

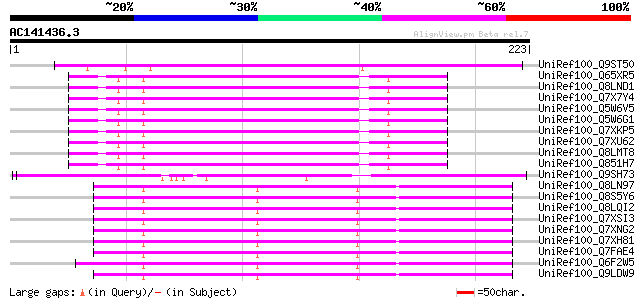
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141436.3 + phase: 0 /pseudo
(223 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ST50 Transposase related protein [Zea mays] 102 7e-21
UniRef100_Q65XR5 Putative polyprotein [Oryza sativa] 99 6e-20
UniRef100_Q8LND1 Putative transposable element [Oryza sativa] 99 8e-20
UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa] 99 8e-20
UniRef100_Q5W6V5 Putative polyprotein [Oryza sativa] 99 8e-20
UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa] 99 8e-20
UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa] 98 1e-19
UniRef100_Q7XU62 OSJNBb0091E11.13 protein [Oryza sativa] 98 1e-19
UniRef100_Q8LMT8 Putative retroelement [Oryza sativa] 97 3e-19
UniRef100_Q851H7 Putative mutator-like transposase [Oryza sativa] 96 5e-19
UniRef100_Q9SH73 F22C12.1 [Arabidopsis thaliana] 92 1e-17
UniRef100_Q8LN97 Putative Mu transposable element [Oryza sativa] 87 3e-16
UniRef100_Q8S5Y6 Putative transposase related protein [Oryza sat... 86 5e-16
UniRef100_Q8LQI2 Putative mutator-like transposase [Oryza sativa] 86 5e-16
UniRef100_Q7XSI3 OSJNBa0073L13.4 protein [Oryza sativa] 86 5e-16
UniRef100_Q7XNG2 OSJNBa0096F01.11 protein [Oryza sativa] 86 5e-16
UniRef100_Q7XH81 Putative mutator-like transposase [Oryza sativa] 86 5e-16
UniRef100_Q7FAE4 OSJNBa0052P16.6 protein [Oryza sativa] 86 5e-16
UniRef100_Q6F2W5 Putative MuDR family transposase [Oryza sativa] 86 5e-16
UniRef100_Q9LDW9 EST C28952(C62945) corresponds to a region of t... 86 7e-16
>UniRef100_Q9ST50 Transposase related protein [Zea mays]
Length = 863
Score = 102 bits (254), Expect = 7e-21
Identities = 64/208 (30%), Positives = 104/208 (49%), Gaps = 7/208 (3%)
Query: 20 MNIPTFEYYRSEI-DVADRKALAWVDNIPK--QKWNQSHDDGR-RWGHMTSNLVESQNNV 75
+ + FE E+ D + + W+ + + QKW ++D+G R+ TSN+ E N+V
Sbjct: 614 LEVSFFEDKLKELKDATNAEGKNWIAGLLREPQKWTSAYDEGGWRFEFQTSNMAELFNSV 673
Query: 76 YKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHH 135
KGIRG+P+ AIV ++YRL A F R +A A G ++ +L N ++N H
Sbjct: 674 LKGIRGMPVNAIVTFTFYRLVAWFNDRHAQAKAMQTRGLRWAPKPTAHLNNAKERANRHE 733
Query: 136 VTQFDRDRYTFSVRE---TIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACS 192
V FD D + VR T E P + V L C CG+ R H PCSH +AA
Sbjct: 734 VQCFDEDLGKYEVRTMGGTTSDGEVRPSRTHVVLLDAFSCGCGKPRQYHFPCSHYVAAAR 793
Query: 193 SFSHDYKTFVDNKFTNECVYAVYNIHFD 220
+ +++ + ++F+ E + ++ F+
Sbjct: 794 HRNFAFESMIPSEFSVESLVRTWSPRFE 821
>UniRef100_Q65XR5 Putative polyprotein [Oryza sativa]
Length = 1378
Score = 99.4 bits (246), Expect = 6e-20
Identities = 58/173 (33%), Positives = 93/173 (53%), Gaps = 17/173 (9%)
Query: 26 EYYRSEIDVADRKALAWVDN--IPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGL 82
E R I RK W+++ + K KW++++D +GRRWG+MT+N+ E N+V G+R L
Sbjct: 480 EALRQRIPEGPRK---WLEDELVDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVGVRKL 536
Query: 83 PITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRD 142
P TAIV ++ + F R EA RV G+ +S ++ + K+N H FD+
Sbjct: 537 PATAIVSFTFMKCNDYFVNRHDEALKRVQLGQRWSTKVDSKMKVQKSKANKHTARCFDKQ 596
Query: 143 RYTFSVRETIEHKEGLPKG-------EYKVDLQNKWCDCGRFRALHLPCSHVI 188
+ T+ V E + G+ +G +KV+++ C C R H+PCSH+I
Sbjct: 597 KKTYEVTE----RGGVTRGGVRFGARAFKVEVEGNSCSCQRPLLYHMPCSHLI 645
>UniRef100_Q8LND1 Putative transposable element [Oryza sativa]
Length = 1592
Score = 99.0 bits (245), Expect = 8e-20
Identities = 58/173 (33%), Positives = 93/173 (53%), Gaps = 17/173 (9%)
Query: 26 EYYRSEIDVADRKALAWVDN--IPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGL 82
E R I RK W+++ + K KW++++D +GRRWG+MT+N+ E N+V G+R L
Sbjct: 670 EALRQRIPEGPRK---WLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVGVRKL 726
Query: 83 PITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRD 142
P+TAIV ++ + F R EA RV G+ +S ++ + K+N H FD+
Sbjct: 727 PVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQRWSTKVDSKMKVQESKANKHTARCFDKQ 786
Query: 143 RYTFSVRETIEHKEGLPKG-------EYKVDLQNKWCDCGRFRALHLPCSHVI 188
+ T+ V E + G+ +G +KV+ + C C R H+PCSH+I
Sbjct: 787 KKTYEVTE----RGGITRGGVRFGARAFKVEGEGNSCSCQRPLLYHMPCSHLI 835
>UniRef100_Q7X7Y4 OSJNBa0014F04.28 protein [Oryza sativa]
Length = 1575
Score = 99.0 bits (245), Expect = 8e-20
Identities = 58/173 (33%), Positives = 93/173 (53%), Gaps = 17/173 (9%)
Query: 26 EYYRSEIDVADRKALAWVDN--IPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGL 82
E R I RK W+++ + K KW++++D +GRRWG+MT+N+ E N+V G+R L
Sbjct: 627 EALRQRIPEGPRK---WLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVGVRKL 683
Query: 83 PITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRD 142
P+TAIV ++ + F R EA RV G+ +S ++ + K+N H FD+
Sbjct: 684 PVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQRWSTKVDSKMKVQESKANKHTARCFDKQ 743
Query: 143 RYTFSVRETIEHKEGLPKG-------EYKVDLQNKWCDCGRFRALHLPCSHVI 188
+ T+ V E + G+ +G +KV+ + C C R H+PCSH+I
Sbjct: 744 KKTYEVTE----RGGITRGGVRFGARAFKVEGEGNSCSCQRPLLYHMPCSHLI 792
>UniRef100_Q5W6V5 Putative polyprotein [Oryza sativa]
Length = 1525
Score = 99.0 bits (245), Expect = 8e-20
Identities = 58/173 (33%), Positives = 93/173 (53%), Gaps = 17/173 (9%)
Query: 26 EYYRSEIDVADRKALAWVDN--IPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGL 82
E R I RK W+++ + K KW++++D +GRRWG+MT+N+ E N+V G+R L
Sbjct: 670 EALRQRIPEGPRK---WLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVGVRKL 726
Query: 83 PITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRD 142
P+TAIV ++ + F R EA RV G+ +S ++ + K+N H FD+
Sbjct: 727 PVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQRWSTKVDSKMKVQESKANKHTARCFDKQ 786
Query: 143 RYTFSVRETIEHKEGLPKG-------EYKVDLQNKWCDCGRFRALHLPCSHVI 188
+ T+ V E + G+ +G +KV+ + C C R H+PCSH+I
Sbjct: 787 KKTYEVTE----RGGITRGGVRFGARAFKVEGEGNSCSCQRPLLYHMPCSHLI 835
>UniRef100_Q5W6G1 Putative polyprotein [Oryza sativa]
Length = 1567
Score = 99.0 bits (245), Expect = 8e-20
Identities = 58/173 (33%), Positives = 93/173 (53%), Gaps = 17/173 (9%)
Query: 26 EYYRSEIDVADRKALAWVDN--IPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGL 82
E R I RK W+++ + K KW++++D +GRRWG+MT+N+ E N+V G+R L
Sbjct: 685 EALRQRIPEGPRK---WLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVGVRKL 741
Query: 83 PITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRD 142
P+TAIV ++ + F R EA RV G+ +S ++ + K+N H FD+
Sbjct: 742 PVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQRWSTKVDSKMKVQESKANKHTARCFDKQ 801
Query: 143 RYTFSVRETIEHKEGLPKG-------EYKVDLQNKWCDCGRFRALHLPCSHVI 188
+ T+ V E + G+ +G +KV+ + C C R H+PCSH+I
Sbjct: 802 KKTYEVTE----RGGITRGGVRFGARAFKVEGEGNSCSCQRPLLYHMPCSHLI 850
>UniRef100_Q7XKP5 OSJNBb0013O03.11 protein [Oryza sativa]
Length = 1495
Score = 98.2 bits (243), Expect = 1e-19
Identities = 58/173 (33%), Positives = 93/173 (53%), Gaps = 17/173 (9%)
Query: 26 EYYRSEIDVADRKALAWVDN--IPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGL 82
E R I RK W+++ + K KW++++D +GRRWG+MT+N+ E N+V G+R L
Sbjct: 595 EAVRQRIPEGPRK---WLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVGVRKL 651
Query: 83 PITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRD 142
P+TAIV ++ + F R EA RV G+ +S ++ + K+N H FD+
Sbjct: 652 PVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQRWSTKIDSKMKVQKNKANEHTARCFDQQ 711
Query: 143 RYTFSVRETIEHKEGLPKG-------EYKVDLQNKWCDCGRFRALHLPCSHVI 188
+ T+ V E + G+ +G +KV+ + C C R H+PCSH+I
Sbjct: 712 KKTYEVTE----RGGITRGGVRFGARAFKVEGEGNSCSCQRPLLYHMPCSHLI 760
>UniRef100_Q7XU62 OSJNBb0091E11.13 protein [Oryza sativa]
Length = 1107
Score = 98.2 bits (243), Expect = 1e-19
Identities = 58/173 (33%), Positives = 92/173 (52%), Gaps = 17/173 (9%)
Query: 26 EYYRSEIDVADRKALAWVDN--IPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGL 82
E R I RK W+++ + K KW++++D +GRRWG+MT+N+ E N+V G+R L
Sbjct: 674 EALRQHIPEGSRK---WLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVGVRKL 730
Query: 83 PITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRD 142
P+TAIV ++ + F R EA RV G+ +S ++ + K+N H FD+
Sbjct: 731 PVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQRWSTKVDSKMKVQKSKANKHTARCFDKQ 790
Query: 143 RYTFSVRETIEHKEGLPKG-------EYKVDLQNKWCDCGRFRALHLPCSHVI 188
+ T+ V E + G+ G +KV+ + C C R H+PCSH+I
Sbjct: 791 KKTYEVTE----RGGITHGGVRFGARAFKVEGEGNSCSCQRPLLYHMPCSHLI 839
>UniRef100_Q8LMT8 Putative retroelement [Oryza sativa]
Length = 1460
Score = 97.1 bits (240), Expect = 3e-19
Identities = 57/173 (32%), Positives = 92/173 (52%), Gaps = 17/173 (9%)
Query: 26 EYYRSEIDVADRKALAWVDN--IPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGL 82
E R I RK W+++ + K KW++++D +GRRWG+MT+N+ E N+V G+ L
Sbjct: 727 EALRQRIPEGPRK---WLEDELLDKDKWSRAYDRNGRRWGYMTTNMAEQFNSVLVGVHKL 783
Query: 83 PITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRD 142
P+TAIV ++ + F R EA RV G+ +S ++ + K+N H FD+
Sbjct: 784 PVTAIVSFTFMKCNDYFVNRHDEALKRVQLGQRWSTKVDSKMKVQKSKANKHTARCFDKQ 843
Query: 143 RYTFSVRETIEHKEGLPKG-------EYKVDLQNKWCDCGRFRALHLPCSHVI 188
+ T+ V E + G+ +G +KV+ + C C R H+PCSH+I
Sbjct: 844 KKTYEVTE----RGGITRGGVRFGARAFKVEGEGNSCSCQRPLLYHMPCSHLI 892
>UniRef100_Q851H7 Putative mutator-like transposase [Oryza sativa]
Length = 1360
Score = 96.3 bits (238), Expect = 5e-19
Identities = 57/173 (32%), Positives = 92/173 (52%), Gaps = 17/173 (9%)
Query: 26 EYYRSEIDVADRKALAWVDN--IPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGL 82
E R I RK W+++ + K KW++++D + RWG+MT+N+ E N+V G+R L
Sbjct: 663 EALRQRIPEGPRK---WLEDELLDKDKWSRAYDRNSCRWGYMTTNMAEQFNSVLVGVRKL 719
Query: 83 PITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRD 142
P+TAIV ++ + F R +EA RV SGE +S ++ + K+N H FD+
Sbjct: 720 PVTAIVAFTFMKCNHYFVNRQNEAVKRVQSGERWSAKVASKMKVQKSKANKHTSRCFDKQ 779
Query: 143 RYTFSVRETIEHKEGLPKG-------EYKVDLQNKWCDCGRFRALHLPCSHVI 188
+ T+ V E K G+ + +KV+ + C C R H+PCSH++
Sbjct: 780 KKTYEVTE----KGGITRSGVRFGARAFKVEGEGNSCSCQRPLLYHMPCSHLV 828
>UniRef100_Q9SH73 F22C12.1 [Arabidopsis thaliana]
Length = 3290
Score = 91.7 bits (226), Expect = 1e-17
Identities = 60/224 (26%), Positives = 104/224 (45%), Gaps = 17/224 (7%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
F+D LK V G F+ Y +EI+ + +A W+D P+ +W Q+HD GRR+
Sbjct: 1556 FQDDYLKNLVYEAGSTSEKEEFDSYMNEIEKKNSEARKWLDQFPQYQWAQAHDSGRRYRV 1615
Query: 64 MTSNLVESQN-----NVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSE 118
MT ++++N ++ + GLP+TA + + F +++ RVN G+ +++
Sbjct: 1616 MT---IDAENLFAFCESFQSL-GLPVTATALLLFDEMRFFFYSGLCDSSGRVNRGDMYTK 1671
Query: 119 NSMKYLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFR 178
M L + S H V ++ + + E L + E+ V L C CG F+
Sbjct: 1672 PVMDQLEKLMTDSIPHVVMPLEKGLFQVT--------EPLQEDEWIVQLSEWSCTCGEFQ 1723
Query: 179 ALHLPCSHVIAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVV 222
PC HV+A C + +VD+ ++ + +Y Y F V
Sbjct: 1724 LKKFPCLHVLAVCEKLKINPLQYVDDCYSLDRLYKTYAATFSPV 1767
Score = 80.9 bits (198), Expect = 2e-14
Identities = 62/225 (27%), Positives = 99/225 (43%), Gaps = 19/225 (8%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
F+D L+ VE G F+ Y ++I + +A W+D IP+ KW +HD G R+G
Sbjct: 3053 FRDYNLESLVEQAGSTNQKEEFDSYMNDIKEKNPEAWKWLDQIPRHKWALAHDSGLRYG- 3111
Query: 64 MTSNLVE-SQNNVYKGIRGLP-----ITAIVKASYYRLAALFAKRGHEAAARVNSGEPFS 117
++E + ++ RG P +T V + L + F K + +N G ++
Sbjct: 3112 ----IIEIDREALFAVCRGFPYCTVAMTGGVMLMFDELRSSFDKSLSSIYSSLNRGVVYT 3167
Query: 118 ENSMKYLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRF 177
E M L + S + +TQ +RD +F V E+ E K E+ V L C C +F
Sbjct: 3168 EPFMDKLEEFMTDSIPYVITQLERD--SFKVSESSE------KEEWIVQLNVSTCTCRKF 3219
Query: 178 RALHLPCSHVIAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVV 222
++ PC H +A + +VD +T E Y F V
Sbjct: 3220 QSYKFPCLHALAVFEKLKINPLQYVDECYTVEQYCKTYAATFSPV 3264
Score = 73.9 bits (180), Expect = 3e-12
Identities = 58/225 (25%), Positives = 96/225 (41%), Gaps = 13/225 (5%)
Query: 2 RSFKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRW 61
R F L ++ G F Y ++I + +A W+D P+ +W +HD+GRR+
Sbjct: 2328 RVFPSFCLGARIRRAGSTSQKDEFVSYMNDIKEKNPEARKWLDQFPQNRWALAHDNGRRY 2387
Query: 62 GHMTSNL--VESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSEN 119
G M N + + N ++ G +T V + L + F K + + +N G+ ++E
Sbjct: 2388 GIMEINTKALFAVCNAFEQA-GHVVTGSVLLLFDELRSKFDKSFSCSRSSLNCGDVYTEP 2446
Query: 120 SMKYLRN--EVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRF 177
M L + S+ VT D + + + L KGE V L + C CG F
Sbjct: 2447 VMDKLEEFRTTFVTYSYIVTPLDNNAFQVAT--------ALDKGECIVQLSDCSCTCGDF 2498
Query: 178 RALHLPCSHVIAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVV 222
+ PC H +A C + +VD+ +T E + Y F V
Sbjct: 2499 QRYKFPCLHALAVCKKLKFNPLQYVDDCYTLERLKRTYATIFSHV 2543
Score = 72.8 bits (177), Expect = 6e-12
Identities = 54/222 (24%), Positives = 93/222 (41%), Gaps = 13/222 (5%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
F+D L+ V+ G F+ Y +I+ + +A W+D P+ +W +HD+GRR+G
Sbjct: 728 FRDYYLEDLVKRAGSTSQKEEFDSYMKDIEKKNSEARKWLDQFPQNQWALAHDNGRRYGI 787
Query: 64 M---TSNLVESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENS 120
M T+ L E N + +T V + L F + + G ++E
Sbjct: 788 MEIETTTLFEDFN--VSHLDNHVLTGYVLRLFDELRHSFDEFFCFSRGSRKCGNVYTEPV 845
Query: 121 MKYLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRAL 180
+ L S ++ V D + + + + E+ V L + C CG F++
Sbjct: 846 TEKLAESRKDSVTYDVMPLDNNAFQVTAPQ--------ENDEWTVQLSDCSCTCGEFQSC 897
Query: 181 HLPCSHVIAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVV 222
PC H +A C + +VD+ +T E +Y Y F V
Sbjct: 898 KFPCLHALAVCKLLKINPLEYVDDCYTLERLYKTYTATFSPV 939
>UniRef100_Q8LN97 Putative Mu transposable element [Oryza sativa]
Length = 1536
Score = 87.0 bits (214), Expect = 3e-16
Identities = 55/188 (29%), Positives = 89/188 (47%), Gaps = 9/188 (4%)
Query: 37 RKALAWVDNIPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKASYYRL 95
R W++N PK+KW+ D DG R+G MT+NL E N V +G+R LP+ AIV+ +
Sbjct: 601 RNFTQWIENEPKEKWSLLFDTDGSRYGIMTTNLAEVYNWVMRGVRVLPLVAIVEFILHGT 660
Query: 96 AALFAKRGHE-AAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTFSV------ 148
A F R + + ++ F KY+ +++ K+ H V + + +
Sbjct: 661 QAYFRDRYKKIGPSMADNNIVFGNVVTKYMEDKIKKARRHRVVAQGTQVHRYEIMCVDRS 720
Query: 149 RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKTFVDNKFTN 208
R I K+ + + K D C C + + HLPCSHV+AA +V N F
Sbjct: 721 RRVIYRKQAVQECVLKAD-GGCTCSCMKPKLHHLPCSHVLAAAGDCGISPNVYVSNYFRK 779
Query: 209 ECVYAVYN 216
E ++ ++
Sbjct: 780 EAIFHTWS 787
>UniRef100_Q8S5Y6 Putative transposase related protein [Oryza sativa]
Length = 1557
Score = 86.3 bits (212), Expect = 5e-16
Identities = 55/188 (29%), Positives = 89/188 (47%), Gaps = 9/188 (4%)
Query: 37 RKALAWVDNIPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKASYYRL 95
R W++N PK+KW+ D DG R+G MT+NL E N V +G+R LP+ AIV+ +
Sbjct: 574 RNFTQWIENEPKEKWSLLFDTDGSRYGIMTTNLAEVYNWVMRGVRVLPLVAIVEFILHGT 633
Query: 96 AALFAKRGHE-AAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTFSV------ 148
A F R + + ++ F KY+ +++ K+ H V + + +
Sbjct: 634 QAYFRDRYKKIGPSMADNNIVFGNVVTKYMEDKIKKARRHRVVAQGTQVHRYEIMCVDRS 693
Query: 149 RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKTFVDNKFTN 208
R I K+ + + K D C C + + HLPCSHV+ A S +V N F
Sbjct: 694 RRGIYRKQAVQECVLKAD-GGCTCSCMKPKLHHLPCSHVLTAASDCGISPNVYVSNYFRK 752
Query: 209 ECVYAVYN 216
E ++ ++
Sbjct: 753 EAIFHTWS 760
>UniRef100_Q8LQI2 Putative mutator-like transposase [Oryza sativa]
Length = 1592
Score = 86.3 bits (212), Expect = 5e-16
Identities = 55/188 (29%), Positives = 89/188 (47%), Gaps = 9/188 (4%)
Query: 37 RKALAWVDNIPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKASYYRL 95
R W++N PK+KW+ D DG R+G MT+NL E N V +G+R LP+ AIV+ +
Sbjct: 601 RNFTQWIENEPKEKWSLLFDTDGSRYGIMTTNLAEVYNWVMRGVRVLPLVAIVEFILHGT 660
Query: 96 AALFAKRGHE-AAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTFSV------ 148
A F R + + ++ F KY+ +++ K+ H V + + +
Sbjct: 661 QAYFRDRYKKIGPSMADNNIVFGNVVTKYMEDKIKKARRHRVVAQGTQVHRYEIMCVDRS 720
Query: 149 RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKTFVDNKFTN 208
R I K+ + + K D C C + + HLPCSHV+AA +V N F
Sbjct: 721 RRGIYRKQAVQECVLKAD-GGCTCSCMKPKLHHLPCSHVLAAAGDCGISPNVYVSNYFRK 779
Query: 209 ECVYAVYN 216
E ++ ++
Sbjct: 780 EAIFHTWS 787
>UniRef100_Q7XSI3 OSJNBa0073L13.4 protein [Oryza sativa]
Length = 1342
Score = 86.3 bits (212), Expect = 5e-16
Identities = 55/188 (29%), Positives = 89/188 (47%), Gaps = 9/188 (4%)
Query: 37 RKALAWVDNIPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKASYYRL 95
R W++N PK+KW+ D DG R+G MT+NL E N V +G+R LP+ AIV+ +
Sbjct: 472 RNFTQWIENEPKEKWSLLFDTDGSRYGIMTTNLAEVYNWVMRGVRVLPLVAIVEFILHGT 531
Query: 96 AALFAKRGHE-AAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTFSV------ 148
A F R + + ++ F KY+ +++ K+ H V + + +
Sbjct: 532 QAYFRDRYKKIGPSMADNNIVFGNVVTKYMEDKIKKAQRHRVVAQGTQVHRYEIMCVDRS 591
Query: 149 RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKTFVDNKFTN 208
R I K+ + + K D C C + + HLPCSHV+AA +V N F
Sbjct: 592 RRGIYRKQAVQECVLKAD-GGCTCSCMKPKLHHLPCSHVLAAAGDCGISPNVYVSNYFRK 650
Query: 209 ECVYAVYN 216
E ++ ++
Sbjct: 651 EAIFHTWS 658
>UniRef100_Q7XNG2 OSJNBa0096F01.11 protein [Oryza sativa]
Length = 1365
Score = 86.3 bits (212), Expect = 5e-16
Identities = 55/188 (29%), Positives = 89/188 (47%), Gaps = 9/188 (4%)
Query: 37 RKALAWVDNIPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKASYYRL 95
R W++N PK+KW+ D DG R+G MT+NL E N V +G+R LP+ AIV+ +
Sbjct: 450 RNFTQWIENEPKEKWSLLFDTDGSRYGIMTTNLAEVYNWVMRGVRVLPLVAIVEFILHGT 509
Query: 96 AALFAKRGHE-AAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTFSV------ 148
A F R + + ++ F KY+ +++ K+ H V + + +
Sbjct: 510 QAYFRDRYKKIGPSMADNNIVFGNVVTKYMEDKIKKARRHRVVAQGTQVHQYEIMCVDRS 569
Query: 149 RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKTFVDNKFTN 208
R I K+ + + K D C C + + HLPCSHV+AA +V N F
Sbjct: 570 RRGIYRKQAVQECVLKAD-GGCTCSCMKPKLHHLPCSHVLAAAGDCGISPNVYVSNYFRK 628
Query: 209 ECVYAVYN 216
E ++ ++
Sbjct: 629 EAIFHTWS 636
>UniRef100_Q7XH81 Putative mutator-like transposase [Oryza sativa]
Length = 1597
Score = 86.3 bits (212), Expect = 5e-16
Identities = 55/188 (29%), Positives = 89/188 (47%), Gaps = 9/188 (4%)
Query: 37 RKALAWVDNIPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKASYYRL 95
R W++N PK+KW+ D DG R+G MT+NL E N V +G+R LP+ AIV+ +
Sbjct: 601 RNFTQWIENEPKEKWSLLFDTDGSRYGIMTTNLAEVYNWVMRGVRVLPLVAIVEFILHGT 660
Query: 96 AALFAKRGHE-AAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTFSV------ 148
A F R + + ++ F KY+ +++ K+ H V + + +
Sbjct: 661 QAYFRDRYKKIGPSMADNNIVFGNVVTKYMEDKIKKARRHRVVAQGTQVHRYEIMCVDRS 720
Query: 149 RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKTFVDNKFTN 208
R I K+ + + K D C C + + HLPCSHV+AA +V N F
Sbjct: 721 RRGIYRKQAVQECVLKAD-GGCTCSCMKPKLHHLPCSHVLAAAGDCGISPNVYVSNYFRK 779
Query: 209 ECVYAVYN 216
E ++ ++
Sbjct: 780 EAIFHTWS 787
>UniRef100_Q7FAE4 OSJNBa0052P16.6 protein [Oryza sativa]
Length = 1489
Score = 86.3 bits (212), Expect = 5e-16
Identities = 55/188 (29%), Positives = 89/188 (47%), Gaps = 9/188 (4%)
Query: 37 RKALAWVDNIPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKASYYRL 95
R W++N PK+KW+ D DG R+G MT+NL E N V +G+R LP+ AIV+ +
Sbjct: 450 RNFTQWIENEPKEKWSLLFDTDGSRYGIMTTNLAEVYNWVMRGVRVLPLVAIVEFILHGT 509
Query: 96 AALFAKRGHE-AAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTFSV------ 148
A F R + + ++ F KY+ +++ K+ H V + + +
Sbjct: 510 QAYFRDRYKKIGPSMADNNIVFGNVVTKYMEDKIKKARRHRVVAQGTQVHQYEIMCVDRS 569
Query: 149 RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKTFVDNKFTN 208
R I K+ + + K D C C + + HLPCSHV+AA +V N F
Sbjct: 570 RRGIYRKQAVQECVLKAD-GGCTCSCMKPKLHHLPCSHVLAAAGDCGISPNVYVSNYFRK 628
Query: 209 ECVYAVYN 216
E ++ ++
Sbjct: 629 EAIFHTWS 636
>UniRef100_Q6F2W5 Putative MuDR family transposase [Oryza sativa]
Length = 1043
Score = 86.3 bits (212), Expect = 5e-16
Identities = 56/196 (28%), Positives = 91/196 (45%), Gaps = 9/196 (4%)
Query: 29 RSEIDVADRKALAWVDNIPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGLPITAI 87
R + + R W++N PK+KW+ D DG R+G MT+NL E N V +G+R LP+ AI
Sbjct: 371 RRRLGTSIRNFTQWIENEPKEKWSLLFDTDGSRYGIMTTNLAEVYNWVMRGVRVLPLVAI 430
Query: 88 VKASYYRLAALFAKRGHE-AAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTF 146
V+ + A F R + + ++ F KY+ +++ K H V + +
Sbjct: 431 VEFILHGTQAYFRDRYKKIGPSMADNNIVFGNVVTKYMEDKIKKVRRHRVVAQGTQVHRY 490
Query: 147 SV------RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKT 200
+ R I K+ + + K D C C + + HLPCSHV+AA
Sbjct: 491 EIMCVDRSRRGIYRKQAVQECVLKAD-GGCTCSCMKPKLHHLPCSHVLAAAGDCGISPNV 549
Query: 201 FVDNKFTNECVYAVYN 216
+V N F E ++ ++
Sbjct: 550 YVSNYFRKEAIFHTWS 565
>UniRef100_Q9LDW9 EST C28952(C62945) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 1591
Score = 85.9 bits (211), Expect = 7e-16
Identities = 55/188 (29%), Positives = 88/188 (46%), Gaps = 9/188 (4%)
Query: 37 RKALAWVDNIPKQKWNQSHD-DGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKASYYRL 95
R W++N PK+KW+ D DG R+G MT+NL E N V +G+R LP+ AIV+ +
Sbjct: 601 RNFTQWIENEPKEKWSLLFDTDGSRYGIMTTNLAEVYNWVMRGVRVLPLVAIVEFILHGT 660
Query: 96 AALFAKRGHE-AAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTFSV------ 148
A F R + + + F KY+ +++ K+ H V + + +
Sbjct: 661 QAYFRDRYKKIGPSMADDNIVFGNVVTKYMEDKIKKARRHRVVAQGTQVHRYEIMCVDRS 720
Query: 149 RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKTFVDNKFTN 208
R I K+ + + K D C C + + HLPCSHV+AA +V N F
Sbjct: 721 RRGIYRKQAVQEYVLKAD-GGCTCSCMKPKLHHLPCSHVLAAAGDCGISPNVYVSNYFRK 779
Query: 209 ECVYAVYN 216
E ++ ++
Sbjct: 780 EAIFHTWS 787
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.135 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 388,056,554
Number of Sequences: 2790947
Number of extensions: 15768831
Number of successful extensions: 33729
Number of sequences better than 10.0: 242
Number of HSP's better than 10.0 without gapping: 196
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 33269
Number of HSP's gapped (non-prelim): 257
length of query: 223
length of database: 848,049,833
effective HSP length: 123
effective length of query: 100
effective length of database: 504,763,352
effective search space: 50476335200
effective search space used: 50476335200
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC141436.3


