

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141923.7 - phase: 0
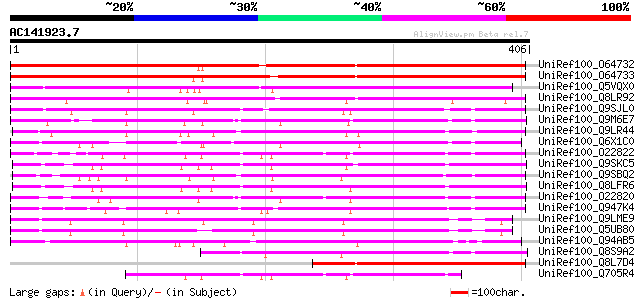
(406 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O64732 Putative glucosyltransferase [Arabidopsis thali... 377 e-103
UniRef100_O64733 Putative glucosyltransferase [Arabidopsis thali... 371 e-101
UniRef100_Q5VQX0 Putative UDP-glucose:salicylic acid glucosyltra... 306 5e-82
UniRef100_Q8LR92 Putative UDP-glucose:salicylic acid glucosyltra... 261 2e-68
UniRef100_Q9SJL0 Putative glucosyltransferase [Arabidopsis thali... 193 6e-48
UniRef100_Q9M6E7 UDP-glucose:salicylic acid glucosyltransferase ... 192 2e-47
UniRef100_Q9LR44 T25N20.21 [Arabidopsis thaliana] 191 4e-47
UniRef100_Q6X1C0 Glucosyltransferase 2 [Crocus sativus] 187 4e-46
UniRef100_O22822 Putative glucosyltransferase [Arabidopsis thali... 187 6e-46
UniRef100_Q9SKC5 Putative glucosyltransferase [Arabidopsis thali... 185 2e-45
UniRef100_Q9SBQ2 Anthocyanin 5-O-glucosyltransferase [Petunia hy... 184 5e-45
UniRef100_Q8LFR6 Putative glucosyltransferase [Arabidopsis thali... 183 9e-45
UniRef100_O22820 Putative glucosyltransferase [Arabidopsis thali... 175 2e-42
UniRef100_Q947K4 Thiohydroximate S-glucosyltransferase [Brassica... 173 9e-42
UniRef100_Q9LME9 T16E15.4 protein [Arabidopsis thaliana] 171 3e-41
UniRef100_Q5UB80 UDP-glucuronosyltransferase [Arabidopsis thaliana] 170 6e-41
UniRef100_Q94AB5 AT3g46660/F12A12_180 [Arabidopsis thaliana] 163 9e-39
UniRef100_Q8S9A2 Glucosyltransferase-7 [Phaseolus angularis] 157 7e-37
UniRef100_Q8L7D4 Putative glucosyltransferase [Arabidopsis thali... 156 1e-36
UniRef100_Q705R4 Glycosyltransferase [Arabidopsis thaliana] 152 2e-35
>UniRef100_O64732 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 440
Score = 377 bits (968), Expect = e-103
Identities = 193/442 (43%), Positives = 273/442 (61%), Gaps = 45/442 (10%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
MP+P RG+INPM+NLCK LV + N+ V+FVVTEEWL FI S+PKP+ I F + N+IPS
Sbjct: 1 MPWPGRGHINPMLNLCKSLVRRDPNLTVTFVVTEEWLGFIGSDPKPNRIHFATLPNIIPS 60
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVANRRNIPAALF 120
EL+ D AF++ V+T++E PFE+LLD L+ PP+ I+ DT + WAV V +RNIP A F
Sbjct: 61 ELVRANDFIAFIDAVLTRLEEPFEQLLDRLNSPPTAIIADTYIIWAVRVGTKRNIPVASF 120
Query: 121 WPMPASIFSVFLHQHIFEQNGHYPVK------------YPG------------------- 149
W A+I S+F++ + +GH+P++ PG
Sbjct: 121 WTTSATILSLFINSDLLASHGHFPIEPSESKLDEIVDYIPGLSPTRLSDLQILHGYSHQV 180
Query: 150 -------FEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKP 202
F ++KA+YLLF S YELE +AID SK P+Y+ GP IP L
Sbjct: 181 FNIFKKSFGELYKAKYLLFPSAYELEPKAIDFFTSKFDFPVYSTGPLIPLEEL-----SV 235
Query: 203 SNTNHSY-YIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEAS 261
N N Y +WLD QP SVLYI+QGSF SVS AQ++EI + + V+F W+AR
Sbjct: 236 GNENRELDYFKWLDEQPESSVLYISQGSFLSVSEAQMEEIVVGVREAGVKFFWVARGGEL 295
Query: 262 RLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQ 321
+LKE +G+++ WCDQLRVL H +IGGFW+HCG+NST E + +GVP LT P++ DQ
Sbjct: 296 KLKEALEGS-LGVVVSWCDQLRVLCHAAIGGFWTHCGYNSTLEGICSGVPLLTFPVFWDQ 354
Query: 322 PFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKI 381
N+KM+VE+W+VG ++ + + L+ D+I +LV FMD + E +++R R+ L +I
Sbjct: 355 FLNAKMIVEEWRVGMGIERKKQMELLIVSDEIKELVKRFMDGESEEGKEMRRRTCDLSEI 414
Query: 382 CLNSIANGGSAHTDFNAFISDV 403
C ++A GGS+ + +AFI D+
Sbjct: 415 CRGAVAKGGSSDANIDAFIKDI 436
>UniRef100_O64733 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 455
Score = 371 bits (953), Expect = e-101
Identities = 195/441 (44%), Positives = 272/441 (61%), Gaps = 44/441 (9%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
MP+P RG+INPMMNLCK LV N+HV+FVVTEEWL FI +PKPD I F + N+IPS
Sbjct: 17 MPYPGRGHINPMMNLCKRLVRRYPNLHVTFVVTEEWLGFIGPDPKPDRIHFSTLPNLIPS 76
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPP-SIIVYDTLLYWAVVVANRRNIPAAL 119
EL+ +D F++ V T++E PFE+LLD L+ PP S+I DT + WAV V +RNIP
Sbjct: 77 ELVRAKDFIGFIDAVYTRLEEPFEKLLDSLNSPPPSVIFADTYVIWAVRVGRKRNIPVVS 136
Query: 120 FWPMPASIFSVFLHQHIFEQNGH------------------------YPVKYPG------ 149
W M A+I S FLH + +GH P + G
Sbjct: 137 LWTMSATILSFFLHSDLLISHGHALFEPSEEEVVDYVPGLSPTKLRDLPPIFDGYSDRVF 196
Query: 150 ------FEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSL-IKNDPKP 202
F+ + A+ LLF++ YELE +AID SKL +P+Y IGP IP L ++ND K
Sbjct: 197 KTAKLCFDELPGARSLLFTTAYELEHKAIDAFTSKLDIPVYAIGPLIPFEELSVQNDNKE 256
Query: 203 SNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASR 262
N YI+WL+ QP GSVLYI+QGSF SVS AQ++EI L S VRFLW+AR +
Sbjct: 257 PN-----YIQWLEEQPEGSVLYISQGSFLSVSEAQMEEIVKGLRESGVRFLWVARGGELK 311
Query: 263 LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQP 322
LKE +G+++ WCDQLRVL H ++GGFW+HCG+NST E + +GVP L P++ DQ
Sbjct: 312 LKEALEGS-LGVVVSWCDQLRVLCHKAVGGFWTHCGFNSTLEGIYSGVPMLAFPLFWDQI 370
Query: 323 FNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKIC 382
N+KM+VEDW+VG R++ K + L+ +++I ++V FMD + E +++R R+ L +I
Sbjct: 371 LNAKMIVEDWRVGMRIERTKKNELLIGREEIKEVVKRFMDRESEEGKEMRRRACDLSEIS 430
Query: 383 LNSIANGGSAHTDFNAFISDV 403
++A GS++ + + F+ +
Sbjct: 431 RGAVAKSGSSNVNIDEFVRHI 451
>UniRef100_Q5VQX0 Putative UDP-glucose:salicylic acid glucosyltransferase [Oryza
sativa]
Length = 482
Score = 306 bits (785), Expect = 5e-82
Identities = 177/449 (39%), Positives = 252/449 (55%), Gaps = 58/449 (12%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+PFP RG++N MMNL +LL + + V+FVVTEEWL +SS P + R+ NVIPS
Sbjct: 22 VPFPGRGHVNAMMNLSRLLAARGAAT-VTFVVTEEWLGLLSSSSAPPGVRLRAIPNVIPS 80
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLLD--------HPPSIIVYDTLLYWAVVVANR 112
E DH F++ V +MEAPFE LLD L P + V D + W V V NR
Sbjct: 81 ENGRAADHAGFLDAVGARMEAPFERLLDRLRLEEEEETAAPVAAFVADFYVPWVVDVGNR 140
Query: 113 RNIPAALFWPMPASIFSVFL-------------HQHIF--------EQNGHY-------- 143
R +P +PM A FS + HQ + ++ HY
Sbjct: 141 RGVPVCSLFPMAAVFFSAYYHFDSLPSWLAKPPHQPVAGATTDNPDQRLEHYISSLASSS 200
Query: 144 --------------PVKY--PGFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIG 187
V+Y I KAQ LLF++IYELE+ ID L+S + P+Y IG
Sbjct: 201 IMLSDLKPLIHSERTVEYILACISSIRKAQCLLFTTIYELEASVIDSLESLVTCPVYPIG 260
Query: 188 PTIPKFSLIKNDPKPSN---TNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAA 244
P IP +L +N+ SN Y WLD QP SVLY++ GSF SVSS+Q+DEIA
Sbjct: 261 PCIPYMTL-ENEHTKSNGEAPGRIDYFAWLDCQPENSVLYVSLGSFVSVSSSQLDEIALG 319
Query: 245 LCASNVRFLWIARSEASRLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKE 304
L S VRFLWI R +++R++E+ G + G+I+ WCDQL+VL HPS+GGF +HCG NST E
Sbjct: 320 LATSEVRFLWILREQSTRVRELVGNTNKGMILPWCDQLKVLCHPSVGGFLTHCGMNSTLE 379
Query: 305 SLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLD 364
++ AGVP LTLP++ DQP N +++VE+WK+G +++ +D L+++++I + V M +
Sbjct: 380 AVFAGVPMLTLPLFFDQPINGRLIVEEWKIGVNLRDSTDKDRLIRREEIARAVKRLMASE 439
Query: 365 GELTRDIRERSKKLQKICLNSIANGGSAH 393
+ IR + + ++I ++ G S+H
Sbjct: 440 EAEMKAIRRHALEWKEISHRAVDKGVSSH 468
>UniRef100_Q8LR92 Putative UDP-glucose:salicylic acid glucosyltransferase [Oryza
sativa]
Length = 470
Score = 261 bits (667), Expect = 2e-68
Identities = 158/455 (34%), Positives = 233/455 (50%), Gaps = 57/455 (12%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSE------PKPDNISFRSG 54
+P+P RG+INPM+ C+LL + + + V+ VVTEEW ++S P + +
Sbjct: 13 VPYPGRGHINPMLAACRLLAAADGELTVTVVVTEEWHGLLASAGVPATLPPAGRVRLATI 72
Query: 55 SNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVANRRN 114
NVIPSE G D F E V KM E+LLD L+ P IV DT L W V R
Sbjct: 73 PNVIPSEHGRGADPAGFFEAVDAKMGVAVEQLLDRLERRPDAIVADTYLAWGVPAGAARG 132
Query: 115 IPAALFWPMPASIFSVFLHQHIF----EQNGHYPVKYPGFE------------------- 151
IP W M A+ F + H++ ++ G + E
Sbjct: 133 IPVCSLWTMAATFFWALYNIHLWPPVDDREGEQDLSRKSLEQYVPGCSSVRLSDVKIFRS 192
Query: 152 W-------------IHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKN 198
W + KAQ +LF+S YELE A+D + +P P+Y +GP+I L
Sbjct: 193 WERSMKLTTEAFVNVRKAQCVLFTSFYELEPCAMDRITQAVPFPVYPVGPSISDMPLDGG 252
Query: 199 DPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARS 258
K + H WLD+QP SVLY++ GS S+ +Q++E+A AL S VRF W+AR
Sbjct: 253 AGKIDDEEHR---AWLDAQPERSVLYVSFGSVVSMWPSQLEEVAVALRDSAVRFFWVARD 309
Query: 259 EASR--LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLP 316
AS L+ I G + GL++ WCDQL VL H S+GGF SHCGWNS E++ AGVP L LP
Sbjct: 310 SASAGDLRRIAGGN--GLVVPWCDQLGVLCHRSVGGFLSHCGWNSLLEAVFAGVPLLALP 367
Query: 317 IYIDQPFNSKMMVEDWKVGCRVKEDVKRD-----TLVKKDKIVKLVHEFMDLDGELTRDI 371
+ DQ +++++ ++W++G + E + + +V +D I MD D +R++
Sbjct: 368 VVWDQVVDARVVADEWRIGVNLSEQRREEDDGGGVVVGRDAIRAAAARLMDPDDGESREM 427
Query: 372 RERSKKLQKICLNSI---ANGGSAHTDFNAFISDV 403
R R+ L++ C ++ +GGS+ N F+ D+
Sbjct: 428 RRRAALLREACRGAVQDGPDGGSSRRSLNGFVKDL 462
>UniRef100_Q9SJL0 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 490
Score = 193 bits (491), Expect = 6e-48
Identities = 123/471 (26%), Positives = 228/471 (48%), Gaps = 82/471 (17%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPD------------- 47
+P+P++G++ P ++L L S+ I +FV T+ IS+ + D
Sbjct: 14 IPYPLQGHVIPFVHLAIKLASHGFTI--TFVNTDSIHHHISTAHQDDAGDIFSAARSSGQ 71
Query: 48 -NISFRSGSNVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLL----DHPPSIIVYDTL 102
+I + + S+ P + +H F E ++ A ++L+ L D P + ++ DT
Sbjct: 72 HDIRYTTVSDGFPLDFDRSLNHDQFFEGILHVFSAHVDDLIAKLSRRDDPPVTCLIADTF 131
Query: 103 LYWAVVVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGH-------------------- 142
W+ ++ ++ N+ FW PA + +++ H + NGH
Sbjct: 132 YVWSSMICDKHNLVNVSFWTEPALVLNLYYHMDLLISNGHFKSLDNRKDVIDYVPGVKAI 191
Query: 143 ----------------------YPVKYPGFEWIHKAQYLLFSSIYELESQAIDVLKSKLP 180
Y + + F+ + +A +++ +++ ELE ++ L++K P
Sbjct: 192 EPKDLMSYLQVSDKDVDTNTVVYRILFKAFKDVKRADFVVCNTVQELEPDSLSALQAKQP 251
Query: 181 LPIYTIGPTIPKFSLIKNDPKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDE 240
+Y IGP S++ S S EWL +P GSVLY++ GS+ V +I E
Sbjct: 252 --VYAIGPVFSTDSVVPT----SLWAESDCTEWLKGRPTGSVLYVSFGSYAHVGKKEIVE 305
Query: 241 IAAALCASNVRFLWIARSE--ASRLKE------ICGAHHMGLIMEWCDQLRVLSHPSIGG 292
IA L S + F+W+ R + S + + + A GL+++WC Q+ V+S+P++GG
Sbjct: 306 IAHGLLLSGISFIWVLRPDIVGSNVPDFLPAGFVDQAQDRGLVVQWCCQMEVISNPAVGG 365
Query: 293 FWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDK 352
F++HCGWNS ES+ G+P L P+ DQ N K++V+DW +G + E + +D+
Sbjct: 366 FFTHCGWNSILESVWCGLPLLCYPLLTDQFTNRKLVVDDWCIGINLCE----KKTITRDQ 421
Query: 353 IVKLVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
+ V M +GE + ++R +K+++ +++ GS+ T+FN F+S+V
Sbjct: 422 VSANVKRLM--NGETSSELRNNVEKVKRHLKDAVTTVGSSETNFNLFVSEV 470
>UniRef100_Q9M6E7 UDP-glucose:salicylic acid glucosyltransferase [Nicotiana tabacum]
Length = 459
Score = 192 bits (487), Expect = 2e-47
Identities = 141/462 (30%), Positives = 225/462 (48%), Gaps = 78/462 (16%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHV----SFVVTEEWLSF-ISSEPKPDNISFRSGS 55
+P+P +G+INPM+ K L S I + SF+ T + LS +S E D + G
Sbjct: 11 LPYPAQGHINPMLQFSKRLQSKGVKITIAATKSFLKTMQELSTSVSVEAISDG--YDDG- 67
Query: 56 NVIPSELICGRDHPAFMEDVMTKMEAPFEELLDLL-------DHPPSIIVYDTLLYWAVV 108
GR+ +T+ + + L L P S IVYD L WAV
Sbjct: 68 ---------GREQAGTFVAYITRFKEVGSDTLSQLIGKLTNCGCPVSCIVYDPFLPWAVE 118
Query: 109 VANRRNIPAALFWPMPASIFSVFLHQH-----IFEQNGHYPVKYPG-------------- 149
V N + A F+ ++ +++ H H + + + PG
Sbjct: 119 VGNNFGVATAAFFTQSCAVDNIYYHVHKGVLKLPPTDVDKEISIPGLLTIEASDVPSFVS 178
Query: 150 --------------FEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSL 195
F + ++L +S YELE + ID + +K+ PI TIGPTIP L
Sbjct: 179 NPESSRILEMLVNQFSNLENTDWVLINSFYELEKEVIDWM-AKI-YPIKTIGPTIPSMYL 236
Query: 196 IKNDPKPSNTNHSYY-------IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCAS 248
K P S + + WL+ QP+ SV+Y++ GS + + Q++E+A L S
Sbjct: 237 DKRLPDDKEYGLSVFKPMTNACLNWLNHQPVSSVVYVSFGSLAKLEAEQMEELAWGLSNS 296
Query: 249 NVRFLWIARS-EASRL-----KEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNST 302
N FLW+ RS E S+L +E+ A GL++ WC QL+VL H SIG F +HCGWNST
Sbjct: 297 NKNFLWVVRSTEESKLPNNFLEEL--ASEKGLVVSWCPQLQVLEHKSIGCFLTHCGWNST 354
Query: 303 KESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMD 362
E++ GVP + +P + DQP N+K++ + W++G R K+D K LV+++ I + + M
Sbjct: 355 LEAISLGVPMIAMPHWSDQPTNAKLVEDVWEMGIRPKQDEK--GLVRREVIEECIKIVM- 411
Query: 363 LDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVM 404
+ + + IRE +KK +++ ++ GGS+ + F+S ++
Sbjct: 412 -EEKKGKKIRENAKKWKELARKAVDEGGSSDRNIEEFVSKLV 452
>UniRef100_Q9LR44 T25N20.21 [Arabidopsis thaliana]
Length = 469
Score = 191 bits (484), Expect = 4e-47
Identities = 127/450 (28%), Positives = 224/450 (49%), Gaps = 58/450 (12%)
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFV--VTEEWLSFISSEPKPDNISFRSGSNVIPS 60
FP +G++NP + + L+ + V+FV V+ S I++ K +N+SF + S+
Sbjct: 11 FPAQGHVNPSLRFARRLIKR-TGARVTFVTCVSVFHNSMIANHNKVENLSFLTFSDGFDD 69
Query: 61 ELICG-RDHPAFMEDVMTKMEAPFEELLDLL---DHPPSIIVYDTLLYWAVVVANRRNIP 116
I D ++ + + ++ D P + ++Y LL WA VA R +P
Sbjct: 70 GGISTYEDRQKRSVNLKVNGDKALSDFIEATKNGDSPVTCLIYTILLNWAPKVARRFQLP 129
Query: 117 AALFWPMPASIFSVFL-----HQHIFE---------------------QNGHYPVKYPGF 150
+AL W PA +F+++ ++ +FE G Y
Sbjct: 130 SALLWIQPALVFNIYYTHFMGNKSVFELPNLSSLEIRDLPSFLTPSNTNKGAYDAFQEMM 189
Query: 151 EWIHKAQY--LLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPKPSNTNHS 208
E++ K +L ++ LE +A+ + + + +GP +P + K S
Sbjct: 190 EFLIKETKPKILINTFDSLEPEALTAFPN---IDMVAVGPLLPTEIFSGSTNKSVKDQSS 246
Query: 209 YYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSEASR------ 262
Y WLDS+ SV+Y++ G+ +S QI+E+A AL FLW+ +++R
Sbjct: 247 SYTLWLDSKTESSVIYVSFGTMVELSKKQIEELARALIEGKRPFLWVITDKSNRETKTEG 306
Query: 263 -----LKEICGAHH----MGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFL 313
+++I G H +G+I+ WC Q+ VLSH ++G F +HCGW+ST ESLV GVP +
Sbjct: 307 EEETEIEKIAGFRHELEEVGMIVSWCSQIEVLSHRAVGCFVTHCGWSSTLESLVLGVPVV 366
Query: 314 TLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRE 373
P++ DQP N+K++ E WK G RV+E+ +D LV++ +I + + M+ E + ++RE
Sbjct: 367 AFPMWSDQPTNAKLLEESWKTGVRVREN--KDGLVERGEIRRCLEAVME---EKSVELRE 421
Query: 374 RSKKLQKICLNSIANGGSAHTDFNAFISDV 403
+KK +++ + + GGS+ + AF+ D+
Sbjct: 422 NAKKWKRLAMEAGREGGSSDKNMEAFVEDI 451
>UniRef100_Q6X1C0 Glucosyltransferase 2 [Crocus sativus]
Length = 460
Score = 187 bits (476), Expect = 4e-46
Identities = 140/459 (30%), Positives = 220/459 (47%), Gaps = 81/459 (17%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRS------G 54
+P P +G+INP++ K L S+N + + V T + SEP P NI S G
Sbjct: 12 LPCPAQGHINPILQFGKRLASHN--LLTTLVNTRFLSNSTKSEPGPVNIQCISDGFDPGG 69
Query: 55 SNVIPSELI--------CGRDHPAFMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWA 106
N PS G+ H +E + ++ P + + WA
Sbjct: 70 MNAAPSRRAYFDRPQSRSGQKHVGLIESLRSR------------GRPGACFGLRPVPLWA 117
Query: 107 VVVANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVKYP------------------ 148
+ VA R + + F+ P ++ +++ +H++E PV P
Sbjct: 118 MNVAERSGLRSVAFFTQPCAVDTIY--RHVWEGRIKVPVAEPVRLPGLPPLEPSDLPCVR 175
Query: 149 -GF----------------EWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIP 191
GF + + KA + +SIYELE+ +D S+LPLP+ +IGPT+P
Sbjct: 176 NGFGRVVNPDLLPLRVNQHKNLDKADMMGRNSIYELEADLLD--GSRLPLPVKSIGPTVP 233
Query: 192 KFSLIKNDPKPSNTNHSYY-------IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAA 244
L P S+ + Y ++WLDS+ SV+Y++ GS S+S Q +EIA+
Sbjct: 234 STYLDNRIPSDSHYGFNLYTPDTTPYLDWLDSKAPNSVIYVSFGSLSSLSPDQTNEIASG 293
Query: 245 LCASNVRFLWIAR-SEASRLKEICGAHHM--GLIMEWCDQLRVLSHPSIGGFWSHCGWNS 301
L A+N F+W+ R SE ++L + GL++ WCDQL +L+H + G F +HCGWNS
Sbjct: 294 LIATNKSFIWVVRTSELAKLPANFTQENASRGLVVTWCDQLDLLAHVATGCFVTHCGWNS 353
Query: 302 TKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFM 361
T E + GVP + +P + DQP N+K + + WKVG R K K V+ ++ + V E M
Sbjct: 354 TMEGVALGVPMVGVPQWSDQPMNAKYVEDVWKVGVRAKTYGK--DFVRGEEFKRCVEEVM 411
Query: 362 DLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFI 400
DGE + IRE + + K+ +S++ GGS+ FI
Sbjct: 412 --DGERSGKIRENAARWCKLAKDSVSEGGSSDKCIKEFI 448
>UniRef100_O22822 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 449
Score = 187 bits (474), Expect = 6e-46
Identities = 137/456 (30%), Positives = 225/456 (49%), Gaps = 73/456 (16%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+P+P +G+I P CK L + + + +T +F+ + PD SG I +
Sbjct: 11 VPYPTQGHITPFRQFCKRL--HFKGLKTTLALT----TFVFNSINPD----LSGPISIAT 60
Query: 61 ELICGRDHPAF---------MEDVMTKMEAPFEELLD---LLDHPPSIIVYDTLLYWAVV 108
+ G DH F ++D T +++ D+P + IVYD L WA+
Sbjct: 61 -ISDGYDHGGFETADSIDDYLKDFKTSGSKTIADIIQKHQTSDNPITCIVYDAFLPWALD 119
Query: 109 VANRRNIPAALFWPMPASIFSVFLHQHI-----------------------FEQNGHYPV 145
VA + A F+ P ++ V+ +I F +G YP
Sbjct: 120 VAREFGLVATPFFTQPCAVNYVYYLSYINNGSLQLPIEELPFLELQDLPSFFSVSGSYPA 179
Query: 146 KYP----GFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSL---IKN 198
+ F KA ++L +S ELE ++ P + TIGPTIP L IK+
Sbjct: 180 YFEMVLQQFINFEKADFVLVNSFQELELHENELWSKACP--VLTIGPTIPSIYLDQRIKS 237
Query: 199 DPKPS-----NTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFL 253
D + + S+ I WLD++P GSV+Y+A GS +++ Q++E+A+A+ SN FL
Sbjct: 238 DTGYDLNLFESKDDSFCINWLDTRPQGSVVYVAFGSMAQLTNVQMEELASAV--SNFSFL 295
Query: 254 WIARSEASR------LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLV 307
W+ RS L+ + L+++W QL+VLS+ +IG F +HCGWNST E+L
Sbjct: 296 WVVRSSEEEKLPSGFLETV--NKEKSLVLKWSPQLQVLSNKAIGCFLTHCGWNSTMEALT 353
Query: 308 AGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGEL 367
GVP + +P + DQP N+K + + WK G RVK + K + K+++I + E M +GE
Sbjct: 354 FGVPMVAMPQWTDQPMNAKYIQDVWKAGVRVKTE-KESGIAKREEIEFSIKEVM--EGER 410
Query: 368 TRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
++++++ KK + + + S+ GGS T+ + F+S V
Sbjct: 411 SKEMKKNVKKWRDLAVKSLNEGGSTDTNIDTFVSRV 446
>UniRef100_Q9SKC5 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 456
Score = 185 bits (469), Expect = 2e-45
Identities = 130/457 (28%), Positives = 224/457 (48%), Gaps = 70/457 (15%)
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPSEL 62
FP++G+INP++ K L+S N N V+F+ T + I + G+ +P
Sbjct: 14 FPIQGHINPLLQFSKRLLSKNVN--VTFLTTSSTHNSILRR------AITGGATALPLSF 65
Query: 63 I-----CGRDHPA------FMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVAN 111
+ DHP+ + + EL+ +D P+ +VYD+ L + + V
Sbjct: 66 VPIDDGFEEDHPSTDTSPDYFAKFQENVSRSLSELISSMDPKPNAVVYDSCLPYVLDVCR 125
Query: 112 RR-NIPAALFWPMPASIFSVFLH---------QHIFEQNGHYPVK--------------Y 147
+ + AA F+ +++ + ++H Q+ P+K
Sbjct: 126 KHPGVAAASFFTQSSTVNATYIHFLRGEFKEFQNDVVLPAMPPLKGNDLPVFLYDNNLCR 185
Query: 148 PGFEWIHKA-------QYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDP 200
P FE I + L +S ELE + + +K++ P + IGP IP L K
Sbjct: 186 PLFELISSQFVNVDDIDFFLVNSFDELEVEVLQWMKNQWP--VKNIGPMIPSMYLDKRLA 243
Query: 201 KPS-------NTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFL 253
N + ++WLDS+P GSV+Y++ GS + Q+ E+AA L + FL
Sbjct: 244 GDKDYGINLFNAQVNECLDWLDSKPPGSVIYVSFGSLAVLKDDQMIEVAAGLKQTGHNFL 303
Query: 254 WIARSEASR------LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLV 307
W+ R ++ +++IC GLI+ W QL+VL+H SIG F +HCGWNST E+L
Sbjct: 304 WVVRETETKKLPSNYIEDICDK---GLIVNWSPQLQVLAHKSIGCFMTHCGWNSTLEALS 360
Query: 308 AGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGEL 367
GV + +P Y DQP N+K + + WKVG RVK D ++ V K++IV+ V E M+ E
Sbjct: 361 LGVALIGMPAYSDQPTNAKFIEDVWKVGVRVKAD--QNGFVPKEEIVRCVGEVMEDMSEK 418
Query: 368 TRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDVM 404
++IR+ +++L + ++++GG++ + + F++ ++
Sbjct: 419 GKEIRKNARRLMEFAREALSDGGNSDKNIDEFVAKIV 455
>UniRef100_Q9SBQ2 Anthocyanin 5-O-glucosyltransferase [Petunia hybrida]
Length = 468
Score = 184 bits (466), Expect = 5e-45
Identities = 139/468 (29%), Positives = 219/468 (46%), Gaps = 81/468 (17%)
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRS---GSNVIP 59
FP +G+INP + K LV I V+F + I ++ + D S + G N IP
Sbjct: 11 FPAQGHINPALQFAKNLVK--MGIEVTFSTS------IYAQSRMDEKSILNAPKGLNFIP 62
Query: 60 SE--LICGRDH---PAFMEDVMTKMEAPFEELLDLL----DHPPSIIVYDTLLYWAVVVA 110
G DH P F + K + + + L P + ++Y L WA VA
Sbjct: 63 FSDGFDEGFDHSKDPVFYMSQLRKCGSETVKKIILTCSENGQPITCLLYSIFLPWAAEVA 122
Query: 111 NRRNIPAALFWPMPASIFSVFLHQ-HIFEQ-------NGHYPVKYPGFEWIHKAQY---- 158
+IP+AL W PA+I ++ H +E+ + ++ ++ PG +
Sbjct: 123 REVHIPSALLWSQPATILDIYYFNFHGYEKAMANESNDPNWSIQLPGLPLLETRDLPSFL 182
Query: 159 ------------------------------LLFSSIYELESQAIDVLKSKLPLPIYTIGP 188
+L ++ ELE +A++ ++ Y IGP
Sbjct: 183 LPYGAKGSLRVALPPFKELIDTLDAETTPKILVNTFDELEPEALNAIEG---YKFYGIGP 239
Query: 189 TIPKFSLIKNDPKPSN------TNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIA 242
IP L NDP ++ N + Y+EWL+S+P SV+YI+ GS + S +Q++EI+
Sbjct: 240 LIPSAFLGGNDPLDASFGGDLFQNSNDYMEWLNSKPNSSVVYISFGSLMNPSISQMEEIS 299
Query: 243 AALCASNVRFLWIARS-------EASRLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWS 295
L FLW+ + E +L I +G I+ WC QL VL HPS+G F S
Sbjct: 300 KGLIDIGRPFLWVIKENEKGKEEENKKLGCIEELEKIGKIVPWCSQLEVLKHPSLGCFVS 359
Query: 296 HCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVK 355
HCGWNS ESL GVP + P + DQ N+K + + WK G RV+ + D +V+ ++I +
Sbjct: 360 HCGWNSALESLACGVPVVAFPQWTDQMTNAKQVEDVWKSGVRVR--INEDGVVESEEIKR 417
Query: 356 LVHEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
+ MD GE ++R+ +KK +++ ++ GGS+H + AFI DV
Sbjct: 418 CIELVMD-GGEKGEELRKNAKKWKELAREAVKEGGSSHKNLKAFIDDV 464
>UniRef100_Q8LFR6 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 456
Score = 183 bits (464), Expect = 9e-45
Identities = 131/454 (28%), Positives = 222/454 (48%), Gaps = 64/454 (14%)
Query: 3 FPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPSEL 62
FP++G+INP++ K L+S N N V+F+ T + I + G+ +P
Sbjct: 14 FPIQGHINPLLQFSKRLLSKNVN--VTFLTTSSTHNSILRR------AITGGATALPLSF 65
Query: 63 I-----CGRDHPA------FMEDVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVAN 111
+ DHP+ + + EL+ +D P+ +VYD+ L + + V
Sbjct: 66 VPIDDGFEEDHPSTDTSPDYFAKFQENVSRSLSELISSMDPKPNAVVYDSCLPYVLDVCR 125
Query: 112 RR-NIPAALFWPMPASIFSVFLH---------QHIFEQNGHYPVK--------------Y 147
+ + AA F+ +++ + ++H Q+ P+K
Sbjct: 126 KHPGVAAASFFTQSSTVNATYIHFLRGEFKEFQNDVVLPAMPPLKGNDLPVFLYDNNLCR 185
Query: 148 PGFEWIHKA-------QYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDP 200
P FE I + L +S ELE + + +K++ P + IGP IP L K
Sbjct: 186 PLFELISSQFVNVDDIDFFLVNSFDELEVEVLQWMKNQWP--VKNIGPMIPSMYLDKRLA 243
Query: 201 KPS-------NTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFL 253
N + ++WLDS+P GSV+Y++ GS + Q+ E+AA L + FL
Sbjct: 244 GDKDYGINLFNAQVNECLDWLDSKPPGSVIYVSFGSLAVLKDDQMIEVAAGLKQTGHNFL 303
Query: 254 WIAR-SEASRLKE--ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGV 310
W+ R +E +L I GLI+ W QL+VL+H SIG F +HCGWNST E+L GV
Sbjct: 304 WVVRETETKKLPSNYIEDIGEKGLIVNWSPQLQVLAHKSIGCFMTHCGWNSTLEALSLGV 363
Query: 311 PFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRD 370
+ +P Y DQP N+K + + WKVG RVK D ++ V K++IV+ V E M+ E ++
Sbjct: 364 ALIGMPAYSDQPTNAKFIEDVWKVGVRVKAD--QNGFVPKEEIVRCVGEVMEDMSEKGKE 421
Query: 371 IRERSKKLQKICLNSIANGGSAHTDFNAFISDVM 404
IR+ +++L + ++++GG++ + + F++ ++
Sbjct: 422 IRKNARRLMEFAREALSDGGNSDKNIDEFVAKIV 455
>UniRef100_O22820 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 449
Score = 175 bits (444), Expect = 2e-42
Identities = 133/454 (29%), Positives = 222/454 (48%), Gaps = 69/454 (15%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+PFP +G+I P+ CK L S F T +FI + I S + +
Sbjct: 11 VPFPSQGHITPIRQFCKRLHSKG------FKTTHTLTTFIFN-----TIHLDPSSPISIA 59
Query: 61 ELICGRDH---------PAFMEDVMT---KMEAPFEELLDLLDHPPSIIVYDTLLYWAVV 108
+ G D P ++++ T K A D+P + IVYD+ + WA+
Sbjct: 60 TISDGYDQGGFSSAGSVPEYLQNFKTFGSKTVADIIRKHQSTDNPITCIVYDSFMPWALD 119
Query: 109 VANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVK---------------------- 146
+A + AA F+ ++ + +I + P+K
Sbjct: 120 LAMDFGLAAAPFFTQSCAVNYINYLSYINNGSLTLPIKDLPLLELQDLPTFVTPTGSHLA 179
Query: 147 -----YPGFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKNDPK 201
F KA ++L +S ++L+ ++L SK+ P+ TIGPT+P L +
Sbjct: 180 YFEMVLQQFTNFDKADFVLVNSFHDLDLHEEELL-SKV-CPVLTIGPTVPSMYLDQQIKS 237
Query: 202 PSNTNHSYY--------IEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFL 253
++ + + + +WLD +P GSV+YIA GS +SS Q++EIA+A+ SN +L
Sbjct: 238 DNDYDLNLFDLKEAALCTDWLDKRPEGSVVYIAFGSMAKLSSEQMEEIASAI--SNFSYL 295
Query: 254 WIAR-SEASRLKE---ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAG 309
W+ R SE S+L L+++W QL+VLS+ +IG F +HCGWNST E L G
Sbjct: 296 WVVRASEESKLPPGFLETVDKDKSLVLKWSPQLQVLSNKAIGCFMTHCGWNSTMEGLSLG 355
Query: 310 VPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTR 369
VP + +P + DQP N+K + + WKVG RVK + K + K+++I + E M +GE ++
Sbjct: 356 VPMVAMPQWTDQPMNAKYIQDVWKVGVRVKAE-KESGICKREEIEFSIKEVM--EGEKSK 412
Query: 370 DIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
+++E + K + + + S++ GGS + N F+S +
Sbjct: 413 EMKENAGKWRDLAVKSLSEGGSTDININEFVSKI 446
>UniRef100_Q947K4 Thiohydroximate S-glucosyltransferase [Brassica napus]
Length = 466
Score = 173 bits (438), Expect = 9e-42
Identities = 139/458 (30%), Positives = 220/458 (47%), Gaps = 70/458 (15%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+P+PV+G++NPM+ K LVS + V+ T S IS+ P G + IP
Sbjct: 20 LPYPVQGHLNPMVQFAKRLVSKG--VKVTIATTTYTASSIST-PSVSVEPISDGHDFIPI 76
Query: 61 ELICGRDHPAFME-------DVMTKMEAPFEELLDLLDHPPSIIVYDTLLYWAVVVANRR 113
+ G A+ E + +T++ + F+ D P +VYD+ L W + VA
Sbjct: 77 G-VPGVSIDAYSESFKLNGSETLTRVISKFKST----DSPIDSLVYDSFLPWGLEVARSN 131
Query: 114 NIPAALFW------------------PMPASIFSV-----------------FLHQHIFE 138
+I AA F+ P+PA S F+ +H
Sbjct: 132 SISAAAFFTNNLTVCSVLRKFASGEFPLPADPASAPYLVRGLPALSYDELPSFVGRHSSS 191
Query: 139 QNGHYPVKYPGFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSL--- 195
H V F A +L + LE+Q +V +S+ + IGP IP L
Sbjct: 192 HAEHGRVLLNQFRNHEDADWLFVNGFEGLETQGCEVGESEA-MKATLIGPMIPSAYLDGR 250
Query: 196 IKNDP-------KPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCAS 248
IK+D KP + +EWLD++ SV++++ GSF + Q+ E+A AL S
Sbjct: 251 IKDDKGYGSSLMKPLSEE---CMEWLDTKLSKSVVFVSFGSFGILFEKQLAEVAKALQES 307
Query: 249 NVRFLWIAR-SEASRLKE--ICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKES 305
N FLW+ + + ++L E + L++ WC+QL VL+H SIG F +HCGWNST E
Sbjct: 308 NFNFLWVIKEAHIAKLPEGFVEATKDRALLVSWCNQLEVLAHESIGCFLTHCGWNSTLEG 367
Query: 306 LVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDG 365
L GVP + +P + DQ ++K + E W+VG R KE+ +VK D++V+ + M +G
Sbjct: 368 LSLGVPMVGVPQWSDQMNDAKFVEEVWRVGYRAKEEA-GGGVVKSDEVVRCLRGVM--EG 424
Query: 366 ELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
E + +IRE SKK + + + +++ GGS+ N F+ +
Sbjct: 425 ESSVEIRESSKKWKDLAVKAMSEGGSSDRSINEFVESL 462
>UniRef100_Q9LME9 T16E15.4 protein [Arabidopsis thaliana]
Length = 450
Score = 171 bits (434), Expect = 3e-41
Identities = 130/431 (30%), Positives = 208/431 (48%), Gaps = 55/431 (12%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKP------DNISFRSG 54
+P P +G+INPM+ + KLL + HV+FV T + + P + F S
Sbjct: 17 VPHPAQGHINPMLKVAKLLHARG--FHVTFVNTVYNHNRLLRSRGPYALDGLPSFRFESI 74
Query: 55 SNVIP-SELICGRDHPAFMEDVMTKMEAPFEELL----DLLDHPP-SIIVYDTLLYWAVV 108
++ +P ++ +D PA M APF+ELL D+ D PP S IV D ++ + +
Sbjct: 75 ADGLPDTDGDKTQDIPALCVSTMKNCLAPFKELLRRINDVDDVPPVSCIVSDGVMSFTLD 134
Query: 109 VANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVKYPGF--------EWIH-KAQYL 159
A N+P +FW A F FLH ++F + G P K + W + A +
Sbjct: 135 AAEELNLPEVIFWTNSACGFMTFLHFYLFIEKGLSPFKDESYMSKEHLDTRWSNPNAPVI 194
Query: 160 LFSSIYELESQAIDVLKSKLPLPIYTIGPT-------IPKFSLIKNDPKPSNTNHSYYIE 212
+ ++ +L+ I ++S L P+YTIGP I + S I + ++
Sbjct: 195 ILNTFDDLDHDLIQSMQSILLPPVYTIGPLHLLANQEIDEVSEIGRMGLNLWKEDTECLD 254
Query: 213 WLDSQPI-GSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSE-------ASRLK 264
WLDS+ SV+++ G +S+ Q+ E A L AS FLW+ R + A +
Sbjct: 255 WLDSKTTPNSVVFVNFGCITVMSAKQLLEFAWGLAASGKEFLWVIRPDLVAGETTAILSE 314
Query: 265 EICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFN 324
+ G+++ WC Q +V+SHP +GGF +HCGWNST ES+ GVP + P + +Q N
Sbjct: 315 FLTETADRGMLVSWCSQEKVISHPMVGGFLTHCGWNSTLESISGGVPIICWPFFAEQQTN 374
Query: 325 SKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICL- 383
K ++W VG + DVKR +++ +V E MD RE+ KK+++ +
Sbjct: 375 CKFCCDEWGVGVEIGGDVKR------EEVETVVRELMD---------REKGKKMREKAVE 419
Query: 384 -NSIANGGSAH 393
+AN + H
Sbjct: 420 WRRLANEATEH 430
>UniRef100_Q5UB80 UDP-glucuronosyltransferase [Arabidopsis thaliana]
Length = 430
Score = 170 bits (431), Expect = 6e-41
Identities = 128/422 (30%), Positives = 204/422 (48%), Gaps = 57/422 (13%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKP------DNISFRSG 54
+P P +G+INPM+ + KLL + HV+FV T + + P + F S
Sbjct: 17 VPHPAQGHINPMLKVAKLLHARG--FHVTFVNTVYNHNRLLRSRGPYALDGLPSFRFESI 74
Query: 55 SNVIP-SELICGRDHPAFMEDVMTKMEAPFEELL----DLLDHPP-SIIVYDTLLYWAVV 108
++ +P ++ +D PA M APF+ELL D+ D PP S IV D ++ + +
Sbjct: 75 ADGLPDTDGDKTQDIPALCVSTMKNCLAPFKELLRRINDVDDVPPVSCIVSDGVMSFTLD 134
Query: 109 VANRRNIPAALFWPMPASIFSVFLHQHIFEQNGHYPVKYPGFEWIHKAQYLLFSSIYELE 168
A N+P +FW A F FLH ++F + G P K ++ ++ +L+
Sbjct: 135 AAEELNLPEVIFWTNSACGFMTFLHFYLFIEKGLSPFKV-----------IILNTFDDLD 183
Query: 169 SQAIDVLKSKLPLPIYTIGPT-------IPKFSLIKNDPKPSNTNHSYYIEWLDSQPI-G 220
I ++S L P+YTIGP I + S I + ++WLDS+
Sbjct: 184 HDLIQSMQSILLPPVYTIGPLHLLANQEIDEVSEIGRMGLNLWKEDTECLDWLDSKTTPN 243
Query: 221 SVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSE-------ASRLKEICGAHHMG 273
SV+++ G +S+ Q+ E A L AS FLW+ R + A + + G
Sbjct: 244 SVVFVNFGCITVMSAKQLLEFAWGLAASGKEFLWVIRPDLVAGETTAILSEFLTETADRG 303
Query: 274 LIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWK 333
+++ WC Q +V+SHP +GGF +HCGWNST ES+ GVP + P + +Q N K ++W
Sbjct: 304 MLVSWCSQEKVISHPMVGGFLTHCGWNSTLESISGGVPIICWPFFAEQQTNCKFCCDEWG 363
Query: 334 VGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKICL--NSIANGGS 391
VG + DVKR +++ +V E MD RE+ KK+++ + +AN +
Sbjct: 364 VGVEIGGDVKR------EEVETVVRELMD---------REKGKKMREKAVEWRRLANEAT 408
Query: 392 AH 393
H
Sbjct: 409 EH 410
>UniRef100_Q94AB5 AT3g46660/F12A12_180 [Arabidopsis thaliana]
Length = 458
Score = 163 bits (412), Expect = 9e-39
Identities = 126/450 (28%), Positives = 207/450 (46%), Gaps = 65/450 (14%)
Query: 1 MPFPVRGNINPMMNLCKLLVSNNSNIHVSFVVTEEWLSFISSEPKPDNISFRSGSNVIPS 60
+PFP +G+I+PMM L K L +I V V ++ F S+ + F + +P
Sbjct: 18 VPFPAQGHISPMMQLAKTLHLKGFSITV---VQTKFNYFSPSDDFTHDFQFVTIPESLPE 74
Query: 61 ELICGRDHPAFMEDVMTKMEAPFEELLDLL----DHPPSIIVYDTLLYWAVVVANRRNIP 116
F+ + + + F++ L L + S ++YD +Y+A A +P
Sbjct: 75 SDFKNLGPIQFLFKLNKECKVSFKDCLGQLVLQQSNEISCVIYDEFMYFAEAAAKECKLP 134
Query: 117 AALFWPMPASIF---SVF-------LHQHIFEQNGH--------YPVKYPGFEWIHKAQY 158
+F A+ F SVF + + E G YP++Y F A
Sbjct: 135 NIIFSTTSATAFACRSVFDKLYANNVQAPLKETKGQQEELVPEFYPLRYKDFPVSRFASL 194
Query: 159 LLFSSIYE------------------LESQAIDVLKSK-LPLPIYTIGPTIPKFSLIKND 199
+Y LES ++ L+ + L +P+Y IGP ++ +
Sbjct: 195 ESIMEVYRNTVDKRTASSVIINTASCLESSSLSFLQQQQLQIPVYPIGP----LHMVASA 250
Query: 200 PKPSNTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARSE 259
P + IEWL+ Q + SV+YI+ GS + +I E+A+ L ASN FLW+ R
Sbjct: 251 PTSLLEENKSCIEWLNKQKVNSVIYISMGSIALMEINEIMEVASGLAASNQHFLWVIRPG 310
Query: 260 ASRLKEICGAH---------HMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGV 310
+ E + G I++W Q VLSHP++GGFWSHCGWNST ES+ GV
Sbjct: 311 SIPGSEWIESMPEEFSKMVLDRGYIVKWAPQKEVLSHPAVGGFWSHCGWNSTLESIGQGV 370
Query: 311 PFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRD 370
P + P DQ N++ + WK+G +V+ ++ R + ++ VK + +D +GE +
Sbjct: 371 PMICRPFSGDQKVNARYLECVWKIGIQVEGELDRGVV---ERAVKRL--MVDEEGE---E 422
Query: 371 IRERSKKLQKICLNSIANGGSAHTDFNAFI 400
+R+R+ L++ S+ +GGS+H F+
Sbjct: 423 MRKRAFSLKEQLRASVKSGGSSHNSLEEFV 452
>UniRef100_Q8S9A2 Glucosyltransferase-7 [Phaseolus angularis]
Length = 274
Score = 157 bits (396), Expect = 7e-37
Identities = 86/263 (32%), Positives = 144/263 (54%), Gaps = 12/263 (4%)
Query: 150 FEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPIYTIGPTIPKFSLIKN-----DPKPSN 204
F I+KA ++L +++Y+++ + +D K P IGP IP F L + D +
Sbjct: 17 FSNINKADWILCNTLYDMDKEIVDGFKEIWP-KFRCIGPNIPSFFLDEQYEDDQDYGVTE 75
Query: 205 TNHSYYIEWLDSQPIGSVLYIAQGSFFSVSSAQIDEIAAALCASNVRFLWIARS--EASR 262
IEWLD +P SV+Y++ GS S Q++EIA L + FLW+ R EA+
Sbjct: 76 LKRDECIEWLDDKPKDSVVYVSFGSIASFEKEQMEEIACCLKECSHYFLWVVRKSEEANL 135
Query: 263 LKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQP 322
K G ++ WC QL+VL+H +IG F +HCGWNST E+L GVP + +P + DQ
Sbjct: 136 PKGFEKKTEKGFVVTWCSQLKVLAHEAIGCFVTHCGWNSTLETLCLGVPTIAIPFWSDQS 195
Query: 323 FNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLVHEFMDLDGELTRDIRERSKKLQKIC 382
N+K+M + WK+G R D K+ +V+++ + + E M + E +++ + + + +
Sbjct: 196 TNAKLMEDVWKMGIRAPFDEKK--VVRREALKHCIREIM--ENEKGNELKNNANQWRTLA 251
Query: 383 LNSIANGGSAHTDFNAFISDVMH 405
+ ++ +GGS+H F++ H
Sbjct: 252 VKAVKSGGSSHKSILEFVNSFFH 274
>UniRef100_Q8L7D4 Putative glucosyltransferase [Arabidopsis thaliana]
Length = 169
Score = 156 bits (394), Expect = 1e-36
Identities = 70/166 (42%), Positives = 111/166 (66%), Gaps = 1/166 (0%)
Query: 238 IDEIAAALCASNVRFLWIARSEASRLKEICGAHHMGLIMEWCDQLRVLSHPSIGGFWSHC 297
++EI + + V+F W+AR +LKE +G+++ WCDQLRVL H +IGGFW+HC
Sbjct: 1 MEEIVVGVREAGVKFFWVARGGELKLKEALEGS-LGVVVSWCDQLRVLCHAAIGGFWTHC 59
Query: 298 GWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVKKDKIVKLV 357
G+NST E + +GVP LT P++ DQ N+KM+VE+W+VG ++ + + L+ D+I +LV
Sbjct: 60 GYNSTLEGICSGVPLLTFPVFWDQFLNAKMIVEEWRVGMGIERKKQMELLIVSDEIKELV 119
Query: 358 HEFMDLDGELTRDIRERSKKLQKICLNSIANGGSAHTDFNAFISDV 403
FMD + E +++R R+ L +IC ++A GGS+ + +AFI D+
Sbjct: 120 KRFMDGESEEGKEMRRRTCDLSEICRGAVAKGGSSDANIDAFIKDI 165
>UniRef100_Q705R4 Glycosyltransferase [Arabidopsis thaliana]
Length = 301
Score = 152 bits (384), Expect = 2e-35
Identities = 101/304 (33%), Positives = 156/304 (51%), Gaps = 48/304 (15%)
Query: 91 DHPPSIIVYDTLLYWAVVVANRRNIPAALFWPMPASIFSVFLHQHI-------------- 136
D P + IVYD L WA+ VA + A F+ P ++ V+ +I
Sbjct: 2 DSPITCIVYDAFLPWALDVAREFGLVATPFFTQPCAVNYVYYLSYINNGSLQLPIEELPF 61
Query: 137 ---------FEQNGHYPVKYP----GFEWIHKAQYLLFSSIYELESQAIDVLKSKLPLPI 183
F +G YP + F KA ++L +S ELE ++ P +
Sbjct: 62 LELQDLPSFFSVSGSYPAYFEMVLQQFINFEKADFVLVNSFQELELHENELWSKACP--V 119
Query: 184 YTIGPTIPKFSL---IKNDPKPS-----NTNHSYYIEWLDSQPIGSVLYIAQGSFFSVSS 235
TIGPTIP L IK+D + + S+ I WLD++P GSV+Y+A GS +++
Sbjct: 120 LTIGPTIPSIYLDQRIKSDTGYDLNLFESKDDSFCINWLDTRPQGSVVYVAFGSMAQLTN 179
Query: 236 AQIDEIAAALCASNVRFLWIARSEASR------LKEICGAHHMGLIMEWCDQLRVLSHPS 289
Q++E+A+A+ SN F+W+ RS L+ + L+++W QL+VLS+ +
Sbjct: 180 VQMEELASAV--SNFSFVWVVRSSEEEKLPSGFLETV--NKEKSLVLKWSPQLQVLSNKA 235
Query: 290 IGGFWSHCGWNSTKESLVAGVPFLTLPIYIDQPFNSKMMVEDWKVGCRVKEDVKRDTLVK 349
IG F +HCGWNST E+L GVP + +P + DQP N+K + + WK G RVK + K + K
Sbjct: 236 IGCFLTHCGWNSTMEALTFGVPMVAMPQWTDQPMNAKYIQDVWKSGVRVKTE-KESGIAK 294
Query: 350 KDKI 353
+++I
Sbjct: 295 REEI 298
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.138 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 712,977,721
Number of Sequences: 2790947
Number of extensions: 30425454
Number of successful extensions: 72203
Number of sequences better than 10.0: 1073
Number of HSP's better than 10.0 without gapping: 1029
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 69783
Number of HSP's gapped (non-prelim): 1450
length of query: 406
length of database: 848,049,833
effective HSP length: 130
effective length of query: 276
effective length of database: 485,226,723
effective search space: 133922575548
effective search space used: 133922575548
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC141923.7


