLTGILNLCALVGSLLAGRTSDII 81
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR +++A++++++G+++ + PN+P+L+ GR G+G+G A+ AP+Y AE A R
Sbjct: 82 GRRYTIVLASILFMLGSILMGWGPNYPVLLSGRCTAGLGVGFALMVAPVYSAEIATASHR 141
Query: 150 GQLVSLKEFFIVIGIVAGYGLGSLL--VDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
G L SL I IGI+ GY + + GWR M GI++ ++++ FG+ +P SPR
Sbjct: 142 GLLASLPHLCISIGILLGYIVNYFFSKLPMHIGWRLMLGIAAVPSLVLAFGILKMPESPR 201
Query: 208 WILLRAIQKKGD--LQTLKDTAIRSLCQLQG-RTFHDSAPQQVDEIMAEFSYLGEENDVT 264
W++++ K+G L+ + ++ + + Q + P+ VD+++ V
Sbjct: 202 WLIMQGRLKEGKEILELVSNSPEEAELRFQDIKAAAGIDPKCVDDVVKMEGKKTHGEGVW 261
Query: 265 LGEMFRGK--CRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADATR 319
+ R R+ L+ + G+ FQ I+ V +GP R G +
Sbjct: 262 KELILRPTPAVRRVLLTALGIHFFQHASGIEAVLLYGP------RIFKKAGITTKDKLFL 315
Query: 320 VSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAA------ 373
V+I +G+ K A +++D++GRR LLL V G+VI+L +LG NA
Sbjct: 316 VTIGVGIMKTTFIFTATLLLDKVGRRKLLLTSVGGMVIALTMLGFGLTMAQNAGGKLAWA 375
Query: 374 -VLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSP 432
VL++V +V + I GP+ W+ +E+FPL+LR +G S+ V VN NA V+ +F
Sbjct: 376 LVLSIVAAYSFVAFFSIGLGPITWVYSSEVFPLKLRAQGASLGVAVNRVMNATVSMSFLS 435
Query: 433 LKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
L + G F++F+ +A + F +F++PETKG +LEEIEA
Sbjct: 436 LTSAITTGGAFFMFAGVAAVAWNFFFFLLPETKGKSLEEIEA 477
>UniRef100_O34718 Metabolite transport protein [Bacillus subtilis]
Length = 473
Score = 228 bits (582), Expect = 2e-58
Identities = 149/458 (32%), Positives = 240/458 (51%), Gaps = 28/458 (6%)
Query: 23 LFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLA 82
L FGGLLFGYD G + A + +L+A GL+TS L+GA +G+V
Sbjct: 16 LVSTFGGLLFGYDTGVLNGALPYMGEPDQ-----LNLNAFTEGLVTSSLLFGAALGAVFG 70
Query: 83 FNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAE 142
++DF GRR+ +L A+++ + + FAPN +++I R V GI +G A P Y+AE
Sbjct: 71 GRMSDFNGRRKNILFLAVIFFISTIGCTFAPNVTVMIISRFVLGIAVGGASVTVPAYLAE 130
Query: 143 TAPTPIRGQLVSLKEFFIVIGIVAGYG----LGSLLVDTVAGWRYMFGISSPVAVIMGFG 198
+P RG++V+ E IV G + + LG+ + D WR+M I+S A+ + FG
Sbjct: 131 MSPVESRGRMVTQNELMIVSGQLLAFVFNAILGTTMGDNSHVWRFMLVIASLPALFLFFG 190
Query: 199 MWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLG 258
M +P SPRW++ + K+ A+R L +++ A ++ EI F
Sbjct: 191 MIRMPESPRWLVSKG---------RKEDALRVLKKIRD---EKRAAAELQEIEFAFKKED 238
Query: 259 EENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADAT 318
+ T ++ R+ + I G+ + QQI V +LR +GF A A
Sbjct: 239 QLEKATFKDLSVPWVRRIVFIGLGIAIVQQITGVNSIMYYGTEILR---NSGFQTEA-AL 294
Query: 319 RVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLAVV 378
+I GV ++ T V + ++ R+GRRP+L+ G+ G +L L+G + + L+ + L V
Sbjct: 295 IGNIANGVISVLATFVGIWLLGRVGRRPMLMTGLIGTTTALLLIGIFSLVLEGSPALPYV 354
Query: 379 GLLL---YVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKD 435
L L ++ Q + P+ WLM++EIFPLRLRG G+ + V + N V+F F L
Sbjct: 355 VLSLTVTFLAFQQGAISPVTWLMLSEIFPLRLRGLGMGVTVFCLWMVNFAVSFTFPILLA 414
Query: 436 LLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+G F+IF + + S++F+ +PETKGL+LE++E
Sbjct: 415 AIGLSTTFFIFVGLGICSVLFVKRFLPETKGLSLEQLE 452
>UniRef100_Q84RI1 Sorbitol transporter [Malus domestica]
Length = 481
Score = 228 bits (581), Expect = 3e-58
Identities = 147/462 (31%), Positives = 240/462 (51%), Gaps = 33/462 (7%)
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
+L GYDIG S A++ I+ + + V++ ++ +LIGS LA +D++
Sbjct: 4 ILLGYDIGVMSGASLFIKEN-------LKISDVQVEIMNGTLNLYSLIGSALAGRTSDWI 56
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR +++A ++ +GAL+ FAPN+ L+ GR V G+G+G A+ AP+Y AE +P R
Sbjct: 57 GRRYTIVLAGTIFFIGALLMGFAPNYAFLMFGRFVAGVGVGYALMIAPVYTAEISPASFR 116
Query: 150 GQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
G L S E F+ IGI+ GY + WR M G+ + +VI+ G+ +P SPR
Sbjct: 117 GFLTSFPEVFVNIGILLGYVSNYAFSKLPIHLNWRIMLGVGAFPSVILAVGVLAMPESPR 176
Query: 208 WILLRAIQKKGDLQTLKDTAIRSL--CQLQGRTFHDSA--PQQVDEIMAEFSYLGEENDV 263
W++++ + GD + + S+ CQL+ ++A P++ ++ + + S V
Sbjct: 177 WLVMQG--RLGDAKRVLQKTSESIEECQLRLDDIKEAAGIPKESNDDVVQVSKRSHGEGV 234
Query: 264 TLGEMFR--GKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADAT 318
+ R L+ + G+ F+Q I +V + P R G +
Sbjct: 235 WKELLLHPTPAVRHILIAALGIHFFEQSSGIDSVVLYSP------RIFEKAGITSYDHKL 288
Query: 319 RVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN------- 371
++ +GV K I VA V +D+ GRRPLLL V+G+V SL LG+ +D
Sbjct: 289 LATVAVGVVKTICILVATVFLDKFGRRPLLLTSVAGMVFSLSCLGASLTIVDQQHGKIMW 348
Query: 372 AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFS 431
A VL + +LL V + I GP+ W+ +EIFPL+LR +G S+ V VN + +++ F
Sbjct: 349 AIVLCITMVLLNVAFFSIGLGPITWVYSSEIFPLQLRAQGCSMGVAVNRVTSGVISMTFI 408
Query: 432 PLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
L + G F++++ IA VF Y + PET+G TLE++E
Sbjct: 409 SLYKAITIGGAFFLYAGIAAVGWVFFYMLYPETQGRTLEDME 450
>UniRef100_Q84QH3 Putative sorbitol transporter [Prunus cerasus]
Length = 538
Score = 227 bits (579), Expect = 5e-58
Identities = 152/462 (32%), Positives = 238/462 (50%), Gaps = 33/462 (7%)
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
+L GYDIG S A I I+ + VEI +L +LIGS A +D++
Sbjct: 47 ILLGYDIGVMSGAVIYIKKD-------LKVSDVEIEVLVGILNLYSLIGSAAAGRTSDWI 99
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR +++A ++ GAL+ FAPN+ L+ GR V GIG+G A+ AP+Y AE +P R
Sbjct: 100 GRRYTIVLAGAIFFAGALLMGFAPNYAFLMFGRFVAGIGVGYALMIAPVYTAEVSPASSR 159
Query: 150 GQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPR 207
G L S E FI GI+ GY G + T GWR M G+ + ++ + G+ +P SPR
Sbjct: 160 GFLTSFPEVFINAGILFGYVSNYGFSKLPTHLGWRLMLGVGAIPSIFLAIGVLAMPESPR 219
Query: 208 WILLRAIQKKGDLQTLKDTAIRSL--CQLQGRTFHDSA--PQQVDEIMAEFSYLGEENDV 263
W++++ + GD + + D SL +L+ ++A P+ ++ + E + +V
Sbjct: 220 WLVMQG--RLGDARKVLDKTSDSLEESKLRLGEIKEAAGIPEHCNDDIVEVKKRSQGQEV 277
Query: 264 TLGEMFR--GKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAADAT 318
+ R R L+ + GL FQQ I V + P R G +
Sbjct: 278 WKQLLLRPTPAVRHILMCAVGLHFFQQASGIDAVVLYSP------RIFEKAGITNPDHVL 331
Query: 319 RVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN------- 371
++ +G K + VA ++DR+GRRPLLL V+G+V +L LG +D+
Sbjct: 332 LCTVAVGFVKTVFILVATFMLDRIGRRPLLLTSVAGMVFTLACLGLGLTIIDHSGEKIMW 391
Query: 372 AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFS 431
A L++ +L YV + I GP+ W+ +EIFPL+LR +G SI V VN + +++ F
Sbjct: 392 AIALSLTMVLAYVAFFSIGMGPITWVYSSEIFPLQLRAQGCSIGVAVNRVVSGVLSMTFI 451
Query: 432 PLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
L + G F++F+AIA F + ++PET+G TLE++E
Sbjct: 452 SLYKAITIGGAFFLFAAIAAVGWTFFFTMLPETQGRTLEDME 493
>UniRef100_Q84KI7 Sorbitol transporter [Prunus cerasus]
Length = 509
Score = 226 bits (577), Expect = 8e-58
Identities = 155/464 (33%), Positives = 240/464 (51%), Gaps = 37/464 (7%)
Query: 30 LLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADFL 89
+L GYDIG S A+I IQ + VE+ +L +LIGS A +D++
Sbjct: 39 ILLGYDIGVMSGASIYIQKD-------LKISDVEVEILIGILNLYSLIGSAAAGRTSDWI 91
Query: 90 GRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPIR 149
GRR ++ A ++ GAL+ A N+ L++GR V GIG+G A+ AP+Y AE +P R
Sbjct: 92 GRRYTIVFAGAIFFTGALLMGLATNYAFLMVGRFVAGIGVGYALMIAPVYNAEVSPASSR 151
Query: 150 GQLVSLKEFFIVIGI----VAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPAS 205
G L S E F+ IGI VA Y L +D GWR M G+ +VI+ G+ +P S
Sbjct: 152 GALTSFPEVFVNIGILLGYVANYAFSGLPID--LGWRLMLGVGVFPSVILAVGVLSMPES 209
Query: 206 PRWILLRAIQKKGDLQTLKDTAIRSL--CQLQGRTFHDSA--PQQV--DEIMAEFSYLGE 259
PRW++++ + G+ + + D SL QL+ ++A P+ D + GE
Sbjct: 210 PRWLVMQG--RLGEAKQVLDKTSDSLEEAQLRLADIKEAAGIPEHCVEDVVQVPKHSHGE 267
Query: 260 ENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGFSLAAD 316
E L R L+ + G FQQ I + + P R G + ++
Sbjct: 268 EVWKELLLHPTPPVRHILIAAIGFHFFQQLSGIDALVLYSP------RIFEKAGITDSST 321
Query: 317 ATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN----- 371
++ +G K I T VA+ +DR+GRRPLLL V+G++ SL LG+ +D+
Sbjct: 322 LLLATVAVGFSKTIFTLVAIGFLDRVGRRPLLLTSVAGMIASLLCLGTSLTIVDHETEKM 381
Query: 372 --AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFA 429
A+VL + +L YVG + I GP+ W+ +EIFPL+LR +G S+ VN + +++ +
Sbjct: 382 MWASVLCLTMVLAYVGFFSIGMGPIAWVYSSEIFPLKLRAQGCSMGTAVNRIMSGVLSMS 441
Query: 430 FSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
F L + G F++++ IA VF Y ++PET+G TLE++E
Sbjct: 442 FISLYKAITMGGTFFLYAGIATVGWVFFYTMLPETQGRTLEDME 485
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.328 0.145 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 742,610,759
Number of Sequences: 2790947
Number of extensions: 31003127
Number of successful extensions: 131568
Number of sequences better than 10.0: 5916
Number of HSP's better than 10.0 without gapping: 3274
Number of HSP's successfully gapped in prelim test: 2644
Number of HSP's that attempted gapping in prelim test: 117985
Number of HSP's gapped (non-prelim): 9701
length of query: 477
length of database: 848,049,833
effective HSP length: 131
effective length of query: 346
effective length of database: 482,435,776
effective search space: 166922778496
effective search space used: 166922778496
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC142222.8


