

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141862.23 + phase: 0
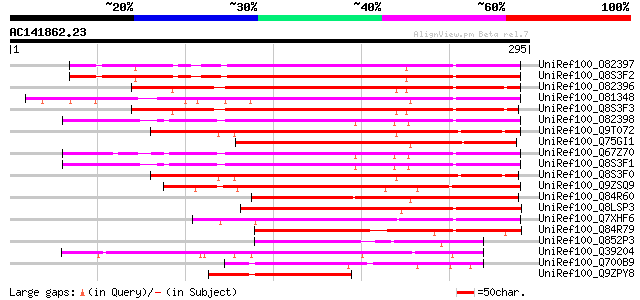
(295 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O82397 Putative bHLH transcription factor [Arabidopsis... 158 2e-37
UniRef100_Q8S3F2 Putative bHLH transcription factor [Arabidopsis... 157 4e-37
UniRef100_O82396 Putative bHLH transcription factor [Arabidopsis... 156 7e-37
UniRef100_O81348 Symbiotic ammonium transporter [Glycine max] 155 1e-36
UniRef100_Q8S3F3 Putative bHLH transcription factor [Arabidopsis... 154 2e-36
UniRef100_O82398 Putative bHLH transcription factor [Arabidopsis... 152 1e-35
UniRef100_Q9T072 Hypothetical protein AT4g37850 [Arabidopsis tha... 152 1e-35
UniRef100_Q75GI1 Putative symbiotic ammonium transport protein [... 151 2e-35
UniRef100_Q67Z70 Putative bHLH transcription factor [Arabidopsis... 151 2e-35
UniRef100_Q8S3F1 Putative bHLH transcription factor [Arabidopsis... 151 2e-35
UniRef100_Q8S3F0 Putative bHLH transcription factor [Arabidopsis... 150 4e-35
UniRef100_Q9ZSQ9 Transporter homolog [Mesembryanthemum crystalli... 142 8e-33
UniRef100_Q84R60 Putative ammonium transporter [Oryza sativa] 119 1e-25
UniRef100_Q8LSP3 Putative bHLH transcription factor [Oryza sativa] 112 2e-23
UniRef100_Q7XHF6 Putative bHLH transcription factor [Oryza sativa] 98 3e-19
UniRef100_Q84R79 Putative ammonium transporter [Oryza sativa] 90 8e-17
UniRef100_Q852P3 F3G1 [Perilla frutescens] 79 2e-13
UniRef100_Q39204 Transcription factor AtMYC2 [Arabidopsis thaliana] 76 1e-12
UniRef100_Q700B9 MYC transcription factor [Solanum tuberosum] 72 1e-11
UniRef100_Q9ZPY8 Putative bHLH transcription factor [Arabidopsis... 72 2e-11
>UniRef100_O82397 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 284
Score = 158 bits (399), Expect = 2e-37
Identities = 101/266 (37%), Positives = 162/266 (59%), Gaps = 24/266 (9%)
Query: 35 QESTYSSHTNINNKRIHSESTQNSSFPTQSPDQSVA----SATPPTKLLKASPKIISFDY 90
+E+ S +++N R + Q+S F Q + + +A P + ++ISFD+
Sbjct: 23 EENNLSPDESLSNSR---RADQSSKFDHQMHFECLREKPKAAVKPMMKINNKQQLISFDF 79
Query: 91 SNNDSKVKKPKTEIGYGENLNFGSVISQGDYYKRENKVSAVNRNPIQAQDHVMAERRRRE 150
S+N + P E + ++ +G + S R+P+ A++HV+AER+RRE
Sbjct: 80 SSN--VISSPAAE-----EIIMDKLVGRGT---KRKTCSHGTRSPVLAKEHVLAERKRRE 129
Query: 151 KLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTL-EEHVADKKVESAVFMKR 209
KLS++FI+LS+LLPGLKK DK TIL+DAI +KQL E+++TL EE A +++ES + +K+
Sbjct: 130 KLSEKFIALSALLPGLKKADKVTILDDAISRMKQLQEQLRTLKEEKEATRQMESMILVKK 189
Query: 210 S-ILFEEDDRSSCDEN----SDQSLSKIEARVSGKDMLIRIHGDKHCGRTATAILNELEK 264
S + F+E+ SC + DQ+L +IEA++S D+LIRI +K G ILN +E
Sbjct: 190 SKVFFDEEPNLSCSPSVHIEFDQALPEIEAKISQNDILIRILCEKSKG-CMINILNTIEN 248
Query: 265 HHLSVQSSSILPFGNNYLDITIVAQV 290
L +++S +LPFG++ LDIT++AQV
Sbjct: 249 FQLRIENSIVLPFGDSTLDITVLAQV 274
>UniRef100_Q8S3F2 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 295
Score = 157 bits (396), Expect = 4e-37
Identities = 100/266 (37%), Positives = 162/266 (60%), Gaps = 24/266 (9%)
Query: 35 QESTYSSHTNINNKRIHSESTQNSSFPTQSPDQSVA----SATPPTKLLKASPKIISFDY 90
+E+ S +++N R + Q+S F Q + + +A P + ++ISFD+
Sbjct: 23 EENNLSPDESLSNSR---RADQSSKFDHQMHFECLREKPKAAVKPMMKINNKQQLISFDF 79
Query: 91 SNNDSKVKKPKTEIGYGENLNFGSVISQGDYYKRENKVSAVNRNPIQAQDHVMAERRRRE 150
S+N + P E + ++ +G + S R+P+ A++HV+AER+RRE
Sbjct: 80 SSN--VISSPAAE-----EIIMDKLVGRGT---KRKTCSHGTRSPVLAKEHVLAERKRRE 129
Query: 151 KLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTL-EEHVADKKVESAVFMKR 209
KLS++FI+LS+LLPGLKK DK TIL+DAI +KQL E+++TL EE A +++ES + +K+
Sbjct: 130 KLSEKFIALSALLPGLKKADKVTILDDAISRMKQLQEQLRTLKEEKEATRQMESMILVKK 189
Query: 210 S-ILFEEDDRSSCDEN----SDQSLSKIEARVSGKDMLIRIHGDKHCGRTATAILNELEK 264
S + F+E+ SC + DQ+L +IEA++S D+LIRI +K G ILN +E
Sbjct: 190 SKVFFDEEPNLSCSPSVHIEFDQALPEIEAKISQNDILIRILCEKSKG-CMINILNTIEN 248
Query: 265 HHLSVQSSSILPFGNNYLDITIVAQV 290
L +++S +LPFG++ LDIT++AQ+
Sbjct: 249 FQLRIENSIVLPFGDSTLDITVLAQM 274
>UniRef100_O82396 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 288
Score = 156 bits (394), Expect = 7e-37
Identities = 99/237 (41%), Positives = 148/237 (61%), Gaps = 24/237 (10%)
Query: 70 ASATPPTKLLKASPKIISFDYS-------NNDSKVKKPKTE-IGYGENLNFGSVISQGDY 121
+S+ PP + S +I+SF+ + N+ + + PK E IG E+ +I
Sbjct: 54 SSSNPPPPKHQPSSRILSFEKTGLHVMNHNSPNLIFSPKDEEIGLPEHKKAELII----- 108
Query: 122 YKRENKVSAVNRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKH 181
+ + ++ R+ AQDH++AER+RREKL+QRF++LS+L+PGLKKMDKA++L DAIKH
Sbjct: 109 -RGTKRAQSLTRSQSNAQDHILAERKRREKLTQRFVALSALIPGLKKMDKASVLGDAIKH 167
Query: 182 LKQLNERVKTLEEHVADKKVESAVFMKRSILFEEDDR-----SSCDEN---SDQSLSKIE 233
+K L E VK EE +K +ES V +K+S L +++ SS D N S +L +IE
Sbjct: 168 IKYLQESVKEYEEQKKEKTMESVVLVKKSSLVLDENHQPSSSSSSDGNRNSSSSNLPEIE 227
Query: 234 ARVSGKDMLIRIHGDKHCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
RVSGKD+LI+I +K G I+ E+EK LS+ +S++LPFG + DI+I+AQV
Sbjct: 228 VRVSGKDVLIKILCEKQKG-NVIKIMGEIEKLGLSITNSNVLPFGPTF-DISIIAQV 282
>UniRef100_O81348 Symbiotic ammonium transporter [Glycine max]
Length = 347
Score = 155 bits (392), Expect = 1e-36
Identities = 115/309 (37%), Positives = 174/309 (56%), Gaps = 39/309 (12%)
Query: 10 HEYQMDSY------AFQFDDMAYFKSFSES----PQESTYSSHTNINN--KRIHSESTQN 57
H++Q++S F D+ SFS++ P+ S +S T I K++ S +
Sbjct: 27 HQWQLNSIDTTSLKGAAFGDILQKHSFSDNSNFNPKTSMETSQTGIERYAKQLGDNSWNH 86
Query: 58 SSFPTQSPDQSVASATPPTKLLKASPKIISFDYSNNDSKVK--KPKTEIG---YGENLNF 112
+ Q+P+ AS + ++SF +N S++ KPK E+ N
Sbjct: 87 NKSQQQTPETQFASCS----------NLLSFVNTNYTSELGLVKPKVEMACPKIDNNALA 136
Query: 113 GSVISQGDY------YKRENKVSAVNRNP--IQAQDHVMAERRRREKLSQRFISLSSLLP 164
+ISQG +K + + P Q QDH++AER+RREKLSQRFI+LS+L+P
Sbjct: 137 DMLISQGTLGNQNYIFKASQETKKIKTRPKLSQPQDHIIAERKRREKLSQRFIALSALVP 196
Query: 165 GLKKMDKATILEDAIKHLKQLNERVKTLEEHVADKK-VESAVFMKRSILFEEDDRSSCDE 223
GLKKMDKA++L +AIK+LKQ+ E+V LEE K+ VES V +K+S L + + SS +
Sbjct: 197 GLKKMDKASVLGEAIKYLKQMQEKVSALEEEQNRKRTVESVVIVKKSQLSSDAEDSSSET 256
Query: 224 NSD--QSLSKIEARVSGKDMLIRIHGDKHCGRTATAILNELEKHHLSVQSSSILPFGNNY 281
++L +IEAR +++LIRIH +K+ G ++E+EK HL V +SS L FG+
Sbjct: 257 GGTFVEALPEIEARFWERNVLIRIHCEKNKG-VIEKTISEIEKLHLKVINSSALTFGSFI 315
Query: 282 LDITIVAQV 290
LDITI+AQ+
Sbjct: 316 LDITIIAQM 324
>UniRef100_Q8S3F3 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 304
Score = 154 bits (390), Expect = 2e-36
Identities = 98/236 (41%), Positives = 147/236 (61%), Gaps = 24/236 (10%)
Query: 70 ASATPPTKLLKASPKIISFDYS-------NNDSKVKKPKTE-IGYGENLNFGSVISQGDY 121
+S+ PP + S +I+SF+ + N+ + + PK E IG E+ +I
Sbjct: 54 SSSNPPPPKHQPSSRILSFEKTGLHVMNHNSPNLIFSPKDEEIGLPEHKKAELII----- 108
Query: 122 YKRENKVSAVNRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKH 181
+ + ++ R+ AQDH++AER+RREKL+QRF++LS+L+PGLKKMDKA++L DAIKH
Sbjct: 109 -RGTKRAQSLTRSQSNAQDHILAERKRREKLTQRFVALSALIPGLKKMDKASVLGDAIKH 167
Query: 182 LKQLNERVKTLEEHVADKKVESAVFMKRSILFEEDDR-----SSCDEN---SDQSLSKIE 233
+K L E VK EE +K +ES V +K+S L +++ SS D N S +L +IE
Sbjct: 168 IKYLQESVKEYEEQKKEKTMESVVLVKKSSLVLDENHQPSSSSSSDGNRNSSSSNLPEIE 227
Query: 234 ARVSGKDMLIRIHGDKHCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQ 289
RVSGKD+LI+I +K G I+ E+EK LS+ +S++LPFG + DI+I+AQ
Sbjct: 228 VRVSGKDVLIKILCEKQKG-NVIKIMGEIEKLGLSITNSNVLPFGPTF-DISIIAQ 281
>UniRef100_O82398 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 322
Score = 152 bits (384), Expect = 1e-35
Identities = 108/279 (38%), Positives = 157/279 (55%), Gaps = 35/279 (12%)
Query: 31 SESPQESTYSSHTNINNKRIHSESTQNSSFPTQSPDQSVASATPPTKLLKASPKIISFDY 90
SES + + K++ + + NS+ + S S S T ++ISF
Sbjct: 37 SESGSGTGFELLAERPTKQMKTNNNMNSTSSSPSSSSSSGSRTS---------QVISF-- 85
Query: 91 SNNDSKVKKPKTEIGYGENLNFGSVISQGDYYKRENKVS-AVNRNPIQAQDHVMAERRRR 149
+ D+K +T + + ++ + KR++ V+ R P ++HV+AER+RR
Sbjct: 86 GSPDTKTNPVETSLNFSNQVSMDQKVGS----KRKDCVNNGGRREPHLLKEHVLAERKRR 141
Query: 150 EKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEH--VADKKVESAVFM 207
+KL++R I+LS+LLPGLKK DKAT+LEDAIKHLKQL ERVK LEE V K +S + +
Sbjct: 142 QKLNERLIALSALLPGLKKTDKATVLEDAIKHLKQLQERVKKLEEERVVTKKMDQSIILV 201
Query: 208 KRSILFEEDD--------------RSSCDENS--DQSLSKIEARVSGKDMLIRIHGDKHC 251
KRS ++ +DD SS DE S Q++ IEARVS +D+LIR+H +K+
Sbjct: 202 KRSQVYLDDDSSSYSSTCSAASPLSSSSDEVSIFKQTMPMIEARVSDRDLLIRVHCEKNK 261
Query: 252 GRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
G IL+ LEK L V +S LPFGN+ L ITI+ +V
Sbjct: 262 G-CMIKILSSLEKFRLEVVNSFTLPFGNSTLVITILTKV 299
>UniRef100_Q9T072 Hypothetical protein AT4g37850 [Arabidopsis thaliana]
Length = 314
Score = 152 bits (383), Expect = 1e-35
Identities = 94/221 (42%), Positives = 144/221 (64%), Gaps = 13/221 (5%)
Query: 81 ASPKIISF-DYSNNDSKVKKPKTEIGYGENLNFGSVIS---QGDYYKREN--KVSAVNRN 134
+S +I+SF DY +ND + + T + + + + + D + R+ + +RN
Sbjct: 63 SSSRILSFEDYGSNDMEHEYSPTYLNSIFSPKLEAQVQPHQKSDEFNRKGTKRAQPFSRN 122
Query: 135 PIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEE 194
AQDH++AER+RREKL+QRF++LS+L+PGLKKMDKA++L DA+KH+K L ERV LEE
Sbjct: 123 QSNAQDHIIAERKRREKLTQRFVALSALVPGLKKMDKASVLGDALKHIKYLQERVGELEE 182
Query: 195 HVADKKVESAVFMKRSILFEEDDR----SSCDEN-SDQSLSKIEARVSGKDMLIRIHGDK 249
++++ES V +K+S L +D+ SSC++ SD L +IE R S +D+LI+I +K
Sbjct: 183 QKKERRLESMVLVKKSKLILDDNNQSFSSSCEDGFSDLDLPEIEVRFSDEDVLIKILCEK 242
Query: 250 HCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
G A I+ E+EK H+ + +SS+L FG LDITI+A+V
Sbjct: 243 QKGHLA-KIMAEIEKLHILITNSSVLNFGPT-LDITIIAKV 281
>UniRef100_Q75GI1 Putative symbiotic ammonium transport protein [Oryza sativa]
Length = 359
Score = 151 bits (382), Expect = 2e-35
Identities = 84/166 (50%), Positives = 122/166 (72%), Gaps = 7/166 (4%)
Query: 129 SAVNRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNER 188
+A +R Q Q+H++AER+RREKLSQRFI+LS ++PGLKKMDKA++L DAIK++KQL ++
Sbjct: 170 AAASRPASQNQEHILAERKRREKLSQRFIALSKIVPGLKKMDKASVLGDAIKYVKQLQDQ 229
Query: 189 VKTLEEHVADKKVESAVFMKRSIL-FEEDDRSSCDENSD-----QSLSKIEARVSGKDML 242
VK LEE + VE+AV +K+S L ++DD SSCDEN D L +IEARVS + +L
Sbjct: 230 VKGLEEEARRRPVEAAVLVKKSQLSADDDDGSSCDENFDGGEATAGLPEIEARVSERTVL 289
Query: 243 IRIHGDKHCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVA 288
++IH + G TA L+E+E L++ ++++LPF ++ LDITI+A
Sbjct: 290 VKIHCENRKGALITA-LSEVETIGLTIMNTNVLPFTSSSLDITIMA 334
>UniRef100_Q67Z70 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 320
Score = 151 bits (381), Expect = 2e-35
Identities = 107/279 (38%), Positives = 160/279 (56%), Gaps = 35/279 (12%)
Query: 31 SESPQESTYSSHTNINNKRIHSESTQNSSFPTQSPDQSVASATPPTKLLKASPKIISFDY 90
SES + + K++ + + NS+ + SP S +S + + ++ISF
Sbjct: 37 SESGSGTGFELLAERPTKQMKTNNNMNST--SSSPSSSCSSGS-------RTSQVISF-- 85
Query: 91 SNNDSKVKKPKTEIGYGENLNFGSVISQGDYYKRENKVS-AVNRNPIQAQDHVMAERRRR 149
+ D+K +T + + ++ + KR++ V+ R P ++HV+AER+RR
Sbjct: 86 GSPDTKTNPVETSLNFSNQVSMDQKVGS----KRKDCVNNGGRREPHLLKEHVLAERKRR 141
Query: 150 EKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEH--VADKKVESAVFM 207
+KL++R I+LS+LLPGLKK DKAT+LEDAIKHLKQL ERVK LEE V K +S + +
Sbjct: 142 QKLNERLIALSALLPGLKKTDKATVLEDAIKHLKQLQERVKKLEEERVVTKKMDQSIILV 201
Query: 208 KRSILFEEDD--------------RSSCDENS--DQSLSKIEARVSGKDMLIRIHGDKHC 251
KRS ++ +DD SS DE S Q++ IEARVS +D+LIR+H +K+
Sbjct: 202 KRSQVYLDDDSSSYSSTCSAASPLSSSSDEVSIFKQTMPMIEARVSDRDLLIRVHCEKNK 261
Query: 252 GRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
G IL+ LEK L V +S LPFGN+ L ITI+ ++
Sbjct: 262 G-CMIKILSSLEKFRLEVVNSFTLPFGNSTLVITILTKM 299
>UniRef100_Q8S3F1 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 320
Score = 151 bits (381), Expect = 2e-35
Identities = 107/279 (38%), Positives = 157/279 (55%), Gaps = 35/279 (12%)
Query: 31 SESPQESTYSSHTNINNKRIHSESTQNSSFPTQSPDQSVASATPPTKLLKASPKIISFDY 90
SES + + K++ + + NS+ + S S S T ++ISF
Sbjct: 37 SESGSGTGFELLAERPTKQMKTNNNMNSTSSSPSSSSSSGSRTS---------QVISF-- 85
Query: 91 SNNDSKVKKPKTEIGYGENLNFGSVISQGDYYKRENKVS-AVNRNPIQAQDHVMAERRRR 149
+ D+K +T + + ++ + KR++ V+ R P ++HV+AER+RR
Sbjct: 86 GSPDTKTNPVETSLNFSNQVSMDQKVGS----KRKDCVNNGGRREPHLLKEHVLAERKRR 141
Query: 150 EKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEH--VADKKVESAVFM 207
+KL++R I+LS+LLPGLKK DKAT+LEDAIKHLKQL ERVK LEE V K +S + +
Sbjct: 142 QKLNERLIALSALLPGLKKTDKATVLEDAIKHLKQLQERVKKLEEERVVTKKMDQSIILV 201
Query: 208 KRSILFEEDD--------------RSSCDENS--DQSLSKIEARVSGKDMLIRIHGDKHC 251
KRS ++ +DD SS DE S Q++ IEARVS +D+LIR+H +K+
Sbjct: 202 KRSQVYLDDDSSSYSSTCSAASPLSSSSDEVSIFKQTMPMIEARVSDRDLLIRVHCEKNK 261
Query: 252 GRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
G IL+ LEK L V +S LPFGN+ L ITI+ ++
Sbjct: 262 G-CMIKILSSLEKFRLEVVNSFTLPFGNSTLVITILTKM 299
>UniRef100_Q8S3F0 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 304
Score = 150 bits (379), Expect = 4e-35
Identities = 93/220 (42%), Positives = 143/220 (64%), Gaps = 13/220 (5%)
Query: 81 ASPKIISF-DYSNNDSKVKKPKTEIGYGENLNFGSVIS---QGDYYKREN--KVSAVNRN 134
+S +I+SF DY +ND + + T + + + + + D + R+ + +RN
Sbjct: 63 SSSRILSFEDYGSNDMEHEYSPTYLNSIFSPKLEAQVQPHQKSDEFNRKGTKRAQPFSRN 122
Query: 135 PIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEE 194
AQDH++AER+RREKL+QRF++LS+L+PGLKKMDKA++L DA+KH+K L ERV LEE
Sbjct: 123 QSNAQDHIIAERKRREKLTQRFVALSALVPGLKKMDKASVLGDALKHIKYLQERVGELEE 182
Query: 195 HVADKKVESAVFMKRSILFEEDDR----SSCDEN-SDQSLSKIEARVSGKDMLIRIHGDK 249
++++ES V +K+S L +D+ SSC++ SD L +IE R S +D+LI+I +K
Sbjct: 183 QKKERRLESMVLVKKSKLILDDNNQSFSSSCEDGFSDLDLPEIEVRFSDEDVLIKILCEK 242
Query: 250 HCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQ 289
G A I+ E+EK H+ + +SS+L FG LDITI+A+
Sbjct: 243 QKGHLA-KIMAEIEKLHILITNSSVLNFGPT-LDITIIAK 280
>UniRef100_Q9ZSQ9 Transporter homolog [Mesembryanthemum crystallinum]
Length = 300
Score = 142 bits (359), Expect = 8e-33
Identities = 93/219 (42%), Positives = 134/219 (60%), Gaps = 20/219 (9%)
Query: 88 FDYSNNDSKVKKPKTEI----GYGENLNFGSVISQGDYYKRENKV----SAVNRNPIQAQ 139
FD S KK +T + G++++ GS ++ Y ++NKV S + +Q
Sbjct: 61 FDDGKPKSNNKKEETAVVVHDDVGDHVHGGSNYTK---YSQQNKVRNNSSKFGSIGLCSQ 117
Query: 140 DHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEHVADK 199
+HV+AER+RREK++QRF +LS+L+PGLKKMDKA+IL DA K+LKQL E+VK LEE A +
Sbjct: 118 EHVLAERKRREKMTQRFHALSALVPGLKKMDKASILGDAAKYLKQLEEQVKLLEEQTASR 177
Query: 200 KVESAVFMKRSIL----FEEDDRSSCDENSDQSLS----KIEARVSGKDMLIRIHGDKHC 251
VES V +K S + + SS +ENS+ SL+ +IEA ++LIRIH K
Sbjct: 178 TVESVVLVKNSNVQDPNLDHGGNSSSNENSNSSLNNPLLEIEAGACNNNVLIRIHAQKD- 236
Query: 252 GRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQV 290
+LNE+E HL+ + + +PFG +DITIVAQ+
Sbjct: 237 QDLVRKVLNEIENLHLTTLNFNTIPFGGYAMDITIVAQM 275
>UniRef100_Q84R60 Putative ammonium transporter [Oryza sativa]
Length = 353
Score = 119 bits (297), Expect = 1e-25
Identities = 62/164 (37%), Positives = 104/164 (62%), Gaps = 13/164 (7%)
Query: 138 AQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEHVA 197
AQ+H++AER+RREK++QRFI LS+++PGLKKMDKATIL DA++++K++ E++ LE+H
Sbjct: 190 AQEHIIAERKRREKINQRFIELSTVIPGLKKMDKATILSDAVRYVKEMQEKLSELEQH-Q 248
Query: 198 DKKVESAVFMKRSILFEEDDRSSCDENSD------------QSLSKIEARVSGKDMLIRI 245
+ VESA+ +K+ + C S SL +IEA++S ++++RI
Sbjct: 249 NGGVESAILLKKPCIATSSSDGGCPAASSAVAGSSSSGTARSSLPEIEAKISHGNVMVRI 308
Query: 246 HGDKHCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQ 289
HG+ + + +L +E HL + ++++PF ITI+A+
Sbjct: 309 HGENNGKGSLVRLLAAVEGLHLGITHTNVMPFSACTAIITIMAK 352
>UniRef100_Q8LSP3 Putative bHLH transcription factor [Oryza sativa]
Length = 451
Score = 112 bits (279), Expect = 2e-23
Identities = 66/166 (39%), Positives = 103/166 (61%), Gaps = 7/166 (4%)
Query: 132 NRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKT 191
+R P AQ+HV+AER+RREKL Q+F++L++++PGLKK DK ++L I ++KQL E+VK
Sbjct: 279 SRPPANAQEHVIAERKRREKLQQQFVALATIVPGLKKTDKISLLGSTIDYVKQLEEKVKA 338
Query: 192 LEEHVADKKVESAVF-MKRSILFEEDDRSSC-----DENSDQSLSKIEARVSGKDMLIRI 245
LEE + F K I ++DD S D +S S +EA + G +L++I
Sbjct: 339 LEEGSRRTAEPTTAFESKCRITVDDDDGGSASSGTDDGSSSSSSPTVEASIHGNTVLLKI 398
Query: 246 HGDKHCGRTATAILNELEKHHLSVQSSSILPFGNNYLDITIVAQVK 291
+ G IL+ELEK LS+ ++S++PF ++ L+ITI A+ +
Sbjct: 399 CCKERRG-LLVMILSELEKQGLSIINTSVVPFTDSCLNITITAKAR 443
>UniRef100_Q7XHF6 Putative bHLH transcription factor [Oryza sativa]
Length = 361
Score = 97.8 bits (242), Expect = 3e-19
Identities = 64/193 (33%), Positives = 110/193 (56%), Gaps = 9/193 (4%)
Query: 105 GYGENLNFGSVISQ---GDYYKRENKVSAVNRNPIQA----QDHVMAERRRREKLSQRFI 157
G +LNF ++ Q G K + +S + R +A Q+HV+AER+RREK+ Q+F
Sbjct: 150 GSSSSLNFSALEQQQDSGPMTKFCSPLSEMKRGGRRATSSMQEHVIAERKRREKMHQQFT 209
Query: 158 SLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEHVADKKVESAVFMKRSILFEEDD 217
+L+S++P + K DK ++L I+++ L ERVK L++ + + + RS DD
Sbjct: 210 TLASIVPEITKTDKVSVLGSTIEYVHHLRERVKILQDIQSMGSTQPPISDARSRAGSGDD 269
Query: 218 RSSCDENSDQSLSKIEARVSGKDMLIRIHGDKHCGRTATAILNELEKHHLSVQSSSILPF 277
D N+++ K+EA + G +L+R+ + G +L ELEK LS +++++PF
Sbjct: 270 EDD-DGNNNEVEIKVEANLQGTTVLLRVVCPEKKG-VLIKLLTELEKLGLSTMNTNVVPF 327
Query: 278 GNNYLDITIVAQV 290
++ L+ITI AQ+
Sbjct: 328 ADSSLNITITAQI 340
>UniRef100_Q84R79 Putative ammonium transporter [Oryza sativa]
Length = 301
Score = 89.7 bits (221), Expect = 8e-17
Identities = 58/156 (37%), Positives = 94/156 (60%), Gaps = 14/156 (8%)
Query: 140 DHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEHVADK 199
+HV+AER+RREK++QRF+ LS+++P LKKMDKATIL DA ++++L E++K LEE A +
Sbjct: 130 EHVVAERKRREKINQRFMELSAVIPKLKKMDKATILSDAASYIRELQEKLKALEEQAAAR 189
Query: 200 KVESAVFMKRSILFEEDDRSSCDENSDQSLSKIEARVSGKD--MLIRIHGDKHCGRTATA 257
E+A+ + N +IE R S + +++RIH + G
Sbjct: 190 VTEAAM---------ATPSPARAMNHLPVPPEIEVRCSPTNNVVMVRIHCENGEG-VIVR 239
Query: 258 ILNELEKHHLSVQSSSILPFGNN--YLDITIVAQVK 291
IL E+E+ HL + +++++PF + + ITI A+ K
Sbjct: 240 ILAEVEEIHLRIINANVMPFLDQGATMIITIAAKAK 275
>UniRef100_Q852P3 F3G1 [Perilla frutescens]
Length = 519
Score = 78.6 bits (192), Expect = 2e-13
Identities = 51/132 (38%), Positives = 76/132 (56%), Gaps = 10/132 (7%)
Query: 140 DHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHLKQLNERVKTLEEHVADK 199
+HVMAERRRREKL+QRFI L S++P + KMDKA+IL D I +LKQL +R++ LE + D
Sbjct: 363 NHVMAERRRREKLNQRFIVLRSMVPFITKMDKASILADTIDYLKQLKKRIQELESKIGDM 422
Query: 200 KVESAVFMKRSILFEEDDRSSCDENSDQSLSKIEARVSGKDMLIR--IHGDKHCGRTATA 257
K KR I + D +S + + +S + +E S K L+ I + G T
Sbjct: 423 K-------KREIRMSDAD-ASVEVSIIESDALVEIECSQKPGLLSDFIQALRGLGIQITT 474
Query: 258 ILNELEKHHLSV 269
+ + + H ++
Sbjct: 475 VQSSINTTHATL 486
>UniRef100_Q39204 Transcription factor AtMYC2 [Arabidopsis thaliana]
Length = 623
Score = 75.9 bits (185), Expect = 1e-12
Identities = 85/287 (29%), Positives = 132/287 (45%), Gaps = 49/287 (17%)
Query: 30 FSESPQESTYSSHTNINNKR-------IHSESTQNSSFPTQSPDQSVASATPPTKLLKAS 82
FS+S Q SS T N +HS+ TQN F + S + T + S
Sbjct: 305 FSKSIQFENGSSSTITENPNLDPTPSPVHSQ-TQNPKFNNTFSRELNFSTSSSTLVKPRS 363
Query: 83 PKIISF-DYSNNDSKVKKPKTEIGYG--EN-------LNFGSVISQGDYYKREN------ 126
+I++F D S P + G EN LN V+S GD E+
Sbjct: 364 GEILNFGDEGKRSSGNPDPSSYSGQTQFENKRKRSMVLNEDKVLSFGDKTAGESDHSDLE 423
Query: 127 ----KVSAVNRNPI-----------QAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDK 171
K AV + P + +HV AER+RREKL+QRF +L +++P + KMDK
Sbjct: 424 ASVVKEVAVEKRPKKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNVSKMDK 483
Query: 172 ATILEDAIKHLKQLNER-VKTLEEHVADK----KVESAVFMKRSILFEEDDRSSCDENSD 226
A++L DAI ++ +L + VKT E + K +V+ + +++ D SSC
Sbjct: 484 ASLLGDAIAYINELKSKVVKTESEKLQIKNQLEEVKLELAGRKASASGGDMSSSCSSIKP 543
Query: 227 QSLSKIEARVSGKDMLIRIHGDKH---CGRTATAILN-ELEKHHLSV 269
+ +IE ++ G D +IR+ K R +A+++ ELE +H S+
Sbjct: 544 VGM-EIEVKIIGWDAMIRVESSKRNHPAARLMSALMDLELEVNHASM 589
>UniRef100_Q700B9 MYC transcription factor [Solanum tuberosum]
Length = 646
Score = 72.4 bits (176), Expect = 1e-11
Identities = 53/157 (33%), Positives = 85/157 (53%), Gaps = 16/157 (10%)
Query: 123 KRENKVSAVNRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKATILEDAIKHL 182
KR K + P+ +HV AER+RREKL+QRF +L +++P + KMDKA++L DAI ++
Sbjct: 459 KRGRKPANGREEPL---NHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAIAYI 515
Query: 183 KQLNERVKT--LEEHVADKKVESAVFMKRSILFEEDDRSSCDENSDQSLS----KIEARV 236
+L +V+ L++ ++ES +++ + + S S+Q L I+ +V
Sbjct: 516 NELKSKVQNSDLDKEELRSQIES---LRKELANKGSSNYSSSPPSNQDLKIVDMDIDVKV 572
Query: 237 SGKDMLIRIHGDK--HCGRTATAILN--ELEKHHLSV 269
G D +IRI K H A L +L+ HH SV
Sbjct: 573 IGWDAMIRIQCSKKNHPAARLMAALKDLDLDVHHASV 609
>UniRef100_Q9ZPY8 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 566
Score = 71.6 bits (174), Expect = 2e-11
Identities = 37/81 (45%), Positives = 56/81 (68%), Gaps = 3/81 (3%)
Query: 114 SVISQGDYYKRENKVSAVNRNPIQAQDHVMAERRRREKLSQRFISLSSLLPGLKKMDKAT 173
SV+ + KR K + P+ +HV AER+RREKL+QRF +L S++P + KMDKA+
Sbjct: 372 SVVDEKRPRKRGRKPANGREEPL---NHVEAERQRREKLNQRFYALRSVVPNISKMDKAS 428
Query: 174 ILEDAIKHLKQLNERVKTLEE 194
+L DAI ++K+L E+VK +E+
Sbjct: 429 LLGDAISYIKELQEKVKIMED 449
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.128 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 461,432,609
Number of Sequences: 2790947
Number of extensions: 18645618
Number of successful extensions: 66342
Number of sequences better than 10.0: 1150
Number of HSP's better than 10.0 without gapping: 351
Number of HSP's successfully gapped in prelim test: 800
Number of HSP's that attempted gapping in prelim test: 65384
Number of HSP's gapped (non-prelim): 1386
length of query: 295
length of database: 848,049,833
effective HSP length: 126
effective length of query: 169
effective length of database: 496,390,511
effective search space: 83889996359
effective search space used: 83889996359
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 74 (33.1 bits)
Medicago: description of AC141862.23


