

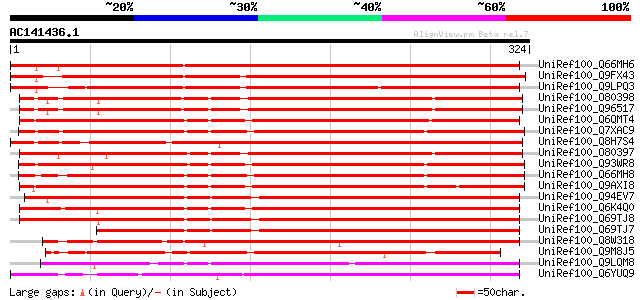
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141436.1 + phase: 0
(324 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q66MH6 MAPKK [Lycopersicon esculentum] 412 e-114
UniRef100_Q9FX43 MAP kinase, putative [Arabidopsis thaliana] 361 1e-98
UniRef100_Q9LPQ3 F15H18.14 [Arabidopsis thaliana] 323 4e-87
UniRef100_O80398 MAP kinase kinase 5 [Arabidopsis thaliana] 285 1e-75
UniRef100_Q96517 MAP kinase kinase alpha protein kinase [Arabido... 283 4e-75
UniRef100_Q6QMT4 Mitogen-activated protein kinase kinase 5 [Petr... 283 6e-75
UniRef100_Q7XAC9 Mitogen-activated protein kinase kinase [Solanu... 279 8e-74
UniRef100_Q8H7S4 Hypothetical protein OSJNBa0081P02.7 [Oryza sat... 277 3e-73
UniRef100_O80397 MAP kinase kinase 4 [Arabidopsis thaliana] 277 3e-73
UniRef100_Q93WR8 MEK map kinase kinsae [Medicago varia] 274 3e-72
UniRef100_Q66MH8 MAPKK [Lycopersicon esculentum] 271 1e-71
UniRef100_Q9AXI8 Mitogen-activated protein kinase 2 [Nicotiana t... 269 7e-71
UniRef100_Q94EV7 MAP kinase kinase [Zea mays] 261 2e-68
UniRef100_Q6K4Q0 Putative MAP kinase kinase [Oryza sativa] 257 3e-67
UniRef100_Q69TJ8 Putative MAP kinase kinase [Oryza sativa] 256 6e-67
UniRef100_Q69TJ7 Putative MAP kinase kinase [Oryza sativa] 248 2e-64
UniRef100_Q8W318 Putative kinase [Oryza sativa] 245 1e-63
UniRef100_Q9M8J5 Putative MAP kinase [Arabidopsis thaliana] 244 2e-63
UniRef100_Q9LQM8 F5D14.7 protein [Arabidopsis thaliana] 225 1e-57
UniRef100_Q6YUQ9 Putative MAP kinase kinase [Oryza sativa] 203 6e-51
>UniRef100_Q66MH6 MAPKK [Lycopersicon esculentum]
Length = 335
Score = 412 bits (1058), Expect = e-114
Identities = 204/328 (62%), Positives = 246/328 (74%), Gaps = 11/328 (3%)
Query: 1 MALVHRRNSPKLRLP--EISDHRPRFPVPLP--------ANIYKQPSTSATTASVAGGDN 50
MALV R LRLP E S+ RPRFP+PLP + +T+ TT + A
Sbjct: 1 MALVRDRRHLNLRLPLPEPSERRPRFPLPLPPSSVPTVNSTASTTTATNTTTTTTASTTT 60
Query: 51 ISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRAT 110
IS + EKL VLGHGNGGTVYKVRHK TS IYALK+ H DSDP RR+ L E++ILRR T
Sbjct: 61 ISISELEKLKVLGHGNGGTVYKVRHKRTSAIYALKVVHGDSDPEIRRQILREISILRR-T 119
Query: 111 DCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYL 170
+ V+K HG + P GD+ ILMEYM++G+LE LK+ TFSE L+ +A+ +L GL YL
Sbjct: 120 ESPYVIKCHGVIDMPGGDIGILMEYMNAGTLENLLKSQLTFSELCLARIAKQVLGGLDYL 179
Query: 171 HARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEV 230
H+ I HRD+KPSN+LVN + EVKIADFGVSK MGRTL+ CNSYVGTCAYMSPERFDP+
Sbjct: 180 HSHKIIHRDLKPSNLLVNREMEVKIADFGVSKIMGRTLDPCNSYVGTCAYMSPERFDPDT 239
Query: 231 YGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSE 290
YGGNYNG++ADIWSLGLTL ELY+G+FPFL GQRPDWA+LMCAICF +PPSLPE S +
Sbjct: 240 YGGNYNGYAADIWSLGLTLMELYMGHFPFLPPGQRPDWATLMCAICFGEPPSLPENTSEK 299
Query: 291 FRNFVECCLKKESGERWSAAQLLTHPFL 318
F +F++CCL+KES +RWSA QLL HPF+
Sbjct: 300 FNDFMKCCLQKESSKRWSAHQLLQHPFI 327
>UniRef100_Q9FX43 MAP kinase, putative [Arabidopsis thaliana]
Length = 310
Score = 361 bits (927), Expect = 1e-98
Identities = 192/325 (59%), Positives = 231/325 (71%), Gaps = 18/325 (5%)
Query: 1 MALVHRRNSPKLRLP--EISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEK 58
MALV R LRLP ISD R + S+SATT +VAG + ISA D EK
Sbjct: 1 MALVRERRQLNLRLPLPPISDRR-----------FSTSSSSATTTTVAGCNGISACDLEK 49
Query: 59 LSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKY 118
L+VLG GNGG VYKVRHK TS IYALK + D DP R+ + E+ ILRR TD VVK
Sbjct: 50 LNVLGCGNGGIVYKVRHKTTSEIYALKTVNGDMDPIFTRQLMREMEILRR-TDSPYVVKC 108
Query: 119 HGSFEKPT-GDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAH 177
HG FEKP G+V ILMEYMD G+LE+ G +E KL+ A+ IL GL+YLHA I H
Sbjct: 109 HGIFEKPVVGEVSILMEYMDGGTLESL---RGGVTEQKLAGFAKQILKGLSYLHALKIVH 165
Query: 178 RDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNG 237
RDIKP+N+L+N KNEVKIADFGVSK + R+L++CNSYVGTCAYMSPERFD E GG+ +
Sbjct: 166 RDIKPANLLLNSKNEVKIADFGVSKILVRSLDSCNSYVGTCAYMSPERFDSESSGGSSDI 225
Query: 238 FSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVEC 297
++ DIWS GL + EL VG+FP L GQRPDWA+LMCA+CF +PP PE S EFR+FVEC
Sbjct: 226 YAGDIWSFGLMMLELLVGHFPLLPPGQRPDWATLMCAVCFGEPPRAPEGCSEEFRSFVEC 285
Query: 298 CLKKESGERWSAAQLLTHPFLCKDM 322
CL+K+S +RW+A QLL HPFL +D+
Sbjct: 286 CLRKDSSKRWTAPQLLAHPFLREDL 310
>UniRef100_Q9LPQ3 F15H18.14 [Arabidopsis thaliana]
Length = 307
Score = 323 bits (828), Expect = 4e-87
Identities = 175/321 (54%), Positives = 217/321 (67%), Gaps = 21/321 (6%)
Query: 1 MALVHRRNSPKLRLP--EISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEK 58
MALV +R LRLP +S H P F S +++TA V + ISA D EK
Sbjct: 1 MALVRKRRQINLRLPVPPLSVHLPWF------------SFASSTAPVIN-NGISASDVEK 47
Query: 59 LSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKY 118
L VLG G+ G VYKV HK T IYALK + D P R+ E+ ILRR TD VV+
Sbjct: 48 LHVLGRGSSGIVYKVHHKTTGEIYALKSVNGDMSPAFTRQLAREMEILRR-TDSPYVVRC 106
Query: 119 HGSFEKP-TGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAH 177
G FEKP G+V ILMEYMD G+LE+ G +E +L+ +R IL GL+YLH+ I H
Sbjct: 107 QGIFEKPIVGEVSILMEYMDGGNLESL---RGAVTEKQLAGFSRQILKGLSYLHSLKIVH 163
Query: 178 RDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNG 237
RDIKP+N+L+N +NEVKIADFGVSK + R+L+ CNSYVGTCAYMSPERFD G N +
Sbjct: 164 RDIKPANLLLNSRNEVKIADFGVSKIITRSLDYCNSYVGTCAYMSPERFDSAA-GENSDV 222
Query: 238 FSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVEC 297
++ DIWS G+ + EL+VG+FP L GQRPDWA+LMC +CF +PP PE S EFR+FV+C
Sbjct: 223 YAGDIWSFGVMILELFVGHFPLLPQGQRPDWATLMCVVCFGEPPRAPEGCSDEFRSFVDC 282
Query: 298 CLKKESGERWSAAQLLTHPFL 318
CL+KES ERW+A+QLL HPFL
Sbjct: 283 CLRKESSERWTASQLLGHPFL 303
>UniRef100_O80398 MAP kinase kinase 5 [Arabidopsis thaliana]
Length = 348
Score = 285 bits (729), Expect = 1e-75
Identities = 157/321 (48%), Positives = 209/321 (64%), Gaps = 19/321 (5%)
Query: 7 RNSPKLRLPEISDHRP---RFPVPLPANIYKQPSTSATTASVAGGDNISAG----DFEKL 59
R P L LP HR P+PLP S+SA +S A NISA + E++
Sbjct: 19 RKRPDLSLP--LPHRDVALAVPLPLPP---PSSSSSAPASSSAISTNISAAKSLSELERV 73
Query: 60 SVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYH 119
+ +G G GGTVYKV H TS +ALK+ + + + T RR+ E+ ILR + D NVVK H
Sbjct: 74 NRIGSGAGGTVYKVIHTPTSRPFALKVIYGNHEDTVRRQICREIEILR-SVDHPNVVKCH 132
Query: 120 GSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRD 179
F+ G++ +L+E+MD GSLE A + E +L+ ++R IL+GL YLH R+I HRD
Sbjct: 133 DMFDH-NGEIQVLLEFMDQGSLEGA----HIWQEQELADLSRQILSGLAYLHRRHIVHRD 187
Query: 180 IKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFS 239
IKPSN+L+N VKIADFGVS+ + +T++ CNS VGT AYMSPER + ++ G Y+G++
Sbjct: 188 IKPSNLLINSAKNVKIADFGVSRILAQTMDPCNSSVGTIAYMSPERINTDLNHGRYDGYA 247
Query: 240 ADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCL 299
D+WSLG+++ E Y+G FPF S Q DWASLMCAIC S PP P TAS EFR+FV CCL
Sbjct: 248 GDVWSLGVSILEFYLGRFPFAVSRQ-GDWASLMCAICMSQPPEAPATASQEFRHFVSCCL 306
Query: 300 KKESGERWSAAQLLTHPFLCK 320
+ + +RWSA QLL HPF+ K
Sbjct: 307 QSDPPKRWSAQQLLQHPFILK 327
>UniRef100_Q96517 MAP kinase kinase alpha protein kinase [Arabidopsis thaliana]
Length = 348
Score = 283 bits (724), Expect = 4e-75
Identities = 156/321 (48%), Positives = 209/321 (64%), Gaps = 19/321 (5%)
Query: 7 RNSPKLRLPEISDHRP---RFPVPLPANIYKQPSTSATTASVAGGDNISAG----DFEKL 59
R P L LP HR P+PLP S+SA +S A NISA + E++
Sbjct: 19 RKRPDLSLP--LPHRDVALAVPLPLPP---PSSSSSAPASSSAISTNISAAKSLSELERV 73
Query: 60 SVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYH 119
+ +G G GGTVYKV H TS +ALK+ + + + T RR+ E+ ILR + D NVVK H
Sbjct: 74 NRIGSGAGGTVYKVIHTPTSRPFALKVIYGNHEDTVRRQICREIEILR-SVDHPNVVKCH 132
Query: 120 GSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRD 179
F+ G++ +L+E+MD GSLE A + E +L+ ++R IL+GL YLH R+I HRD
Sbjct: 133 DMFDH-NGEIQVLLEFMDQGSLEGA----HIWQEQELADLSRQILSGLAYLHRRHIVHRD 187
Query: 180 IKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFS 239
IKPS++L+N VKIADFGVS+ + +T++ CNS VGT AYMSPER + ++ G Y+G++
Sbjct: 188 IKPSDLLINSAKNVKIADFGVSRILAQTMDPCNSSVGTIAYMSPERINTDLNHGRYDGYA 247
Query: 240 ADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCL 299
D+WSLG+++ E Y+G FPF S Q DWASLMCAIC S PP P TAS EFR+FV CCL
Sbjct: 248 GDVWSLGVSILEFYLGRFPFAVSRQ-GDWASLMCAICMSQPPEAPATASQEFRHFVSCCL 306
Query: 300 KKESGERWSAAQLLTHPFLCK 320
+ + +RWSA QLL HPF+ K
Sbjct: 307 QSDPPKRWSAQQLLQHPFILK 327
>UniRef100_Q6QMT4 Mitogen-activated protein kinase kinase 5 [Petroselinum crispum]
Length = 356
Score = 283 bits (723), Expect = 6e-75
Identities = 144/312 (46%), Positives = 205/312 (65%), Gaps = 8/312 (2%)
Query: 7 RNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGN 66
R P L LP + P VPLP PS++ ++ A I+ + ++++ +G G+
Sbjct: 18 RRRPDLTLP-LPQRDPAIAVPLPLPPNSAPSSTTAASAAAAIQPINFSELDRINRIGSGS 76
Query: 67 GGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPT 126
GGTVYKV H+ T +YALK+ + + + + RR+ E+ ILR D NVVK H ++
Sbjct: 77 GGTVYKVAHRPTGKLYALKVIYGNHEESVRRQICREIEILRDV-DNLNVVKCHDMYDH-A 134
Query: 127 GDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNIL 186
G++ +L+EYMD GSLE T +E+ L+ + R IL+G+ YLH + I HRDIKPSN+L
Sbjct: 135 GEIQVLLEYMDRGSLEGIHIT----NEAALADLTRQILSGIHYLHRKRIVHRDIKPSNLL 190
Query: 187 VNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLG 246
VN + +VKIADFGVS+ + +T++ CNS VGT AYMSPER + ++ G Y+G++ DIWSLG
Sbjct: 191 VNSRKQVKIADFGVSRVLAQTMDPCNSSVGTIAYMSPERINTDLNQGRYDGYAGDIWSLG 250
Query: 247 LTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGER 306
+++ E Y+G FPF S + DWASLMCAIC S PP P TAS +FR F+ CCL+++ R
Sbjct: 251 VSILEFYMGRFPFAVS-RGGDWASLMCAICMSQPPEAPPTASRQFREFISCCLQRDPHRR 309
Query: 307 WSAAQLLTHPFL 318
W+A QLL HPF+
Sbjct: 310 WTANQLLRHPFV 321
>UniRef100_Q7XAC9 Mitogen-activated protein kinase kinase [Solanum tuberosum]
Length = 374
Score = 279 bits (713), Expect = 8e-74
Identities = 146/316 (46%), Positives = 206/316 (64%), Gaps = 10/316 (3%)
Query: 6 RRNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHG 65
RR L LP+ D P+PLP PS+S++++S + + E+++ +G G
Sbjct: 36 RRTDLTLPLPQ-RDVALAVPLPLPPT--SAPSSSSSSSSSPLPTPLHFSELERVNRIGSG 92
Query: 66 NGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKP 125
GGTVYKV H+ T +YALK+ + + + + R + E+ ILR D NVV+ H F+
Sbjct: 93 TGGTVYKVLHRPTGRLYALKVIYGNHEDSVRLQMCREIEILRDV-DNPNVVRCHDMFDH- 150
Query: 126 TGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNI 185
G++ +L+E+MD GSLE E LS + R +L+GL YLH R I HRDIKPSN+
Sbjct: 151 NGEIQVLLEFMDKGSLEGIHIPL----EQPLSDLTRQVLSGLYYLHRRKIVHRDIKPSNL 206
Query: 186 LVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSL 245
L+N + EVKIADFGVS+ + +T++ CNS VGT AYMSPER + ++ G Y+G++ DIWSL
Sbjct: 207 LINSRREVKIADFGVSRVLAQTMDPCNSSVGTIAYMSPERINTDLNHGQYDGYAGDIWSL 266
Query: 246 GLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGE 305
G+++ E Y+G FPF G++ DWASLMCAIC S PP P TAS EFR F+ CCL+++
Sbjct: 267 GVSILEFYLGRFPF-SVGRQGDWASLMCAICMSQPPEAPPTASREFREFIACCLQRDPAR 325
Query: 306 RWSAAQLLTHPFLCKD 321
RW+AAQLL HPF+ ++
Sbjct: 326 RWTAAQLLRHPFITQN 341
>UniRef100_Q8H7S4 Hypothetical protein OSJNBa0081P02.7 [Oryza sativa]
Length = 339
Score = 277 bits (708), Expect = 3e-73
Identities = 154/327 (47%), Positives = 200/327 (61%), Gaps = 18/327 (5%)
Query: 1 MALVH-RRNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKL 59
MALV RR+ P L LP DH P P P +QP+ + +T+S ++ DFE++
Sbjct: 1 MALVRQRRHLPHLTLP--LDHFALRPPPAPQQ-QQQPAVAPSTSS-----DVRLSDFERI 52
Query: 60 SVLGHGNGGTVYKVRHKL-TSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKY 118
SVLGHGNGGTVYK RH+ L + + + + R E ILR A D +VV+
Sbjct: 53 SVLGHGNGGTVYKARHRRGCPAQQPLALKLFAAGDLSAAR---EAEILRLAADAPHVVRL 109
Query: 119 HGSFEKPTGDV----CILMEYMDSGSLETALKTTGT-FSESKLSTVARDILNGLTYLHAR 173
H G V + +E M GSL L+ G E ++ VAR L GL LHA
Sbjct: 110 HAVVPSAAGGVEEPAALALELMPGGSLAGLLRRLGRPMGERPIAAVARQALLGLEALHAL 169
Query: 174 NIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGG 233
I HRD+KPSN+L+ EVKIADFG K + R L+ C SYVGT AYMSPERFDPE Y G
Sbjct: 170 RIVHRDLKPSNLLLGADGEVKIADFGAGKVLRRRLDPCASYVGTAAYMSPERFDPEAYSG 229
Query: 234 NYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRN 293
+Y+ ++AD+WSLG+ + ELY+G+FP L GQRPDWA+LMCAICF + P +P AS EFR+
Sbjct: 230 DYDPYAADVWSLGVAILELYLGHFPLLPVGQRPDWAALMCAICFGEAPEMPAAASEEFRD 289
Query: 294 FVECCLKKESGERWSAAQLLTHPFLCK 320
FV CL+K++G R S +LL HPF+ +
Sbjct: 290 FVSRCLEKKAGRRASVGELLEHPFIAE 316
>UniRef100_O80397 MAP kinase kinase 4 [Arabidopsis thaliana]
Length = 366
Score = 277 bits (708), Expect = 3e-73
Identities = 151/325 (46%), Positives = 209/325 (63%), Gaps = 18/325 (5%)
Query: 7 RNSPKLRLP-EISDHRPRFPVPLP-------ANIYKQPSTSATTASVAGGDNISAGDFEK 58
R P L LP D P+PLP + PS+ + +S +I A ++
Sbjct: 19 RRRPDLTLPLPQRDVSLAVPLPLPPTSGGSGGSSGSAPSSGGSASSTNTNSSIEAKNYSD 78
Query: 59 L---SVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNV 115
L + +G G GGTVYKV H+ +S +YALK+ + + + T RR+ E+ ILR NV
Sbjct: 79 LVRGNRIGSGAGGTVYKVIHRPSSRLYALKVIYGNHEETVRRQICREIEILRDVNH-PNV 137
Query: 116 VKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNI 175
VK H F++ G++ +L+E+MD GSLE A + E +L+ ++R IL+GL YLH+R+I
Sbjct: 138 VKCHEMFDQ-NGEIQVLLEFMDKGSLEGA----HVWKEQQLADLSRQILSGLAYLHSRHI 192
Query: 176 AHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNY 235
HRDIKPSN+L+N VKIADFGVS+ + +T++ CNS VGT AYMSPER + ++ G Y
Sbjct: 193 VHRDIKPSNLLINSAKNVKIADFGVSRILAQTMDPCNSSVGTIAYMSPERINTDLNQGKY 252
Query: 236 NGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFV 295
+G++ DIWSLG+++ E Y+G FPF S Q DWASLMCAIC S PP P TAS EFR+F+
Sbjct: 253 DGYAGDIWSLGVSILEFYLGRFPFPVSRQ-GDWASLMCAICMSQPPEAPATASPEFRHFI 311
Query: 296 ECCLKKESGERWSAAQLLTHPFLCK 320
CCL++E G+R SA QLL HPF+ +
Sbjct: 312 SCCLQREPGKRRSAMQLLQHPFILR 336
>UniRef100_Q93WR8 MEK map kinase kinsae [Medicago varia]
Length = 368
Score = 274 bits (700), Expect = 3e-72
Identities = 146/320 (45%), Positives = 206/320 (63%), Gaps = 12/320 (3%)
Query: 6 RRNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDN----ISAGDFEKLSV 61
R++ L LP+ D P+PLP + S S +GG + I + E+L+
Sbjct: 33 RKDHLTLPLPQ-RDTNLAVPLPLPPSGGSGGSGGGGNGSGSGGASQQLVIPFSELERLNR 91
Query: 62 LGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGS 121
+G G+GGTVYKV HK+ YALK+ + + + RR+ E+ ILR D NVVK H
Sbjct: 92 IGSGSGGTVYKVVHKINGRAYALKVIYGHHEESVRRQIHREIQILRDVDD-VNVVKCHEM 150
Query: 122 FEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIK 181
++ ++ +L+EYMD GSLE E++L+ VAR IL GL YLH R+I HRDIK
Sbjct: 151 YDH-NAEIQVLLEYMDGGSLEGKHIP----QENQLADVARQILRGLAYLHRRHIVHRDIK 205
Query: 182 PSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSAD 241
PSN+L+N + +VKIADFGV + + +T++ CNS VGT AYMSPER + ++ G Y+ ++ D
Sbjct: 206 PSNLLINSRKQVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDINDGQYDAYAGD 265
Query: 242 IWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKK 301
IWSLG+++ E Y+G FPF G++ DWASLMCAIC S PP P TAS EFR+FV CL++
Sbjct: 266 IWSLGVSILEFYMGRFPF-AVGRQGDWASLMCAICMSQPPEAPTTASPEFRDFVSRCLQR 324
Query: 302 ESGERWSAAQLLTHPFLCKD 321
+ RW+A++LL+HPFL ++
Sbjct: 325 DPSRRWTASRLLSHPFLVRN 344
>UniRef100_Q66MH8 MAPKK [Lycopersicon esculentum]
Length = 359
Score = 271 bits (694), Expect = 1e-71
Identities = 143/316 (45%), Positives = 202/316 (63%), Gaps = 15/316 (4%)
Query: 6 RRNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHG 65
RR L LP+ VPLP P TS++++S + + E+++ +G G
Sbjct: 28 RRTDLTLPLPQ---RDVALAVPLPL-----PPTSSSSSSSPLPTPLHFSELERVNRIGSG 79
Query: 66 NGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKP 125
GGTVYKV H+ T +YALK+ + + + + R + E+ ILR D NVV+ H F+
Sbjct: 80 TGGTVYKVLHRPTGRLYALKVIYGNHEDSVRLQMCREIEILRDV-DNPNVVRCHDMFDH- 137
Query: 126 TGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNI 185
G++ +L+E+MD GSLE E LS + R +L+GL YLH R I HRDIKPSN+
Sbjct: 138 NGEIQVLLEFMDKGSLEGIHIPL----EQPLSDLTRQVLSGLYYLHRRKIVHRDIKPSNL 193
Query: 186 LVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSL 245
L+N + EVKIADFGVS+ + +T++ CNS VGT AYMSPER + ++ G Y+G++ DIWSL
Sbjct: 194 LINSRREVKIADFGVSRVLAQTMDPCNSSVGTIAYMSPERINTDLNHGQYDGYAGDIWSL 253
Query: 246 GLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGE 305
G+++ E Y+G FPF G++ DWASLMCAIC S PP P +AS EFR F+ CCL+++
Sbjct: 254 GVSILEFYLGRFPF-SVGRQGDWASLMCAICMSQPPEAPPSASREFREFIACCLQRDPAR 312
Query: 306 RWSAAQLLTHPFLCKD 321
RW+A QLL HPF+ ++
Sbjct: 313 RWTAGQLLRHPFITQN 328
>UniRef100_Q9AXI8 Mitogen-activated protein kinase 2 [Nicotiana tabacum]
Length = 372
Score = 269 bits (688), Expect = 7e-71
Identities = 144/319 (45%), Positives = 206/319 (64%), Gaps = 13/319 (4%)
Query: 7 RNSPKLR----LPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVL 62
RN P+ R LP + P VPLP PS+S++++S ++ + E+++ +
Sbjct: 30 RNRPRRRTDLTLP-LPQRDPALAVPLPLPPTSAPSSSSSSSSSPLPTPLNFSELERINRI 88
Query: 63 GHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSF 122
G G GGTVYKV H+ T +YALK+ + + + + R + E+ ILR D NVV+ H F
Sbjct: 89 GSGAGGTVYKVLHRPTGRLYALKVIYGNHEDSVRLQMCREIEILRDV-DNPNVVRCHDMF 147
Query: 123 EKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKP 182
+ G++ +L+E+MD GSLE ES LS + R +L+GL YLH R I HRDIKP
Sbjct: 148 DH-NGEIQVLLEFMDKGSLEGIHIP----KESALSDLTRQVLSGLYYLHRRKIVHRDIKP 202
Query: 183 SNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADI 242
SN+L+N + EVKIADFGVS+ + +T++ CNS VGT AYMSPER + ++ G Y+G++ DI
Sbjct: 203 SNLLINSRREVKIADFGVSRVLAQTMDPCNSSVGTIAYMSPERINTDLNHGQYDGYAGDI 262
Query: 243 WSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKE 302
WSLG+++ E Y+G FPF G+ DWASLMCAIC S + P AS EFR+F+ CCL+++
Sbjct: 263 WSLGVSILEFYLGRFPF-SVGRSGDWASLMCAICMSH-GTAPANASREFRDFIACCLQRD 320
Query: 303 SGERWSAAQLLTHPFLCKD 321
RW+A QLL HPF+ ++
Sbjct: 321 PARRWTAVQLLRHPFITQN 339
>UniRef100_Q94EV7 MAP kinase kinase [Zea mays]
Length = 406
Score = 261 bits (667), Expect = 2e-68
Identities = 139/312 (44%), Positives = 196/312 (62%), Gaps = 9/312 (2%)
Query: 10 PKLRLPEISDHRP--RFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGNG 67
P R P S R R P P P ++ S + + E++ +G G G
Sbjct: 87 PSARSPPPSPSRSLSRQPPPPPGGPPRRASPRPPPRRSSSSSRPPLAELERVRRVGSGAG 146
Query: 68 GTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTG 127
GTV+ VRH+ T YALK+ + + D RR+ E+ ILR A + VV+ HG +E+ G
Sbjct: 147 GTVWMVRHRGTGRPYALKVLYGNHDDAVRRQIAREIAILRTA-EHPAVVRCHGMYERG-G 204
Query: 128 DVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILV 187
++ IL+EYMD GSL+ F L+ V R +L+G+ YLH R+I HRDIKPSN+L+
Sbjct: 205 ELQILLEYMDGGSLDGRRIAAEPF----LADVGRQVLSGIAYLHRRHIVHRDIKPSNLLI 260
Query: 188 NIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGL 247
+ VKIADFGV + + +T++ CNS VGT AYMSPER + ++ G+Y+G++ DIWS GL
Sbjct: 261 DSARRVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDLNDGSYDGYAGDIWSFGL 320
Query: 248 TLFELYVGYFPFLQS-GQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGER 306
++ E Y+G FPF ++ G++ DWA+LMCAIC++DPP P TAS EFR F+ CCL+K +R
Sbjct: 321 SILEFYLGRFPFGENLGRQGDWAALMCAICYNDPPEPPPTASREFRGFIACCLQKNPAKR 380
Query: 307 WSAAQLLTHPFL 318
+AAQLL HPF+
Sbjct: 381 LTAAQLLQHPFV 392
>UniRef100_Q6K4Q0 Putative MAP kinase kinase [Oryza sativa]
Length = 369
Score = 257 bits (656), Expect = 3e-67
Identities = 142/331 (42%), Positives = 203/331 (60%), Gaps = 27/331 (8%)
Query: 7 RNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISA------------- 53
R P L LP VPLP + PS++ ++ S +G ++
Sbjct: 28 RRRPDLTLPLPQRDLTSLAVPLPLPL--PPSSAPSSTSSSGSSSLGGVPTPPNSVGSAPP 85
Query: 54 -----GDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRR 108
+ E++ +G G GGTV+ VRH+ T YALK+ + + D RR+ E+ ILR
Sbjct: 86 APPPLSELERVRRIGSGAGGTVWMVRHRPTGRPYALKVLYGNHDDAVRRQITREIAILRT 145
Query: 109 ATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLT 168
A + VV+ HG +E+ G++ IL+EYMD GSLE SE+ L+ VAR +L+G+
Sbjct: 146 A-EHPAVVRCHGMYEQ-AGELQILLEYMDGGSLEGRRIA----SEAFLADVARQVLSGIA 199
Query: 169 YLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDP 228
YLH R+I HRDIKPSN+L++ VKIADFGV + + +T++ CNS VGT AYMSPER +
Sbjct: 200 YLHRRHIVHRDIKPSNLLIDSGRRVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINT 259
Query: 229 EVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQS-GQRPDWASLMCAICFSDPPSLPETA 287
++ G Y+G++ DIWS GL++ E Y+G FP ++ G++ DWA+LMCAIC+SD P+ P A
Sbjct: 260 DLNDGAYDGYAGDIWSFGLSILEFYMGRFPLGENLGKQGDWAALMCAICYSDSPAPPPNA 319
Query: 288 SSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
S EF++F+ CCL+K R SAAQLL H F+
Sbjct: 320 SPEFKSFISCCLQKNPARRPSAAQLLQHRFV 350
>UniRef100_Q69TJ8 Putative MAP kinase kinase [Oryza sativa]
Length = 342
Score = 256 bits (654), Expect = 6e-67
Identities = 146/322 (45%), Positives = 199/322 (61%), Gaps = 16/322 (4%)
Query: 7 RNSPKLRLPEISDHRPR-FPVPLPANIYKQPSTSATTASVAGGDNISAG--------DFE 57
R P L LP P VPLP +TSA A A SAG + E
Sbjct: 12 RRRPDLTLPMPQRDAPTSLAVPLPLPPAATTTTSAPPAGGAMHPLASAGAAPPPPLEELE 71
Query: 58 KLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVK 117
++ +G G GGTV+ VRH+ T YALK+ + + D RR+ E+ ILR A + VV+
Sbjct: 72 RVRRVGSGAGGTVWMVRHRGTGKEYALKVLYGNHDDAVRRQIAREIAILRTA-EHPAVVR 130
Query: 118 YHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAH 177
H +E+ G++ IL+EYMD GSL+ F L+ VAR +L+G+ YLH R+I H
Sbjct: 131 CHDMYERG-GELQILLEYMDGGSLDGRRIADERF----LADVARQVLSGIAYLHRRHIVH 185
Query: 178 RDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNG 237
RDIKPSN+L++ VKIADFGV + + +T++ CNS VGT AYMSPER + ++ G Y+G
Sbjct: 186 RDIKPSNLLIDSARRVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDLNDGAYDG 245
Query: 238 FSADIWSLGLTLFELYVGYFPFLQS-GQRPDWASLMCAICFSDPPSLPETASSEFRNFVE 296
++ DIWS GL++ E Y+G FPF ++ G++ DWA+LMCAIC+SDPP P S EFR+FV
Sbjct: 246 YAGDIWSFGLSILEFYMGKFPFGENLGKQGDWAALMCAICYSDPPEPPAAVSPEFRSFVG 305
Query: 297 CCLKKESGERWSAAQLLTHPFL 318
CL+K +R SAAQL+ HPF+
Sbjct: 306 YCLQKNPAKRPSAAQLMQHPFV 327
>UniRef100_Q69TJ7 Putative MAP kinase kinase [Oryza sativa]
Length = 314
Score = 248 bits (633), Expect = 2e-64
Identities = 128/265 (48%), Positives = 180/265 (67%), Gaps = 7/265 (2%)
Query: 55 DFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTN 114
+ E++ +G G GGTV+ VRH+ T YALK+ + + D RR+ E+ ILR A +
Sbjct: 41 ELERVRRVGSGAGGTVWMVRHRGTGKEYALKVLYGNHDDAVRRQIAREIAILRTA-EHPA 99
Query: 115 VVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARN 174
VV+ H +E+ G++ IL+EYMD GSL+ F L+ VAR +L+G+ YLH R+
Sbjct: 100 VVRCHDMYERG-GELQILLEYMDGGSLDGRRIADERF----LADVARQVLSGIAYLHRRH 154
Query: 175 IAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGN 234
I HRDIKPSN+L++ VKIADFGV + + +T++ CNS VGT AYMSPER + ++ G
Sbjct: 155 IVHRDIKPSNLLIDSARRVKIADFGVGRILNQTMDPCNSSVGTIAYMSPERINTDLNDGA 214
Query: 235 YNGFSADIWSLGLTLFELYVGYFPFLQS-GQRPDWASLMCAICFSDPPSLPETASSEFRN 293
Y+G++ DIWS GL++ E Y+G FPF ++ G++ DWA+LMCAIC+SDPP P S EFR+
Sbjct: 215 YDGYAGDIWSFGLSILEFYMGKFPFGENLGKQGDWAALMCAICYSDPPEPPAAVSPEFRS 274
Query: 294 FVECCLKKESGERWSAAQLLTHPFL 318
FV CL+K +R SAAQL+ HPF+
Sbjct: 275 FVGYCLQKNPAKRPSAAQLMQHPFV 299
>UniRef100_Q8W318 Putative kinase [Oryza sativa]
Length = 345
Score = 245 bits (626), Expect = 1e-63
Identities = 141/306 (46%), Positives = 189/306 (61%), Gaps = 19/306 (6%)
Query: 21 RPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSI 80
R + P P+ A STS+T AS A ++ DFE+++VLG GNGGTVYKVRH+ T
Sbjct: 32 RRQCPPPVAA-----ASTSSTPASRASQFRLA--DFERVAVLGRGNGGTVYKVRHRETCA 84
Query: 81 IYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHG---SFEKPTGDVCILMEYMD 137
+YALK+ H A E +IL R T VV+ H + +GDV +L+E +D
Sbjct: 85 LYALKVQHSAGGGEL---AGVEADILSR-TASPFVVRCHAVLPASASASGDVALLLELVD 140
Query: 138 SGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIAD 197
GSL + G F E+ ++ VA L+GL LHAR + HRDIKP N+LV++ EVKIAD
Sbjct: 141 GGSLASVAARAGAFPEAAVAEVAAQALSGLACLHARRVVHRDIKPGNLLVSVDGEVKIAD 200
Query: 198 FGVSKFM----GRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELY 253
FG++K + G A Y GT AYMSPERFD E++G + F+AD+W LG+T+ EL
Sbjct: 201 FGIAKVVPPRRGGEHRAAYEYEGTAAYMSPERFDSELHGDGADPFAADVWGLGVTVLELL 260
Query: 254 VGYFPFLQSGQRPDWASLMCAICFSDPPSLPE-TASSEFRNFVECCLKKESGERWSAAQL 312
+ +P L +GQ+P WA+LMCAICF + P LP+ AS E R F+ CL K+ +R SAA L
Sbjct: 261 MARYPLLPAGQKPSWAALMCAICFGELPPLPDGAASPELRAFLAACLHKDHTKRPSAAHL 320
Query: 313 LTHPFL 318
LTH F+
Sbjct: 321 LTHQFV 326
>UniRef100_Q9M8J5 Putative MAP kinase [Arabidopsis thaliana]
Length = 293
Score = 244 bits (623), Expect = 2e-63
Identities = 136/288 (47%), Positives = 183/288 (63%), Gaps = 25/288 (8%)
Query: 23 RFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIY 82
RFP+ +PA SAT +S A + S + +++SVLG GNGGTV+KV+ K TS IY
Sbjct: 27 RFPI-IPAT-----KVSATVSSCAS-NTFSVANLDRISVLGSGNGGTVFKVKDKTTSEIY 79
Query: 83 ALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLE 142
ALK + D T+ R E+ ILR V K H F+ P+G+V ILM+YMD GSLE
Sbjct: 80 ALKKVKENWDSTSLR----EIEILRMVNS-PYVAKCHDIFQNPSGEVSILMDYMDLGSLE 134
Query: 143 TALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSK 202
+ T E +L+ ++R +L G YLH I HRDIKP+N+L + K EVKIADFGVSK
Sbjct: 135 SLRGVT----EKQLALMSRQVLEGKNYLHEHKIVHRDIKPANLLRSSKEEVKIADFGVSK 190
Query: 203 FMGRTLEACNSYVGTCAYMSPERFDPEVYG----GNYNGFSADIWSLGLTLFELYVGYFP 258
+ R+L CNS+VGT AYMSPER D E G N ++ DIWS GLT+ E+ VGY+P
Sbjct: 191 IVVRSLNKCNSFVGTFAYMSPERLDSEADGVTEEDKSNVYAGDIWSFGLTMLEILVGYYP 250
Query: 259 FLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGER 306
L PD A+++CA+CF +PP PE S + ++F++CCL+K++ ER
Sbjct: 251 ML-----PDQAAIVCAVCFGEPPKAPEECSDDLKSFMDCCLRKKASER 293
>UniRef100_Q9LQM8 F5D14.7 protein [Arabidopsis thaliana]
Length = 305
Score = 225 bits (574), Expect = 1e-57
Identities = 127/303 (41%), Positives = 178/303 (57%), Gaps = 13/303 (4%)
Query: 20 HRPRFPVPLPANIYKQPSTSATTASVAGGDNI---SAGDFEKLSVLGHGNGGTVYKVRHK 76
H+ + +P IY + S ++S + + + D EKLSVLG G+GGTVYK RH+
Sbjct: 9 HQEPLTLSIPPLIYHGTAFSVASSSSSSPETSPIQTLNDLEKLSVLGQGSGGTVYKTRHR 68
Query: 77 LTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYM 136
T +YALK+ P E +IL+R + + ++K + F D+C +ME M
Sbjct: 69 RTKTLYALKVLR----PNLNTTVTVEADILKRI-ESSFIIKCYAVFVS-LYDLCFVMELM 122
Query: 137 DSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIA 196
+ GSL AL FSE +S++A IL GL YL I H DIKPSN+L+N K EVKIA
Sbjct: 123 EKGSLHDALLAQQVFSEPMVSSLANRILQGLRYLQKMGIVHGDIKPSNLLINKKGEVKIA 182
Query: 197 DFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYG-GNYNGFSADIWSLGLTLFELYVG 255
DFG S+ + N GTCAYMSPER D E +G G GF+ D+WSLG+ + E Y+G
Sbjct: 183 DFGASRIVAGGDYGSN---GTCAYMSPERVDLEKWGFGGEVGFAGDVWSLGVVVLECYIG 239
Query: 256 YFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTH 315
+P + G +PDWA+L CAIC ++ +P + S EFR+FV CL+K+ +R + +LL H
Sbjct: 240 RYPLTKVGDKPDWATLFCAICCNEKVDIPVSCSLEFRDFVGRCLEKDWRKRDTVEELLRH 299
Query: 316 PFL 318
F+
Sbjct: 300 SFV 302
>UniRef100_Q6YUQ9 Putative MAP kinase kinase [Oryza sativa]
Length = 340
Score = 203 bits (516), Expect = 6e-51
Identities = 123/323 (38%), Positives = 172/323 (53%), Gaps = 20/323 (6%)
Query: 1 MALVHRRNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLS 60
MA + R +L +P P P A PST + +A + E LS
Sbjct: 1 MAKLRERRQLRLSVPASPPPFPHLDHPFAA----LPSTPPGSPVLA--------ELEMLS 48
Query: 61 VLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHG 120
V+G G GGTVY+ RH+ T AL + D R A + + A D +VV+ HG
Sbjct: 49 VVGRGAGGTVYRARHRRTGA--ALAVKEMRDDGAALREAGAHLRVAAAAPDHPSVVRLHG 106
Query: 121 -SFEKPTGD---VCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIA 176
P V +++EY+ GSL L G E ++ V R +L GL++LH +A
Sbjct: 107 VCVGHPVAGNRFVYLVLEYLPEGSLSDVL-VRGALPEPAIAGVTRCVLRGLSHLHRLGVA 165
Query: 177 HRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYN 236
H D+KPSN+LV + E+KIADFG S+ + EA + GT AYMSPE+ PE +GG
Sbjct: 166 HGDVKPSNLLVGHRGEIKIADFGASRVVTGRDEAHHQSPGTWAYMSPEKLHPEGFGGGGG 225
Query: 237 G-FSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFV 295
FS D+WSLG+ L E + G FP + +G+RPDW +L+ A+CF+ P +P AS EF FV
Sbjct: 226 ADFSGDVWSLGVVLLECHAGRFPLVAAGERPDWPALVLAVCFAAAPEVPVAASPEFGGFV 285
Query: 296 ECCLKKESGERWSAAQLLTHPFL 318
CL+K+ R + +LL HPF+
Sbjct: 286 RRCLEKDWRRRATVEELLGHPFV 308
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 571,223,271
Number of Sequences: 2790947
Number of extensions: 24477888
Number of successful extensions: 105814
Number of sequences better than 10.0: 19387
Number of HSP's better than 10.0 without gapping: 14945
Number of HSP's successfully gapped in prelim test: 4443
Number of HSP's that attempted gapping in prelim test: 63823
Number of HSP's gapped (non-prelim): 22189
length of query: 324
length of database: 848,049,833
effective HSP length: 127
effective length of query: 197
effective length of database: 493,599,564
effective search space: 97239114108
effective search space used: 97239114108
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC141436.1


