

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141323.12 + phase: 0 /pseudo
(133 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
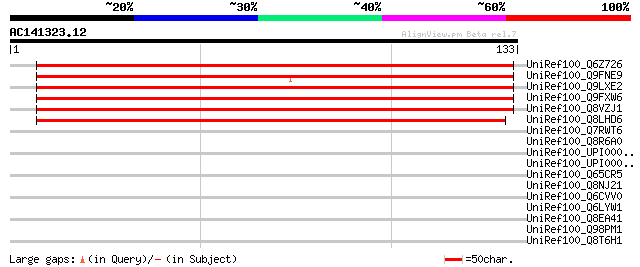
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6Z726 Hypothetical protein P0575F10.5 [Oryza sativa] 187 6e-47
UniRef100_Q9FNE9 Similarity to unknown protein [Arabidopsis thal... 176 1e-43
UniRef100_Q9LXE2 Hypothetical protein F17I14_20 [Arabidopsis tha... 164 3e-40
UniRef100_Q9FXW6 Arabidopsis thaliana genomic DNA, chromosome 5,... 164 3e-40
UniRef100_Q8VZJ1 AT5g09790/F17I14_20 [Arabidopsis thaliana] 164 3e-40
UniRef100_Q8LHD6 P0458E05.13 protein [Oryza sativa] 162 2e-39
UniRef100_Q7RWT6 Predicted protein [Neurospora crassa] 32 3.1
UniRef100_Q8R6A0 Exonuclease SBCC [Fusobacterium nucleatum] 31 5.3
UniRef100_UPI00002C3504 UPI00002C3504 UniRef100 entry 31 7.0
UniRef100_UPI00002B4BF1 UPI00002B4BF1 UniRef100 entry 31 7.0
UniRef100_Q65CR5 Adenylosuccinate synthetase [Bacillus lichenifo... 31 7.0
UniRef100_Q8NJ21 Alpha-aminoadipate reductase [Kluyveromyces lac... 31 7.0
UniRef100_Q6CVV0 Similarity [Kluyveromyces lactis] 31 7.0
UniRef100_Q6LYW1 SAM (And some other nucleotide) binding motif [... 31 7.0
UniRef100_Q8EA41 Hypothetical protein SO4068 [Shewanella oneiden... 30 9.1
UniRef100_Q98PM1 Hypothetical protein MYPU_7010 [Mycoplasma pulm... 30 9.1
UniRef100_Q8T6H1 ABC transporter ABCC.8 [Dictyostelium discoideum] 30 9.1
>UniRef100_Q6Z726 Hypothetical protein P0575F10.5 [Oryza sativa]
Length = 361
Score = 187 bits (474), Expect = 6e-47
Identities = 88/125 (70%), Positives = 106/125 (84%)
Query: 8 VLSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVEADKSIKDLTVIREYVGDIDFLK 67
VL KED ETLNL + MM RGE P L+VV+D EG+TVEAD+ IKDLT+I EYVGD+D+L
Sbjct: 205 VLPKEDVETLNLCKRMMARGEWPPLLVVYDPVEGFTVEADRFIKDLTIITEYVGDVDYLT 264
Query: 68 NREYDDGDRIMTLLSASNPSQSLVVFPDKRSNIAPFITGIDNHTPEGNKKQNMKCVRFNI 127
RE+DDGD +MTLLSA+ PS+SLV+ PDKRSNIA FI GI+NHTP+G KKQN+KCVRF++
Sbjct: 265 RREHDDGDSMMTLLSAATPSRSLVICPDKRSNIARFINGINNHTPDGRKKQNLKCVRFDV 324
Query: 128 GGECR 132
GGECR
Sbjct: 325 GGECR 329
>UniRef100_Q9FNE9 Similarity to unknown protein [Arabidopsis thaliana]
Length = 349
Score = 176 bits (446), Expect = 1e-43
Identities = 88/126 (69%), Positives = 103/126 (80%), Gaps = 1/126 (0%)
Query: 8 VLSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVEADKSIKDLTVIREYVGDIDFLK 67
VLSKE ETL L + MM+ GECP LMVVFD EG+TVEAD+ IKD T+I EYVGD+D+L
Sbjct: 192 VLSKEGVETLALCKKMMDLGECPPLMVVFDPYEGFTVEADRFIKDWTIITEYVGDVDYLS 251
Query: 68 NREYD-DGDRIMTLLSASNPSQSLVVFPDKRSNIAPFITGIDNHTPEGNKKQNMKCVRFN 126
NRE D DGD +MTLL AS+PSQ LV+ PD+RSNIA FI+GI+NH+PEG KKQN+KCVRFN
Sbjct: 252 NREDDYDGDSMMTLLHASDPSQCLVICPDRRSNIARFISGINNHSPEGRKKQNLKCVRFN 311
Query: 127 IGGECR 132
I GE R
Sbjct: 312 INGEAR 317
>UniRef100_Q9LXE2 Hypothetical protein F17I14_20 [Arabidopsis thaliana]
Length = 379
Score = 164 bits (416), Expect = 3e-40
Identities = 81/125 (64%), Positives = 94/125 (74%)
Query: 8 VLSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVEADKSIKDLTVIREYVGDIDFLK 67
VL KED ETL +SM RGECP L+VVFD EGYTVEAD IKDLT I EY GD+D+LK
Sbjct: 223 VLCKEDLETLEQCQSMYRRGECPPLVVVFDPLEGYTVEADGPIKDLTFIAEYTGDVDYLK 282
Query: 68 NREYDDGDRIMTLLSASNPSQSLVVFPDKRSNIAPFITGIDNHTPEGNKKQNMKCVRFNI 127
NRE DD D IMTLL + +PS++LV+ PDK NI+ FI GI+NH P KKQN KCVR++I
Sbjct: 283 NREKDDCDSIMTLLLSEDPSKTLVICPDKFGNISRFINGINNHNPVAKKKQNCKCVRYSI 342
Query: 128 GGECR 132
GECR
Sbjct: 343 NGECR 347
>UniRef100_Q9FXW6 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MTH16
[Arabidopsis thaliana]
Length = 378
Score = 164 bits (416), Expect = 3e-40
Identities = 81/125 (64%), Positives = 94/125 (74%)
Query: 8 VLSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVEADKSIKDLTVIREYVGDIDFLK 67
VL KED ETL +SM RGECP L+VVFD EGYTVEAD IKDLT I EY GD+D+LK
Sbjct: 222 VLCKEDLETLEQCQSMYRRGECPPLVVVFDPLEGYTVEADGPIKDLTFIAEYTGDVDYLK 281
Query: 68 NREYDDGDRIMTLLSASNPSQSLVVFPDKRSNIAPFITGIDNHTPEGNKKQNMKCVRFNI 127
NRE DD D IMTLL + +PS++LV+ PDK NI+ FI GI+NH P KKQN KCVR++I
Sbjct: 282 NREKDDCDSIMTLLLSEDPSKTLVICPDKFGNISRFINGINNHNPVAKKKQNCKCVRYSI 341
Query: 128 GGECR 132
GECR
Sbjct: 342 NGECR 346
>UniRef100_Q8VZJ1 AT5g09790/F17I14_20 [Arabidopsis thaliana]
Length = 352
Score = 164 bits (416), Expect = 3e-40
Identities = 81/125 (64%), Positives = 94/125 (74%)
Query: 8 VLSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVEADKSIKDLTVIREYVGDIDFLK 67
VL KED ETL +SM RGECP L+VVFD EGYTVEAD IKDLT I EY GD+D+LK
Sbjct: 196 VLCKEDLETLEQCQSMYRRGECPPLVVVFDPLEGYTVEADGPIKDLTFIAEYTGDVDYLK 255
Query: 68 NREYDDGDRIMTLLSASNPSQSLVVFPDKRSNIAPFITGIDNHTPEGNKKQNMKCVRFNI 127
NRE DD D IMTLL + +PS++LV+ PDK NI+ FI GI+NH P KKQN KCVR++I
Sbjct: 256 NREKDDCDSIMTLLLSEDPSKTLVICPDKFGNISRFINGINNHNPVAKKKQNCKCVRYSI 315
Query: 128 GGECR 132
GECR
Sbjct: 316 NGECR 320
>UniRef100_Q8LHD6 P0458E05.13 protein [Oryza sativa]
Length = 385
Score = 162 bits (409), Expect = 2e-39
Identities = 76/123 (61%), Positives = 96/123 (77%)
Query: 8 VLSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVEADKSIKDLTVIREYVGDIDFLK 67
+L KED ET+ L R+M +RGECP L+VVFD EG+TV+AD IKD+T I EY GD+DFL+
Sbjct: 229 ILPKEDKETIELCRTMQKRGECPPLLVVFDSREGFTVQADADIKDMTFIAEYTGDVDFLE 288
Query: 68 NREYDDGDRIMTLLSASNPSQSLVVFPDKRSNIAPFITGIDNHTPEGNKKQNMKCVRFNI 127
NR DDGD IMTLL +PS+ LV+ PDKR NI+ FI GI+NHT +G KK+N+KCVR++I
Sbjct: 289 NRANDDGDSIMTLLLTEDPSKRLVICPDKRGNISRFINGINNHTLDGKKKKNIKCVRYDI 348
Query: 128 GGE 130
GE
Sbjct: 349 DGE 351
>UniRef100_Q7RWT6 Predicted protein [Neurospora crassa]
Length = 994
Score = 32.0 bits (71), Expect = 3.1
Identities = 21/75 (28%), Positives = 31/75 (41%), Gaps = 1/75 (1%)
Query: 38 HAEGYTVEADKSIKDLTVIREYVGDIDFLKNREYDDGDRIMTLLSASNPSQSLVVFPDKR 97
H E E D D TV E +GD D + DDG+ + + S +++ D R
Sbjct: 610 HEEAEAEEQDDHEIDRTVGAEELGD-DLTLRQTLDDGNFSLDITEGSTEGNAVLGRDDNR 668
Query: 98 SNIAPFITGIDNHTP 112
+ PF+ H P
Sbjct: 669 TGKYPFLFSSPTHNP 683
>UniRef100_Q8R6A0 Exonuclease SBCC [Fusobacterium nucleatum]
Length = 921
Score = 31.2 bits (69), Expect = 5.3
Identities = 28/127 (22%), Positives = 55/127 (43%), Gaps = 18/127 (14%)
Query: 9 LSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVEADKSIKDLTV--------IREYV 60
+ KED E NL +++ E+ + + VVF+ + E +KSIKDL + ++E
Sbjct: 495 IKKEDLE--NLKKNIKEKTQILVEKVVFEDKKKQYFELEKSIKDLEISLKNEEINLKEIE 552
Query: 61 GDI--------DFLKNREYDDGDRIMTLLSASNPSQSLVVFPDKRSNIAPFITGIDNHTP 112
DI ++N+E+ + + + + +KR N+ + ++
Sbjct: 553 LDIKNLDIDIQKLIENQEFQNSQMLREKKTELEVELRNLNLDEKRENLKNILENLEIEKE 612
Query: 113 EGNKKQN 119
+ K QN
Sbjct: 613 KILKNQN 619
>UniRef100_UPI00002C3504 UPI00002C3504 UniRef100 entry
Length = 444
Score = 30.8 bits (68), Expect = 7.0
Identities = 18/61 (29%), Positives = 31/61 (50%), Gaps = 2/61 (3%)
Query: 22 SMMERGECPLLMVVFDHAEGYTVEADKSIKDLTVIREYVGDIDFLK--NREYDDGDRIMT 79
S+ +R + P ++++ D T A S+ + TV+ E VGD L N E DG+ +
Sbjct: 102 SITDRNDAPAVILLTDGVVPETAAAGTSVGEFTVLDEDVGDTHTLSLVNGEGSDGNALFA 161
Query: 80 L 80
+
Sbjct: 162 I 162
>UniRef100_UPI00002B4BF1 UPI00002B4BF1 UniRef100 entry
Length = 201
Score = 30.8 bits (68), Expect = 7.0
Identities = 26/92 (28%), Positives = 45/92 (48%), Gaps = 6/92 (6%)
Query: 27 GECPLLMVVFDHAEGYTV-----EADKSIKDLTVIREYVGDIDFLKNREYDDGDRIMTLL 81
GE P L ++ EG T+ ADK+I + E +GD+D + ++ D I+
Sbjct: 9 GENPNLNMISKLLEGATLFGVDGGADKAIAAGFEVTEVLGDLDSVNIADWKDRSNILADD 68
Query: 82 SASNPSQSLVVFPDKRSNIAPFITGIDNHTPE 113
S+S+ ++S + D R + GID +P+
Sbjct: 69 SSSDLAKSFGLLLD-RGFTEVDVVGIDGGSPD 99
>UniRef100_Q65CR5 Adenylosuccinate synthetase [Bacillus licheniformis]
Length = 430
Score = 30.8 bits (68), Expect = 7.0
Identities = 24/76 (31%), Positives = 43/76 (56%), Gaps = 6/76 (7%)
Query: 5 ISHVLSKEDFETLNLSRSMMERGECPLLMVVFDHAEGYTVE--ADKSIKDLTVIREYVGD 62
I+ +L ++ FE L+R++ E+ LL ++D AEG+ +E D+ + I++YV D
Sbjct: 147 IADLLDRDVFEE-KLARNLEEKNR--LLEKMYD-AEGFKIEDILDEYYEYGQQIKQYVCD 202
Query: 63 IDFLKNREYDDGDRIM 78
+ N DDG R++
Sbjct: 203 TSVVLNDALDDGRRVL 218
>UniRef100_Q8NJ21 Alpha-aminoadipate reductase [Kluyveromyces lactis]
Length = 1384
Score = 30.8 bits (68), Expect = 7.0
Identities = 19/68 (27%), Positives = 30/68 (43%), Gaps = 16/68 (23%)
Query: 56 IREYVGDIDFLKNREYDDGDRIMT-------------LLSASNPSQSLVVFPDK---RSN 99
IR + G++D +KN +GD I T +L PS+ P+ + N
Sbjct: 903 IRAFAGEVDRVKNSSSSEGDEIKTADYANDAKKLVDSMLPTEYPSREAFADPESLSAKKN 962
Query: 100 IAPFITGI 107
I F+TG+
Sbjct: 963 INVFVTGV 970
>UniRef100_Q6CVV0 Similarity [Kluyveromyces lactis]
Length = 1385
Score = 30.8 bits (68), Expect = 7.0
Identities = 19/68 (27%), Positives = 30/68 (43%), Gaps = 16/68 (23%)
Query: 56 IREYVGDIDFLKNREYDDGDRIMT-------------LLSASNPSQSLVVFPDK---RSN 99
IR + G++D +KN +GD I T +L PS+ P+ + N
Sbjct: 904 IRAFAGEVDRVKNSSSSEGDEIKTADYANDAKKLVDSMLPTEYPSREAFADPESLSAKKN 963
Query: 100 IAPFITGI 107
I F+TG+
Sbjct: 964 INVFVTGV 971
>UniRef100_Q6LYW1 SAM (And some other nucleotide) binding motif [Methanococcus
maripaludis]
Length = 265
Score = 30.8 bits (68), Expect = 7.0
Identities = 12/28 (42%), Positives = 19/28 (67%)
Query: 42 YTVEADKSIKDLTVIREYVGDIDFLKNR 69
YT+ + +K++TV+ + G ID LKNR
Sbjct: 55 YTMPLAREVKEITVVEQSTGMIDILKNR 82
>UniRef100_Q8EA41 Hypothetical protein SO4068 [Shewanella oneidensis]
Length = 702
Score = 30.4 bits (67), Expect = 9.1
Identities = 14/29 (48%), Positives = 22/29 (75%), Gaps = 2/29 (6%)
Query: 5 ISHVLSKEDFETLNL--SRSMMERGECPL 31
I+H+L E+++TLN+ S S+ E+GE PL
Sbjct: 375 ITHILVTENYKTLNIAWSSSLKEKGEVPL 403
>UniRef100_Q98PM1 Hypothetical protein MYPU_7010 [Mycoplasma pulmonis]
Length = 214
Score = 30.4 bits (67), Expect = 9.1
Identities = 16/47 (34%), Positives = 24/47 (51%)
Query: 63 IDFLKNREYDDGDRIMTLLSASNPSQSLVVFPDKRSNIAPFITGIDN 109
I ++KN Y D I T+ S+ NP+ + F D SN++ G N
Sbjct: 92 ISWVKNNRYSYVDLIPTIKSSENPNSDSLNFIDGNSNVSQNFFGTYN 138
>UniRef100_Q8T6H1 ABC transporter ABCC.8 [Dictyostelium discoideum]
Length = 1593
Score = 30.4 bits (67), Expect = 9.1
Identities = 29/95 (30%), Positives = 40/95 (41%), Gaps = 10/95 (10%)
Query: 29 CPLLMVVFDHAEGYTVEADKSI--KDLTVIREYVGDIDFLKNREYDDGDRIMTLLSASNP 86
C ++ HA Y D+ I KD ++ E GD + L + G L+S
Sbjct: 768 CNKTRILVTHAVHYLPYVDRIILMKDGRIVEE--GDFNTL----IEAGSHFTELMSHDEQ 821
Query: 87 SQSLVV--FPDKRSNIAPFITGIDNHTPEGNKKQN 119
Q L PDK S+ I G DN E N++QN
Sbjct: 822 QQQLQQQQAPDKSSDSNEQIGGGDNKESENNEEQN 856
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.137 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 216,572,744
Number of Sequences: 2790947
Number of extensions: 7922669
Number of successful extensions: 18032
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 18023
Number of HSP's gapped (non-prelim): 17
length of query: 133
length of database: 848,049,833
effective HSP length: 109
effective length of query: 24
effective length of database: 543,836,610
effective search space: 13052078640
effective search space used: 13052078640
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC141323.12


