

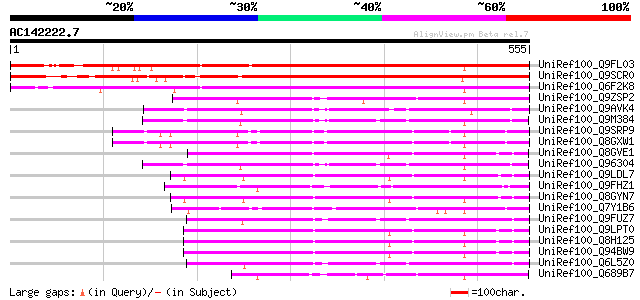
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.7 + phase: 0
(555 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FL03 SCARECROW gene regulator [Arabidopsis thaliana] 594 e-168
UniRef100_Q9SCR0 Scarecrow-like 7 [Arabidopsis thaliana] 510 e-143
UniRef100_Q6F2K8 Putative GRAS family transcription factor [Oryz... 453 e-126
UniRef100_Q9ZSP2 Lateral suppressor protein [Lycopersicon escule... 206 1e-51
UniRef100_Q9AVK4 SCARECROW [Pisum sativum] 198 3e-49
UniRef100_Q9M384 SCARECROW1 [Arabidopsis thaliana] 192 3e-47
UniRef100_Q9SRP9 RGA1-like protein [Arabidopsis thaliana] 191 4e-47
UniRef100_Q8GXW1 Putative RGA1 [Arabidopsis thaliana] 191 4e-47
UniRef100_Q8GVE1 Chitin-inducible gibberellin-responsive protein... 191 5e-47
UniRef100_Q96304 SCARECROW [Arabidopsis thaliana] 190 8e-47
UniRef100_Q9LDL7 SCARECROW gene regulator-like [Arabidopsis thal... 187 7e-46
UniRef100_Q9FHZ1 SCARECROW gene regulator-like protein [Arabidop... 187 7e-46
UniRef100_Q8GYN7 Putative SCARECROW gene regulator [Arabidopsis ... 187 7e-46
UniRef100_Q7Y1B6 GAI-like protein [Lycopersicon esculentum] 186 1e-45
UniRef100_Q9FUZ7 SCARECROW [Zea mays] 186 2e-45
UniRef100_Q9LPT0 Putative transcription factor [Arabidopsis thal... 186 2e-45
UniRef100_Q8H125 Putative scarecrow protein [Arabidopsis thaliana] 186 2e-45
UniRef100_Q94BW9 F17J6.12/F17J6.12 [Arabidopsis thaliana] 184 6e-45
UniRef100_Q6L5Z0 SCARECROW [Oryza sativa] 183 1e-44
UniRef100_Q689B7 Lateral suppressor-like protein [Daucus carota] 181 5e-44
>UniRef100_Q9FL03 SCARECROW gene regulator [Arabidopsis thaliana]
Length = 584
Score = 594 bits (1532), Expect = e-168
Identities = 336/604 (55%), Positives = 409/604 (67%), Gaps = 69/604 (11%)
Query: 1 MAYMCADSGNLMAIAQQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNN 60
MAYMC DSGNLMAIAQQVI QKQQQEQ +QQ HQ H I GI NPLS L+PW N +
Sbjct: 1 MAYMCTDSGNLMAIAQQVIKQKQQQEQQQQQHHQDHQ----IFGI-NPLS--LNPWPNTS 53
Query: 61 IPVSSSTSLPPLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTT----FRFSDF-- 114
+ S S F DPFQV G ++ DP F FP+ H+TT FR SDF
Sbjct: 54 LGFGLSGSA---------FPDPFQVTGGGDSNDPGFPFPNLDHHHATTTGGGFRLSDFGG 104
Query: 115 -------DSDDWMDTLMSAADSHSD------FHLNP---------FSSCPTRLSPQPD-- 150
+SD+WM+TL+S DS +D +H NP F + P+RLS QP
Sbjct: 105 GTGGGEFESDEWMETLISGGDSVADGPDCDTWHDNPDYVIYGPDPFDTYPSRLSVQPSDL 164
Query: 151 -------------LLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTLTEI 197
L P ++PL IP K D T ++ PLLK + +
Sbjct: 165 NRVIDTSSPLPPPTLWPPSSPLSIPPLTHESPTKEDPETNDSEDDDFDLEPPLLKAIYDC 224
Query: 198 ASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQSSIASSNSSSTT 257
A + ++ PN+A +TL + +S+S+ G+P +RV+FYF++AL+N+++ S SS+SSST
Sbjct: 225 ARISDSD-PNEASKTLLQIRESVSELGDPTERVAFYFTEALSNRLSPNSPATSSSSSST- 282
Query: 258 WEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFA 317
E+L LSYK LNDACPYSKFAHLTANQAILEATE SN IHIVDFGIVQGIQW ALLQA A
Sbjct: 283 -EDLILSYKTLNDACPYSKFAHLTANQAILEATEKSNKIHIVDFGIVQGIQWPALLQALA 341
Query: 318 TRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDE 377
TR+SGKP +R+SGIPA +LG SP S+ ATGNRL +FAK+L LNF+F PILTPI LL+
Sbjct: 342 TRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRLRDFAKVLDLNFDFIPILTPIHLLNG 401
Query: 378 SSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGF 437
SSF + PDE LAVNFMLQLY LLDE V+ ALRLAKSLNP++VTLGEYE SL RVGF
Sbjct: 402 SSFRVDPDEVLAVNFMLQLYKLLDETPTIVDTALRLAKSLNPRVVTLGEYEVSL-NRVGF 460
Query: 438 VERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV------RERMEDK 491
R + A +++A FESLEPN+ DS ER +VE L GRRI G+IG RERME+K
Sbjct: 461 ANRVKNALQFYSAVFESLEPNLGRDSEERVRVERELFGRRISGLIGPEKTGIHRERMEEK 520
Query: 492 EQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTV 551
EQW+VLMEN GFESV LS+YA+SQAKILLWNY+YS+LYS+VES+P F+SLAW D+PLLT+
Sbjct: 521 EQWRVLMENAGFESVKLSNYAVSQAKILLWNYNYSNLYSIVESKPGFISLAWNDLPLLTL 580
Query: 552 SSWR 555
SSWR
Sbjct: 581 SSWR 584
>UniRef100_Q9SCR0 Scarecrow-like 7 [Arabidopsis thaliana]
Length = 542
Score = 510 bits (1313), Expect = e-143
Identities = 296/583 (50%), Positives = 377/583 (63%), Gaps = 69/583 (11%)
Query: 1 MAYMCADSGNLMAIAQQVINQKQQQE-QHEQQQHQHHHHHQNILGITNPLSVSLHPWHNN 59
MAYMC DSGNLMAIAQQ+I QKQQQ+ QH+QQ+ Q + PW N
Sbjct: 1 MAYMCTDSGNLMAIAQQLIKQKQQQQSQHQQQEEQEQEPN---------------PWPNP 45
Query: 60 NIPVSSSTSLPPLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDSDDW 119
+ + P + F+DPFQV DP F+FP + +FDSD+W
Sbjct: 46 SFG---------FTLPGSGFSDPFQV-----TNDPGFHFPHLEHHQNAAVASEEFDSDEW 91
Query: 120 MDTLMSAADS---HSDFHL---NPFSSCPTRLSPQPDLLLPA----TAPLQIPTQP---- 165
M++L++ D+ + DF + +PF S P+RLS P L +A Q+P P
Sbjct: 92 MESLINGGDASQTNPDFPIYGHDPFVSFPSRLSA-PSYLNRVNKDDSASQQLPPPPASTA 150
Query: 166 ---PTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQ 222
P+P F + P+ K + + A +KP +TL + +S+S+
Sbjct: 151 IWSPSPPSPQHPPPPPPQPDFDLN-QPIFKAIHDYA-----RKPETKPDTLIRIKESVSE 204
Query: 223 NGNPNQRVSFYFSQALTNKITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTA 282
+G+P QRV +YF++AL++K T + S+SSS++ E+ LSYK LNDACPYSKFAHLTA
Sbjct: 205 SGDPIQRVGYYFAEALSHKETE----SPSSSSSSSLEDFILSYKTLNDACPYSKFAHLTA 260
Query: 283 NQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPV 342
NQAILEAT SNNIHIVDFGI QGIQW+ALLQA ATRSSGKP +RISGIPA +LG SP
Sbjct: 261 NQAILEATNQSNNIHIVDFGIFQGIQWSALLQALATRSSGKPTRIRISGIPAPSLGDSPG 320
Query: 343 SSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDE 402
S+ ATGNRL +FA +L LNFEF P+LTPI+LL+ SSF + PDE L VNFML+LY LLDE
Sbjct: 321 PSLIATGNRLRDFAAILDLNFEFYPVLTPIQLLNGSSFRVDPDEVLVVNFMLELYKLLDE 380
Query: 403 NTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALD 462
+V ALRLA+SLNP+IVTLGEYE SL RV F R + + +++A FESLEPN+ D
Sbjct: 381 TATTVGTALRLARSLNPRIVTLGEYEVSL-NRVEFANRVKNSLRFYSAVFESLEPNLDRD 439
Query: 463 SPERFQVESLLLGRRIDGVI---------GVR-ERMEDKEQWKVLMENCGFESVGLSHYA 512
S ER +VE +L GRRI ++ G R ME+KEQW+VLME GFE V S+YA
Sbjct: 440 SKERLRVERVLFGRRIMDLVRSDDDNNKPGTRFGLMEEKEQWRVLMEKAGFEPVKPSNYA 499
Query: 513 ISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+SQAK+LLWNY+YS+LYSLVES+P F+SLAW +VPLLTVSSWR
Sbjct: 500 VSQAKLLLWNYNYSTLYSLVESEPGFISLAWNNVPLLTVSSWR 542
>UniRef100_Q6F2K8 Putative GRAS family transcription factor [Oryza sativa]
Length = 578
Score = 453 bits (1165), Expect = e-126
Identities = 268/586 (45%), Positives = 348/586 (58%), Gaps = 39/586 (6%)
Query: 1 MAYMCADSGNLMAIAQQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNN 60
MAYMCADSGNLMAIAQQVI Q+QQQ+Q QQQ HHHHH P S+ P H+
Sbjct: 1 MAYMCADSGNLMAIAQQVIQQQQQQQQ--QQQRHHHHHHLP----PPPPPQSMAPHHHQQ 54
Query: 61 IPVSSSTSLP--PLSFPPTDFTDPFQVGSGPENTDPA--------FNFPSPLDPHSTTFR 110
+P P + P + P Q+ G PA F + S
Sbjct: 55 KHHHHHQQMPAMPQAPPSSHGQIPGQLAYGGGAAWPAGEHFFADAFGASAGDAVFSDLAA 114
Query: 111 FSDFDSDDWMDTLMSAAD-SHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQ 169
+DFDSD WM++L+ A SD F++ P + P ATA T P P+
Sbjct: 115 AADFDSDGWMESLIGDAPFQDSDLERLIFTTPPPPVPSPPPTHAAATATATAATAAPRPE 174
Query: 170 RKNDN------------TTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLN 217
++ +S+ +S + P+L++L + T P A L +
Sbjct: 175 AAPALLPQPAAATPVACSSPSPSSADASCSAPILQSLLSCSRAAATD-PGLAAAELASVR 233
Query: 218 KSISQNGNPNQRVSFYFSQALTNKITAQSSIASSNSSSTTW--EELTLSYKALNDACPYS 275
+ + G+P++R++FYF+ AL+ ++ + S + +ELTL YK LNDACPYS
Sbjct: 234 AAATDAGDPSERLAFYFADALSRRLACGTGAPPSAEPDARFASDELTLCYKTLNDACPYS 293
Query: 276 KFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAM 335
KFAHLTANQAILEAT + IHIVDFGIVQGIQWAALLQA ATR GKP +RI+G+P+
Sbjct: 294 KFAHLTANQAILEATGAATKIHIVDFGIVQGIQWAALLQALATRPEGKPTRIRITGVPSP 353
Query: 336 ALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQ 395
LG P +S++AT RL +FAKLLG++FEF P+L P+ L++S F ++PDEA+AVNFMLQ
Sbjct: 354 LLGPQPAASLAATNTRLRDFAKLLGVDFEFVPLLRPVHELNKSDFLVEPDEAVAVNFMLQ 413
Query: 396 LYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESL 455
LY+LL ++ V + LRLAKSL+P +VTLGEYE SL R GFV+RF A +Y+ + FESL
Sbjct: 414 LYHLLGDSDELVRRVLRLAKSLSPAVVTLGEYEVSL-NRAGFVDRFANALSYYRSLFESL 472
Query: 456 EPNMALDSPERFQVESLLLGRRIDGVIGVR------ERMEDKEQWKVLMENCGFESVGLS 509
+ M DSPER +VE + G RI +G ERM +W+ LME CGFE V LS
Sbjct: 473 DVAMTRDSPERVRVERWMFGERIQRAVGPEEGADRTERMAGSSEWQTLMEWCGFEPVPLS 532
Query: 510 HYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+YA SQA +LLWNY YSLVE PAFLSLAW+ PLLTVS+WR
Sbjct: 533 NYARSQADLLLWNYDSKYKYSLVELPPAFLSLAWEKRPLLTVSAWR 578
>UniRef100_Q9ZSP2 Lateral suppressor protein [Lycopersicon esculentum]
Length = 428
Score = 206 bits (524), Expect = 1e-51
Identities = 143/406 (35%), Positives = 219/406 (53%), Gaps = 36/406 (8%)
Query: 175 TTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYF 234
TT TT+ +S + + L A LI + A LT L+ + S G+ +R+ F
Sbjct: 34 TTITTTTITTSPAIQIRQLLISCAELISQSDFSAAKRLLTILSTNSSPFGDSTERLVHQF 93
Query: 235 SQALTNKI------------TAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTA 282
++AL+ ++ T + + +SSS++ + SY +LN P+ +F LTA
Sbjct: 94 TRALSLRLNRYISSTTNHFMTPVETTPTDSSSSSSLALIQSSYLSLNQVTPFIRFTQLTA 153
Query: 283 NQAILEATEGSNN-IHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSP 341
NQAILEA G++ IHIVDF I G+QW L+QA A R ++RI+G +
Sbjct: 154 NQAILEAINGNHQAIHIVDFDINHGVQWPPLMQALADRYPAP--TLRITGT------GND 205
Query: 342 VSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDE-------SSFCIQPDEALAVNFML 394
+ ++ TG+RL++FA LGL F+F P+ D SS + PDE LA+N +
Sbjct: 206 LDTLRRTGDRLAKFAHSLGLRFQFHPLYIANNNHDHDEDPSIISSIVLLPDETLAINCVF 265
Query: 395 QLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFES 454
L+ LL ++ + L KS+NPKIVT+ E EA+ + F++RF A +Y+ A F+S
Sbjct: 266 YLHRLL-KDREKLRIFLHRVKSMNPKIVTIAEKEANHNHPL-FLQRFIEALDYYTAVFDS 323
Query: 455 LEPNMALDSPERFQVESLLLGRRIDGVIGV-----RERMEDKEQWKVLMENCGFESVGLS 509
LE + S ER VE + GR I ++ + +ER E W+V++ +CGF +V LS
Sbjct: 324 LEATLPPGSRERMTVEQVWFGREIVDIVAMEGDKRKERHERFRSWEVMLRSCGFSNVALS 383
Query: 510 HYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+A+SQAK+LL + S Y L S +F L W++ PL ++SSWR
Sbjct: 384 PFALSQAKLLLRLHYPSEGYQLGVSSNSFF-LGWQNQPLFSISSWR 428
>UniRef100_Q9AVK4 SCARECROW [Pisum sativum]
Length = 819
Score = 198 bits (504), Expect = 3e-49
Identities = 138/421 (32%), Positives = 216/421 (50%), Gaps = 26/421 (6%)
Query: 144 RLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIET 203
+L P+ L PAT + ++K + + LL L + A +
Sbjct: 403 QLQQHPEDLAPATTTTTTSAELALARKKKEEIKEQKKKDEEGLH--LLTLLLQCAEAVSA 460
Query: 204 QKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQS----SIASSNSSSTTWE 259
+ QA + L +++ + G QRV+ YFS+A++ ++ + + +S + +
Sbjct: 461 ENLEQANKMLLEISQLSTPFGTSAQRVAAYFSEAISARLVSSCLGIYATLPVSSHTPHNQ 520
Query: 260 ELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATR 319
++ +++ N P+ KF+H TANQAI EA E +HI+D I+QG+QW L A+R
Sbjct: 521 KVASAFQVFNGISPFVKFSHFTANQAIQEAFEREERVHIIDLDIMQGLQWPGLFHILASR 580
Query: 320 SSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESS 379
G P VR++G LGTS + ++ ATG RLS+FA LGL FEF P+ + +D
Sbjct: 581 PGGPP-YVRLTG-----LGTS-METLEATGKRLSDFANKLGLPFEFFPVAEKVGNIDVEK 633
Query: 380 FCIQPDEALAVNFML-QLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFV 438
+ EA+AV+++ LY++ +TN+ L L + L PK+VT+ E L+ F+
Sbjct: 634 LNVSKSEAVAVHWLQHSLYDVTGSDTNT----LWLLQRLAPKVVTV--VEQDLSNAGSFL 687
Query: 439 ERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVRERMEDKE----QW 494
RF A +Y++A F+SL + +S ER VE LL R I V+ V E W
Sbjct: 688 GRFVEAIHYYSALFDSLGSSYGEESEERHVVEQQLLSREIRNVLAVGGPSRSGEIKFHNW 747
Query: 495 KVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 554
+ ++ CGF V L+ A +QA +LL + S Y+LVE L L WKD+ LLT S+W
Sbjct: 748 REKLQQCGFRGVSLAGNAATQASLLLGMFP-SEGYTLVEDN-GILKLGWKDLCLLTASAW 805
Query: 555 R 555
R
Sbjct: 806 R 806
>UniRef100_Q9M384 SCARECROW1 [Arabidopsis thaliana]
Length = 653
Score = 192 bits (487), Expect = 3e-47
Identities = 138/421 (32%), Positives = 215/421 (50%), Gaps = 27/421 (6%)
Query: 143 TRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIE 202
T PQP+ + +Q T +RK + + LL L + A +
Sbjct: 247 TDAPPQPETVTATVPAVQTNTAEALRERKEEIKRQKQDEEGLH----LLTLLLQCAEAVS 302
Query: 203 TQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQ-----SSIASSNSSSTT 257
+A + L +++ + G QRV+ YFS+A++ ++ +++ S T
Sbjct: 303 ADNLEEANKLLLEISQLSTPYGTSAQRVAAYFSEAMSARLLNSCLGIYAALPSRWMPQTH 362
Query: 258 WEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFA 317
++ +++ N P KF+H TANQAI EA E +++HI+D I+QG+QW L A
Sbjct: 363 SLKMVSAFQVFNGISPLVKFSHFTANQAIQEAFEKEDSVHIIDLDIMQGLQWPGLFHILA 422
Query: 318 TRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDE 377
+R G P+ VR++G LGTS + ++ ATG RLS+FA LGL FEF P+ + LD
Sbjct: 423 SRPGGPPH-VRLTG-----LGTS-MEALQATGKRLSDFADKLGLPFEFCPLAEKVGNLDT 475
Query: 378 SSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGF 437
++ EA+AV++ L + L + T S L L + L PK+VT+ E L+ F
Sbjct: 476 ERLNVRKREAVAVHW---LQHSLYDVTGSDAHTLWLLQRLAPKVVTV--VEQDLSHAGSF 530
Query: 438 VERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV----RERMEDKEQ 493
+ RF A +Y++A F+SL + +S ER VE LL + I V+ V R E
Sbjct: 531 LGRFVEAIHYYSALFDSLGASYGEESEERHVVEQQLLSKEIRNVLAVGGPSRSGEVKFES 590
Query: 494 WKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSS 553
W+ M+ CGF+ + L+ A +QA +LL + S Y+LV+ L L WKD+ LLT S+
Sbjct: 591 WREKMQQCGFKGISLAGNAATQATLLLGMFP-SDGYTLVDDN-GTLKLGWKDLSLLTASA 648
Query: 554 W 554
W
Sbjct: 649 W 649
>UniRef100_Q9SRP9 RGA1-like protein [Arabidopsis thaliana]
Length = 547
Score = 191 bits (486), Expect = 4e-47
Identities = 143/470 (30%), Positives = 241/470 (50%), Gaps = 38/470 (8%)
Query: 111 FSDFDSDDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDL-LLPATAPL----QIPTQP 165
++ D +W+++++S ++ + L+ SC R + DL +P + ++ +
Sbjct: 89 YNPSDLSNWVESMLSELNNPASSDLDTTRSCVDR--SEYDLRAIPGLSAFPKEEEVFDEE 146
Query: 166 PTPQR--------KNDNTTTETTSSFSSSTNP-LLKTLTEIASLIETQKPNQAIETLTHL 216
+ +R +D +T S T L+ L A I + N A + +
Sbjct: 147 ASSKRIRLGSWCESSDESTRSVVLVDSQETGVRLVHALVACAEAIHQENLNLADALVKRV 206
Query: 217 NKSISQNGNPNQRVSFYFSQALTNKI----TAQSSIASSNSSSTTWEELTLSYKALNDAC 272
+V+ YF+QAL +I TA++ + ++ + S +EE+ + ++C
Sbjct: 207 GTLAGSQAGAMGKVATYFAQALARRIYRDYTAETDVCAAVNPS--FEEVLEMH--FYESC 262
Query: 273 PYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGI 332
PY KFAH TANQAILEA + +H++D G+ QG+QW AL+QA A R G P S R++GI
Sbjct: 263 PYLKFAHFTANQAILEAVTTARRVHVIDLGLNQGMQWPALMQALALRPGGPP-SFRLTGI 321
Query: 333 PAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILT-PIELLDESSFCIQPD-EALAV 390
T S+ G +L++FA+ +G+ FEF + + L+ F +P+ E L V
Sbjct: 322 GPPQ--TENSDSLQQLGWKLAQFAQNMGVEFEFKGLAAESLSDLEPEMFETRPESETLVV 379
Query: 391 NFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAA 450
N + +L+ LL + S+EK L K++ P IVT+ E EA+ + F++RF A +Y+++
Sbjct: 380 NSVFELHRLL-ARSGSIEKLLNTVKAIKPSIVTVVEQEAN-HNGIVFLDRFNEALHYYSS 437
Query: 451 FFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFES 505
F+SLE + +L S +R E + LGR+I V+ ER E QW++ M++ GF+
Sbjct: 438 LFDSLEDSYSLPSQDRVMSE-VYLGRQILNVVAAEGSDRVERHETAAQWRIRMKSAGFDP 496
Query: 506 VGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+ L A QA +LL Y+ Y VE L + W+ PL+T S+W+
Sbjct: 497 IHLGSSAFKQASMLLSLYATGDGYR-VEENDGCLMIGWQTRPLITTSAWK 545
>UniRef100_Q8GXW1 Putative RGA1 [Arabidopsis thaliana]
Length = 547
Score = 191 bits (486), Expect = 4e-47
Identities = 143/470 (30%), Positives = 241/470 (50%), Gaps = 38/470 (8%)
Query: 111 FSDFDSDDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDL-LLPATAPL----QIPTQP 165
++ D +W+++++S ++ + L+ SC R + DL +P + ++ +
Sbjct: 89 YNPSDLSNWVESMLSELNNPASSDLDTTRSCVDR--SEYDLRAIPGLSAFPKEEEVFDEE 146
Query: 166 PTPQR--------KNDNTTTETTSSFSSSTNP-LLKTLTEIASLIETQKPNQAIETLTHL 216
+ +R +D +T S T L+ L A I + N A + +
Sbjct: 147 ASSKRIRLGSWCESSDESTRSVVLVDSQETGVRLVHALVACAEAIHQENLNLADALVKRV 206
Query: 217 NKSISQNGNPNQRVSFYFSQALTNKI----TAQSSIASSNSSSTTWEELTLSYKALNDAC 272
+V+ YF+QAL +I TA++ + ++ + S +EE+ + ++C
Sbjct: 207 GTLTGSQAGAMGKVATYFAQALARRIYRDYTAETDVCAAVNPS--FEEVLEMH--FYESC 262
Query: 273 PYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGI 332
PY KFAH TANQAILEA + +H++D G+ QG+QW AL+QA A R G P S R++GI
Sbjct: 263 PYLKFAHFTANQAILEAVTTARRVHVIDLGLNQGMQWPALMQALALRPGGPP-SFRLTGI 321
Query: 333 PAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILT-PIELLDESSFCIQPD-EALAV 390
T S+ G +L++FA+ +G+ FEF + + L+ F +P+ E L V
Sbjct: 322 GPPQ--TENSDSLQQLGWKLAQFAQNMGVEFEFKGLAAESLSDLEPEMFETRPESETLVV 379
Query: 391 NFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAA 450
N + +L+ LL + S+EK L K++ P IVT+ E EA+ + F++RF A +Y+++
Sbjct: 380 NSVFELHRLL-ARSGSIEKLLNTVKAIKPSIVTVVEQEAN-HNGIVFLDRFNEALHYYSS 437
Query: 451 FFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFES 505
F+SLE + +L S +R E + LGR+I V+ ER E QW++ M++ GF+
Sbjct: 438 LFDSLEDSYSLPSQDRVMSE-VYLGRQILNVVAAEGSDRVERHETAAQWRIRMKSAGFDP 496
Query: 506 VGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+ L A QA +LL Y+ Y VE L + W+ PL+T S+W+
Sbjct: 497 IHLGSSAFKQASMLLSLYATGDGYR-VEENDGCLMIGWQTRPLITTSAWK 545
>UniRef100_Q8GVE1 Chitin-inducible gibberellin-responsive protein [Oryza sativa]
Length = 544
Score = 191 bits (485), Expect = 5e-47
Identities = 133/375 (35%), Positives = 192/375 (50%), Gaps = 16/375 (4%)
Query: 191 LKTLTEIASLIETQKPNQAIETLT-HLNKSISQNGNPNQRVSFYFSQALTNKITAQS-SI 248
LK L + +K + AI+ + L K +S +G P +R+ Y + L ++ + SI
Sbjct: 174 LKELLIACARAVEEKNSFAIDMMIPELRKIVSVSGEPLERLGAYMVEGLVARLASSGISI 233
Query: 249 ASSNSSSTTWEELTLSYKA-LNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGI 307
+ LSY L +ACPY KF +++AN AI EA +G + IHI+DF I QG
Sbjct: 234 YKALKCKEPKSSDLLSYMHFLYEACPYFKFGYMSANGAIAEAVKGEDRIHIIDFHISQGA 293
Query: 308 QWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTP 367
QW +LLQA A R G P +VRI+GI + + G RLS A L + FEF P
Sbjct: 294 QWISLLQALAARPGGPP-TVRITGIDDSVSAYARGGGLELVGRRLSHIASLCKVPFEFHP 352
Query: 368 ILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDEN---TNSVEKALRLAKSLNPKIVTL 424
+ ++ + + P EALAVNF L+L+++ DE+ N ++ LR+ KSL+PK++TL
Sbjct: 353 LAISGSKVEAAHLGVIPGEALAVNFTLELHHIPDESVSTANHRDRLLRMVKSLSPKVLTL 412
Query: 425 GEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV 484
E E++ T F +RF +Y+ A FES++ + D ER +E L R I +I
Sbjct: 413 VEMESN-TNTAPFPQRFAETLDYYTAIFESIDLTLPRDDRERINMEQHCLAREIVNLIAC 471
Query: 485 R-----ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFL 539
ER E +WK + GF LS + + LL SYS Y L E A L
Sbjct: 472 EGEERAERYEPFGKWKARLTMAGFRPSPLSSLVNATIRTLL--QSYSDNYKLAERDGA-L 528
Query: 540 SLAWKDVPLLTVSSW 554
L WK PL+ S+W
Sbjct: 529 YLGWKSRPLVVSSAW 543
>UniRef100_Q96304 SCARECROW [Arabidopsis thaliana]
Length = 653
Score = 190 bits (483), Expect = 8e-47
Identities = 137/421 (32%), Positives = 214/421 (50%), Gaps = 27/421 (6%)
Query: 143 TRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIE 202
T PQP+ + +Q T +RK + + LL L + A +
Sbjct: 247 TDAPPQPETVTATVPAVQTNTAEALRERKEEIKRQKQDEEGLH----LLTLLLQCAEAVS 302
Query: 203 TQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQ-----SSIASSNSSSTT 257
+A + L +++ + G QRV+ YFS+A++ ++ +++ S T
Sbjct: 303 ADNLEEANKLLLEISQLSTPYGTSAQRVAAYFSEAMSARLLNSCLGIYAALPSRWMPQTH 362
Query: 258 WEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFA 317
++ +++ N P KF+H TANQAI EA E +++HI+D I+QG+QW L A
Sbjct: 363 SLKMVSAFQVFNGISPLVKFSHFTANQAIQEAFEKEDSVHIIDLDIMQGLQWPGLFHILA 422
Query: 318 TRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDE 377
+R G P+ VR++G LGTS + ++ ATG RLS+F LGL FEF P+ + LD
Sbjct: 423 SRPGGPPH-VRLTG-----LGTS-MEALQATGKRLSDFTDKLGLPFEFCPLAEKVGNLDT 475
Query: 378 SSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGF 437
++ EA+AV++ L + L + T S L L + L PK+VT+ E L+ F
Sbjct: 476 ERLNVRKREAVAVHW---LQHSLYDVTGSDAHTLWLLQRLAPKVVTV--VEQDLSHAGSF 530
Query: 438 VERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV----RERMEDKEQ 493
+ RF A +Y++A F+SL + +S ER VE LL + I V+ V R E
Sbjct: 531 LGRFVEAIHYYSALFDSLGASYGEESEERHVVEQQLLSKEIRNVLAVGGPSRSGEVKFES 590
Query: 494 WKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSS 553
W+ M+ CGF+ + L+ A +QA +LL + S Y+LV+ L L WKD+ LLT S+
Sbjct: 591 WREKMQQCGFKGISLAGNAATQATLLLGMFP-SDGYTLVDDN-GTLKLGWKDLSLLTASA 648
Query: 554 W 554
W
Sbjct: 649 W 649
>UniRef100_Q9LDL7 SCARECROW gene regulator-like [Arabidopsis thaliana]
Length = 490
Score = 187 bits (475), Expect = 7e-46
Identities = 131/399 (32%), Positives = 191/399 (47%), Gaps = 21/399 (5%)
Query: 173 DNTTTETTSSFSS-----STNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPN 227
D+T ++ + + S S L L A + A + L + +S +G P
Sbjct: 97 DSTASQEINGWRSTLEAISRRDLRADLVSCAKAMSENDLMMAHSMMEKLRQMVSVSGEPI 156
Query: 228 QRVSFYFSQALTNKITAQSSI---ASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQ 284
QR+ Y + L ++ + S A + EL L + CPY KF +++AN
Sbjct: 157 QRLGAYLLEGLVAQLASSGSSIYKALNRCPEPASTELLSYMHILYEVCPYFKFGYMSANG 216
Query: 285 AILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSS 344
AI EA + N +HI+DF I QG QW L+QAFA R G P +RI+GI M +
Sbjct: 217 AIAEAMKEENRVHIIDFQIGQGSQWVTLIQAFAARPGGPPR-IRITGIDDMTSAYARGGG 275
Query: 345 ISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT 404
+S GNRL++ AK + FEF + + + + ++P EALAVNF L+++ DE+
Sbjct: 276 LSIVGNRLAKLAKQFNVPFEFNSVSVSVSEVKPKNLGVRPGEALAVNFAFVLHHMPDESV 335
Query: 405 ---NSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMAL 461
N ++ LR+ KSL+PK+VTL E E++ T F RF NY+AA FES++ +
Sbjct: 336 STENHRDRLLRMVKSLSPKVVTLVEQESN-TNTAAFFPRFMETMNYYAAMFESIDVTLPR 394
Query: 462 DSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQA 516
D +R VE L R + +I ER E +W+ GF LS S
Sbjct: 395 DHKQRINVEQHCLARDVVNIIACEGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNSTI 454
Query: 517 KILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
K LL N YS Y L E A L L W L+ +W+
Sbjct: 455 KSLLRN--YSDKYRLEERDGA-LYLGWMHRDLVASCAWK 490
>UniRef100_Q9FHZ1 SCARECROW gene regulator-like protein [Arabidopsis thaliana]
Length = 405
Score = 187 bits (475), Expect = 7e-46
Identities = 127/399 (31%), Positives = 207/399 (51%), Gaps = 23/399 (5%)
Query: 166 PTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGN 225
P+ ++ ET + ++ LL L + A + T +A L+ +++ S G+
Sbjct: 16 PSSAKRRIEFPEETLENDGAAAIKLLSLLLQCAEYVATDHLREASTLLSEISEICSPFGS 75
Query: 226 PNQRVSFYFSQALTNKITAQSSIASSNSSSTTWEELTL--------SYKALNDACPYSKF 277
+RV YF+QAL ++ SS S S + + LT+ + + N P KF
Sbjct: 76 SPERVVAYFAQALQTRVI--SSYLSGACSPLSEKPLTVVQSQKIFSALQTYNSVSPLIKF 133
Query: 278 AHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMAL 337
+H TANQAI +A +G +++HI+D ++QG+QW AL A+R K S+RI+G + +
Sbjct: 134 SHFTANQAIFQALDGEDSVHIIDLDVMQGLQWPALFHILASRPR-KLRSIRITGFGSSS- 191
Query: 338 GTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPI-ELLDESSFCIQPDEALAVNFMLQL 396
+++TG RL++FA L L FEF PI I L+D S + EA+ V++M
Sbjct: 192 -----DLLASTGRRLADFASSLNLPFEFHPIEGIIGNLIDPSQLATRQGEAVVVHWMQ-- 244
Query: 397 YNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLE 456
+ L D N++E L + + L P ++T+ E E S F+ RF A +Y++A F++L
Sbjct: 245 HRLYDVTGNNLE-TLEILRRLKPNLITVVEQELSYDDGGSFLGRFVEALHYYSALFDALG 303
Query: 457 PNMALDSPERFQVESLLLGRRIDGVIGVRERMEDKEQWKVLMENCGFESVGLSHYAISQA 516
+ +S ERF VE ++LG I ++ + +WK + GF V L +QA
Sbjct: 304 DGLGEESGERFTVEQIVLGTEIRNIVAHGGGRRKRMKWKEELSRVGFRPVSLRGNPATQA 363
Query: 517 KILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+LL ++ Y+LVE + L L WKD+ LLT S+W+
Sbjct: 364 GLLLGMLPWNG-YTLVE-ENGTLRLGWKDLSLLTASAWK 400
>UniRef100_Q8GYN7 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 411
Score = 187 bits (475), Expect = 7e-46
Identities = 131/399 (32%), Positives = 191/399 (47%), Gaps = 21/399 (5%)
Query: 173 DNTTTETTSSFSS-----STNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPN 227
D+T ++ + + S S L L A + A + L + +S +G P
Sbjct: 18 DSTASQEINGWRSTLEAISRRDLRADLVSCAKAMSENDLMMAHSMMEKLRQMVSVSGEPI 77
Query: 228 QRVSFYFSQALTNKITAQSSI---ASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQ 284
QR+ Y + L ++ + S A + EL L + CPY KF +++AN
Sbjct: 78 QRLGAYLLEGLVAQLASSGSSIYKALNRCPEPASTELLSYMHILYEVCPYFKFGYMSANG 137
Query: 285 AILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSS 344
AI EA + N +HI+DF I QG QW L+QAFA R G P +RI+GI M +
Sbjct: 138 AIAEAMKEENRVHIIDFQIGQGSQWVTLIQAFAARPGGPPR-IRITGIDDMTSAYARGGG 196
Query: 345 ISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT 404
+S GNRL++ AK + FEF + + + + ++P EALAVNF L+++ DE+
Sbjct: 197 LSIVGNRLAKLAKQFNVPFEFNSVSVSVSEVKPKNLGVRPGEALAVNFAFVLHHMPDESV 256
Query: 405 ---NSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMAL 461
N ++ LR+ KSL+PK+VTL E E++ T F RF NY+AA FES++ +
Sbjct: 257 STENHRDRLLRMVKSLSPKVVTLVEQESN-TNTAAFFPRFMETMNYYAAMFESIDVTLPR 315
Query: 462 DSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQA 516
D +R VE L R + +I ER E +W+ GF LS S
Sbjct: 316 DHKQRINVEQHCLARDVVNIIACEGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNSTI 375
Query: 517 KILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
K LL N YS Y L E A L L W L+ +W+
Sbjct: 376 KSLLRN--YSDKYRLEERDGA-LYLGWMHRDLVASCAWK 411
>UniRef100_Q7Y1B6 GAI-like protein [Lycopersicon esculentum]
Length = 588
Score = 186 bits (473), Expect = 1e-45
Identities = 140/417 (33%), Positives = 215/417 (50%), Gaps = 48/417 (11%)
Query: 174 NTTTETTSSFSSSTNP------------LLKTLTEIASLIETQKPNQAIETLTHLN-KSI 220
+TT+ + + SS+T P L+ TL A ++ + A + + H+ ++
Sbjct: 171 STTSSSMVTDSSATRPVVLVDSQETGVRLVHTLMACAEAVQQENLTLADQLVRHIGILAV 230
Query: 221 SQNGNPNQRVSFYFSQALTNKITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHL 280
SQ+G ++V+ YF++AL +I S SS T + L + + + CPY KFAH
Sbjct: 231 SQSG-AMRKVATYFAEALARRIYKIYPQDSMESSYT--DVLQMHFY---ETCPYLKFAHF 284
Query: 281 TANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGI-PAMALGT 339
TANQAILEA G N +H++DF + QG+QW AL+QA A R G P + R++GI P T
Sbjct: 285 TANQAILEAFTGCNKVHVIDFSLKQGMQWPALMQALALRPGGPP-AFRLTGIGPPQPDNT 343
Query: 340 SPVSSISATGNRLSEFAKLLGLNFEFTP-ILTPIELLDESSFCIQPD--EALAVNFMLQL 396
+ + G +L++ A+ +G+ FEF + + LD + I+P EA+A+N + +L
Sbjct: 344 DALQQV---GWKLAQLAETIGVEFEFRGFVANSLADLDATILDIRPSETEAVAINSVFEL 400
Query: 397 YNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLE 456
+ LL ++EK L K +NPKIVTL E EA+ V F++RF A +Y++ F+SLE
Sbjct: 401 HRLL-SRPGAIEKVLNSIKQINPKIVTLVEQEANHNAGV-FIDRFNEALHYYSTMFDSLE 458
Query: 457 ---------PNMALDSP----ERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLM 498
P L P + + + LGR+I V+ ER E QW+V M
Sbjct: 459 SSGSSSSASPTGILPQPPVNNQDLVMSEVYLGRQICNVVACEGSDRVERHETLNQWRVRM 518
Query: 499 ENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+ GF+ V L A QA +LL ++ Y VE L L W PL+ S+W+
Sbjct: 519 NSSGFDPVHLGSNAFKQASMLLALFAGGDGYR-VEENDGCLMLGWHTRPLIATSAWK 574
>UniRef100_Q9FUZ7 SCARECROW [Zea mays]
Length = 668
Score = 186 bits (472), Expect = 2e-45
Identities = 128/376 (34%), Positives = 196/376 (52%), Gaps = 24/376 (6%)
Query: 190 LLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQSS-- 247
LL L + A + + A +TL + + + G QRV+ YF++A++ ++ +
Sbjct: 299 LLTLLLQCAEAVNADNLDDAHQTLLEIAELATPFGTSTQRVAAYFAEAMSARLVSSCLGL 358
Query: 248 ----IASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGI 303
S +++ + +++ N P+ KF+H TANQAI EA E +HI+D I
Sbjct: 359 YAPLPPGSPAAARLHGRVAAAFQVFNGISPFVKFSHFTANQAIQEAFEREERVHIIDLDI 418
Query: 304 VQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNF 363
+QG+QW L A+R G P VR++G+ A + ++ ATG RLS+FA LGL F
Sbjct: 419 MQGLQWPGLFHILASRPGGPPR-VRLTGLGA------SMEALEATGKRLSDFADTLGLPF 471
Query: 364 EFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVT 423
EF + +D + EA+AV++ L++ L + T S L L + L PK+VT
Sbjct: 472 EFCAVAEKAGNVDPEKLGVTRREAVAVHW---LHHSLYDVTGSDSNTLWLIQRLAPKVVT 528
Query: 424 LGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGV-- 481
+ E L+ F+ RF A +Y++A F+SL+ + DSPER VE LL R I V
Sbjct: 529 M--VEQDLSHSGSFLARFVEAIHYYSALFDSLDASYGEDSPERHVVEQQLLSREIRNVLA 586
Query: 482 IGVRERMEDKE--QWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFL 539
+G R D + W+ + GF + L+ A +QA +LL + S Y+LVE A L
Sbjct: 587 VGGPARTGDVKFGSWREKLAQSGFRAASLAGSAAAQASLLLGMFP-SDGYTLVEENGA-L 644
Query: 540 SLAWKDVPLLTVSSWR 555
L WKD+ LLT S+WR
Sbjct: 645 KLGWKDLCLLTASAWR 660
>UniRef100_Q9LPT0 Putative transcription factor [Arabidopsis thaliana]
Length = 526
Score = 186 bits (471), Expect = 2e-45
Identities = 129/380 (33%), Positives = 187/380 (48%), Gaps = 15/380 (3%)
Query: 186 STNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQ 245
S L L E A +E ++ L + +S +G P QR+ Y + L ++ +
Sbjct: 152 SRGDLKGVLYECAKAVENYDLEMTDWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASS 211
Query: 246 SS--IASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGI 303
S + T EL L +ACPY KF + +AN AI EA + + +HI+DF I
Sbjct: 212 GSSIYKALRCKDPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQI 271
Query: 304 VQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNF 363
QG QW +L++A R G PN VRI+GI + + G RL + A++ G+ F
Sbjct: 272 SQGGQWVSLIRALGARPGGPPN-VRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPF 330
Query: 364 EFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPK 420
EF ++ ++ EALAVNF L L+++ DE+ N ++ LRL K L+P
Sbjct: 331 EFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPN 390
Query: 421 IVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDG 480
+VTL E EA+ T F+ RF N++ A FES++ +A D ER VE L R +
Sbjct: 391 VVTLVEQEAN-TNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVN 449
Query: 481 VIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQ 535
+I ER E +W+ GF+ LS Y + K LL SYS Y+L E
Sbjct: 450 LIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLL--ESYSEKYTLEERD 507
Query: 536 PAFLSLAWKDVPLLTVSSWR 555
A L L WK+ PL+T +WR
Sbjct: 508 GA-LYLGWKNQPLITSCAWR 526
>UniRef100_Q8H125 Putative scarecrow protein [Arabidopsis thaliana]
Length = 597
Score = 186 bits (471), Expect = 2e-45
Identities = 129/380 (33%), Positives = 187/380 (48%), Gaps = 15/380 (3%)
Query: 186 STNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQ 245
S L L E A +E ++ L + +S +G P QR+ Y + L ++ +
Sbjct: 223 SRGDLKGVLYECAKAVENYDLEMTDWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASS 282
Query: 246 SS--IASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGI 303
S + T EL L +ACPY KF + +AN AI EA + + +HI+DF I
Sbjct: 283 GSSIYKALRCKDPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQI 342
Query: 304 VQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNF 363
QG QW +L++A R G PN VRI+GI + + G RL + A++ G+ F
Sbjct: 343 SQGGQWVSLIRALGARPGGPPN-VRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPF 401
Query: 364 EFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPK 420
EF ++ ++ EALAVNF L L+++ DE+ N ++ LRL K L+P
Sbjct: 402 EFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPN 461
Query: 421 IVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDG 480
+VTL E EA+ T F+ RF N++ A FES++ +A D ER VE L R +
Sbjct: 462 VVTLVEQEAN-TNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVN 520
Query: 481 VIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQ 535
+I ER E +W+ GF+ LS Y + K LL SYS Y+L E
Sbjct: 521 LIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLL--ESYSEKYTLEERD 578
Query: 536 PAFLSLAWKDVPLLTVSSWR 555
A L L WK+ PL+T +WR
Sbjct: 579 GA-LYLGWKNQPLITSCAWR 597
>UniRef100_Q94BW9 F17J6.12/F17J6.12 [Arabidopsis thaliana]
Length = 526
Score = 184 bits (467), Expect = 6e-45
Identities = 128/380 (33%), Positives = 187/380 (48%), Gaps = 15/380 (3%)
Query: 186 STNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQ 245
S L L E A +E ++ L + +S +G P QR+ Y + L ++ +
Sbjct: 152 SRGDLKGVLYECAKAVENYDLEMTDWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASS 211
Query: 246 SS--IASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGI 303
S + T EL L +ACPY KF + +AN AI EA + + +HI+DF I
Sbjct: 212 GSSIYKALRCKDPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQI 271
Query: 304 VQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNF 363
QG QW +L++A R G PN VRI+GI + + G RL + A++ G+ F
Sbjct: 272 SQGGQWVSLIRALGARPGGPPN-VRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPF 330
Query: 364 EFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPK 420
EF ++ ++ EALAVNF L L+++ DE+ N ++ LRL K L+P
Sbjct: 331 EFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPN 390
Query: 421 IVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDG 480
+VTL E EA+ T F+ RF N++ A FES++ +A D ER VE L R +
Sbjct: 391 VVTLVEQEAN-TNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVN 449
Query: 481 VIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQ 535
+I ER E +W+ GF+ LS Y + + LL SYS Y+L E
Sbjct: 450 LIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIEGLL--ESYSEKYTLEERD 507
Query: 536 PAFLSLAWKDVPLLTVSSWR 555
A L L WK+ PL+T +WR
Sbjct: 508 GA-LYLGWKNQPLITSCAWR 526
>UniRef100_Q6L5Z0 SCARECROW [Oryza sativa]
Length = 660
Score = 183 bits (464), Expect = 1e-44
Identities = 128/376 (34%), Positives = 195/376 (51%), Gaps = 24/376 (6%)
Query: 190 LLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQSSIA 249
LL L + A + ++A L + + + G QRV+ YF++A++ ++ +
Sbjct: 292 LLTLLLQCAESVNADNLDEAHRALLEIAELATPFGTSTQRVAAYFAEAMSARLVSSCLGL 351
Query: 250 ------SSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGI 303
S +++ + +++ N P+ KF+H TANQAI EA E +HI+D I
Sbjct: 352 YAPLPNPSPAAARLHGRVAAAFQVFNGISPFVKFSHFTANQAIQEAFEREERVHIIDLDI 411
Query: 304 VQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNF 363
+QG+QW L A+R G P VR++G+ A + ++ ATG RLS+FA LGL F
Sbjct: 412 MQGLQWPGLFHILASRPGGPPR-VRLTGLGA------SMEALEATGKRLSDFADTLGLPF 464
Query: 364 EFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVT 423
EF P+ LD + EA+AV++ L + L + T S L L + L PK+VT
Sbjct: 465 EFCPVADKAGNLDPEKLGVTRREAVAVHW---LRHSLYDVTGSDSNTLWLIQRLAPKVVT 521
Query: 424 LGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGV-- 481
+ E L+ F+ RF A +Y++A F+SL+ + + DSPER VE LL R I V
Sbjct: 522 M--VEQDLSHSGSFLARFVEAIHYYSALFDSLDASYSEDSPERHVVEQQLLSREIRNVLA 579
Query: 482 IGVRERMEDKE--QWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFL 539
+G R D + W+ + GF L+ A +QA +LL + S Y+L+E A L
Sbjct: 580 VGGPARTGDVKFGSWREKLAQSGFRVSSLAGSAAAQAVLLLGMFP-SDGYTLIEENGA-L 637
Query: 540 SLAWKDVPLLTVSSWR 555
L WKD+ LLT S+WR
Sbjct: 638 KLGWKDLCLLTASAWR 653
>UniRef100_Q689B7 Lateral suppressor-like protein [Daucus carota]
Length = 431
Score = 181 bits (459), Expect = 5e-44
Identities = 122/336 (36%), Positives = 189/336 (55%), Gaps = 34/336 (10%)
Query: 238 LTNKITAQ--SSIASSNSSSTTWEELTL---SYKALNDACPYSKFAHLTANQAILEATEG 292
LT KI+ SS S + S +++ ++ +Y +LN P+ +F HLTANQAILE+ EG
Sbjct: 110 LTPKISPALPSSAGSGSGSQQVFDDESVVQSAYLSLNQITPFIRFTHLTANQAILESVEG 169
Query: 293 SNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRL 352
+ IHI+DF I+ G+QW L+QA A + P +RI+G ++ + TG+RL
Sbjct: 170 HHAIHILDFNIMHGVQWPPLMQAMAEKF--PPPMLRITGT------GDNLTILRRTGDRL 221
Query: 353 SEFAKLLGLNFEFTPILTPIELLDESSFC---------IQPDEALAVNFMLQLYNLLDEN 403
++FA LGL F+F P+L + +ESS +QPD+ LAVN +L L+ L +
Sbjct: 222 AKFAHTLGLRFQFHPVL--LLENEESSITSFFASFAAYLQPDQTLAVNCVLYLHRL---S 276
Query: 404 TNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDS 463
+ L K+LNP+++TL E EA+ + F++RF A +++ A F+SLE + +S
Sbjct: 277 LERLSLCLHQIKALNPRVLTLSEREANHNLPI-FLQRFVEALDHYTALFDSLEATLPPNS 335
Query: 464 PERFQVESLLLGRRIDGVIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKI 518
+R +VE + GR I +I RER E W++++ GF ++ LS +A+SQAK+
Sbjct: 336 RQRIEVEQIWFGREIADIIASEGETRRERHERFRAWELMLRGSGFHNLALSPFALSQAKL 395
Query: 519 LLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 554
LL Y S Y L +F W++ L +VSSW
Sbjct: 396 LLRLYYPSEGYKLHILNDSFF-WGWQNQHLFSVSSW 430
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.129 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 921,922,730
Number of Sequences: 2790947
Number of extensions: 40027170
Number of successful extensions: 292879
Number of sequences better than 10.0: 1521
Number of HSP's better than 10.0 without gapping: 678
Number of HSP's successfully gapped in prelim test: 914
Number of HSP's that attempted gapping in prelim test: 265416
Number of HSP's gapped (non-prelim): 12613
length of query: 555
length of database: 848,049,833
effective HSP length: 133
effective length of query: 422
effective length of database: 476,853,882
effective search space: 201232338204
effective search space used: 201232338204
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC142222.7


