

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141323.10 + phase: 0 /pseudo
(337 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
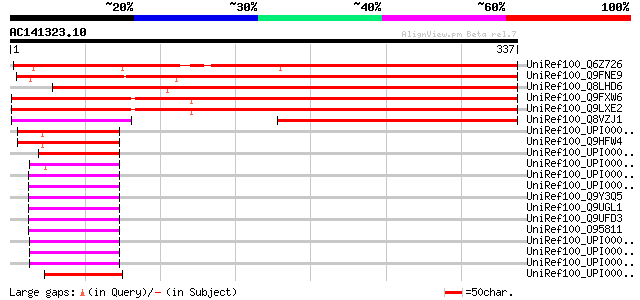
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6Z726 Hypothetical protein P0575F10.5 [Oryza sativa] 371 e-101
UniRef100_Q9FNE9 Similarity to unknown protein [Arabidopsis thal... 369 e-101
UniRef100_Q8LHD6 P0458E05.13 protein [Oryza sativa] 310 4e-83
UniRef100_Q9FXW6 Arabidopsis thaliana genomic DNA, chromosome 5,... 306 5e-82
UniRef100_Q9LXE2 Hypothetical protein F17I14_20 [Arabidopsis tha... 305 1e-81
UniRef100_Q8VZJ1 AT5g09790/F17I14_20 [Arabidopsis thaliana] 250 4e-65
UniRef100_UPI00003C19B7 UPI00003C19B7 UniRef100 entry 72 2e-11
UniRef100_Q9HFW4 Regulator Ustilago maydis 1 protein [Ustilago m... 72 2e-11
UniRef100_UPI0000438D8A UPI0000438D8A UniRef100 entry 72 3e-11
UniRef100_UPI00003A9969 UPI00003A9969 UniRef100 entry 72 3e-11
UniRef100_UPI00003685B7 UPI00003685B7 UniRef100 entry 71 4e-11
UniRef100_UPI00003685B6 UPI00003685B6 UniRef100 entry 71 4e-11
UniRef100_Q9Y3Q5 PLU-1 protein [Homo sapiens] 71 4e-11
UniRef100_Q9UGL1 RB-binding protein [Homo sapiens] 71 4e-11
UniRef100_Q9UFD3 Hypothetical protein DKFZp434N0335 [Homo sapiens] 71 4e-11
UniRef100_O95811 Retinoblastoma binding protein 2 homolog 1 [Hom... 71 4e-11
UniRef100_UPI0000428D78 UPI0000428D78 UniRef100 entry 70 6e-11
UniRef100_UPI000021E349 UPI000021E349 UniRef100 entry 70 6e-11
UniRef100_UPI00001CF564 UPI00001CF564 UniRef100 entry 70 6e-11
UniRef100_UPI00001CE950 UPI00001CE950 UniRef100 entry 70 6e-11
>UniRef100_Q6Z726 Hypothetical protein P0575F10.5 [Oryza sativa]
Length = 361
Score = 371 bits (953), Expect = e-101
Identities = 205/366 (56%), Positives = 243/366 (66%), Gaps = 41/366 (11%)
Query: 3 SPLRRRRIPAPK----KQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTP 58
+PLRRR P + + + +DDDV C+ C SG+S +LLLCD CD G H+FCL P
Sbjct: 5 TPLRRRTRARPAATRAEGGSGGDGDDDDVRCEACGSGESAAELLLCDGCDRGLHIFCLRP 64
Query: 59 ILPSVPKSSWFCPSC---------SHNPKIPK-FPLVQTKIIDFFKIQRTSDASQILNHE 108
ILP VP WFCPSC SH K PK FPLVQTKI+DFFKIQR A+ E
Sbjct: 65 ILPRVPAGDWFCPSCASPSPHSKKSHAAKKPKQFPLVQTKIVDFFKIQRGPAAALAAAAE 124
Query: 109 EEKAE**LGCFKKEKKVVGICAK**FEEEIRTDGVVGDRVDSY*NRVQ**AYVYAWNGAK 168
+ + K+++KV GI + + + + D R++ A + A
Sbjct: 125 SSEGK------KRKRKVGGIR----LVSKKKRKLLPFNPSDDPARRLRQMASLATALTAT 174
Query: 169 GC*FSCS*TW-----------------GNAVLSKEDTETLNLCRSMMERGECPPLMVVYD 211
G FS T+ G VL KED ETLNLC+ MM RGE PPL+VVYD
Sbjct: 175 GAVFSNELTYVPGMAPRAANRAALESGGMQVLPKEDVETLNLCKRMMARGEWPPLLVVYD 234
Query: 212 PVEGFTIEADKSIKDLTIITEYVGDVDFLKNREHDDGDSIMTLLSASNPSQSLVICPDKR 271
PVEGFT+EAD+ IKDLTIITEYVGDVD+L REHDDGDS+MTLLSA+ PS+SLVICPDKR
Sbjct: 235 PVEGFTVEADRFIKDLTIITEYVGDVDYLTRREHDDGDSMMTLLSAATPSRSLVICPDKR 294
Query: 272 SNIARFINGINNHTPEGKKKQNLKCVRYNVDGECRVLLIANRDIAKGERLYYDYNGLEHE 331
SNIARFINGINNHTP+G+KKQNLKCVR++V GECRVLL+ANRDI+KGERLYYDYNG EHE
Sbjct: 295 SNIARFINGINNHTPDGRKKQNLKCVRFDVGGECRVLLVANRDISKGERLYYDYNGSEHE 354
Query: 332 YPTEHF 337
YPT HF
Sbjct: 355 YPTHHF 360
>UniRef100_Q9FNE9 Similarity to unknown protein [Arabidopsis thaliana]
Length = 349
Score = 369 bits (947), Expect = e-101
Identities = 194/346 (56%), Positives = 246/346 (71%), Gaps = 14/346 (4%)
Query: 5 LRRRRIPA-------PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLT 57
+RRRR A P+ S +++D D VC++C+SGK P KLLLCD CD G+HLFCL
Sbjct: 4 VRRRRTQASNPRSEPPQHMSDHDSDSDWDTVCEECSSGKQPAKLLLCDKCDKGFHLFCLR 63
Query: 58 PILPSVPKSSWFCPSCSHNPKIPK-FPLVQTKIIDFFKIQRTSDASQILNHEE----EKA 112
PIL SVPK SWFCPSCS + +IPK FPL+QTKIIDFF+I+R+ D+SQI + + ++
Sbjct: 64 PILVSVPKGSWFCPSCSKH-QIPKSFPLIQTKIIDFFRIKRSPDSSQISSSSDSIGKKRK 122
Query: 113 E**LGCFKKEKKVVGICAK**FEEEIRTDGVVGDRVDSY*NRVQ**AYVYAWNGAKGC*F 172
+ L KK+++++ + + + + + + + +
Sbjct: 123 KTSLVMSKKKRRLLPYNPSNDPQRRLEQMASLATALRASNTKFSNELTYVSGKAPRSANQ 182
Query: 173 SCS*TWGNAVLSKEDTETLNLCRSMMERGECPPLMVVYDPVEGFTIEADKSIKDLTIITE 232
+ G VLSKE ETL LC+ MM+ GECPPLMVV+DP EGFT+EAD+ IKD TIITE
Sbjct: 183 AAFEKGGMQVLSKEGVETLALCKKMMDLGECPPLMVVFDPYEGFTVEADRFIKDWTIITE 242
Query: 233 YVGDVDFLKNREHD-DGDSIMTLLSASNPSQSLVICPDKRSNIARFINGINNHTPEGKKK 291
YVGDVD+L NRE D DGDS+MTLL AS+PSQ LVICPD+RSNIARFI+GINNH+PEG+KK
Sbjct: 243 YVGDVDYLSNREDDYDGDSMMTLLHASDPSQCLVICPDRRSNIARFISGINNHSPEGRKK 302
Query: 292 QNLKCVRYNVDGECRVLLIANRDIAKGERLYYDYNGLEHEYPTEHF 337
QNLKCVR+N++GE RVLL+ANRDI+KGERLYYDYNG EHEYPTEHF
Sbjct: 303 QNLKCVRFNINGEARVLLVANRDISKGERLYYDYNGYEHEYPTEHF 348
>UniRef100_Q8LHD6 P0458E05.13 protein [Oryza sativa]
Length = 385
Score = 310 bits (794), Expect = 4e-83
Identities = 154/315 (48%), Positives = 205/315 (64%), Gaps = 6/315 (1%)
Query: 29 CQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNPK-IPKFPLVQT 87
C C SG+ +LLLCD CD G H FCL PI VP WFCP C+ K + +FP+ QT
Sbjct: 70 CDVCGSGERDEELLLCDGCDRGRHTFCLRPIAARVPTGPWFCPPCAPRSKPVKRFPMTQT 129
Query: 88 KIIDFFKIQRTSDASQ-----ILNHEEEKAE**LGCFKKEKKVVGICAK**FEEEIRTDG 142
KI+DFF+IQ+ ++ ++ + +++ + L KK ++++ + ++
Sbjct: 130 KIVDFFRIQKGAEDAEAEKYGLFQDVKKRRKRSLVMHKKRRRILPYVPTEDKVQRLKQMA 189
Query: 143 VVGDRVDSY*NRVQ**AYVYAWNGAKGC*FSCS*TWGNAVLSKEDTETLNLCRSMMERGE 202
+ + S + + C + G +L KED ET+ LCR+M +RGE
Sbjct: 190 SLATAMTSSKMKFSNELTYMPGMAGRSCNQATLEEGGMQILPKEDKETIELCRTMQKRGE 249
Query: 203 CPPLMVVYDPVEGFTIEADKSIKDLTIITEYVGDVDFLKNREHDDGDSIMTLLSASNPSQ 262
CPPL+VV+D EGFT++AD IKD+T I EY GDVDFL+NR +DDGDSIMTLL +PS+
Sbjct: 250 CPPLLVVFDSREGFTVQADADIKDMTFIAEYTGDVDFLENRANDDGDSIMTLLLTEDPSK 309
Query: 263 SLVICPDKRSNIARFINGINNHTPEGKKKQNLKCVRYNVDGECRVLLIANRDIAKGERLY 322
LVICPDKR NI+RFINGINNHT +GKKK+N+KCVRY++DGE VLL+A RDIA GE+LY
Sbjct: 310 RLVICPDKRGNISRFINGINNHTLDGKKKKNIKCVRYDIDGESHVLLVACRDIACGEKLY 369
Query: 323 YDYNGLEHEYPTEHF 337
YDYNG EHEYPT HF
Sbjct: 370 YDYNGYEHEYPTHHF 384
>UniRef100_Q9FXW6 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MTH16
[Arabidopsis thaliana]
Length = 378
Score = 306 bits (784), Expect = 5e-82
Identities = 156/340 (45%), Positives = 214/340 (62%), Gaps = 6/340 (1%)
Query: 2 VSPLRRRRIPAPKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILP 61
++ + + +P +++ ++ +V C+KC SG+ +LLLCD CD G+H+ CL PI+
Sbjct: 40 MAEIMAKSVPVVEQEEEEDEDSYSNVTCEKCGSGEGDDELLLCDKCDRGFHMKCLRPIVV 99
Query: 62 SVPKSSWFCPSCSHNPKIPKFPLVQTKIIDFFKIQRTSDASQILNHEEEKAE**LGCF-- 119
VP +W C CS + + L Q KI+ FF+I++ + + L +E + C
Sbjct: 100 RVPIGTWLCVDCSDQRPVRR--LSQKKILHFFRIEKHTHQTDKLELSQETRKRRRSCSLT 157
Query: 120 --KKEKKVVGICAK**FEEEIRTDGVVGDRVDSY*NRVQ**AYVYAWNGAKGC*FSCS*T 177
K+ +K++ + ++ + G + + + + + S
Sbjct: 158 VKKRRRKLLPLVPSEDPDQRLAQMGTLASALTALGIKYSDGLNYVPGMAPRSANQSKLEK 217
Query: 178 WGNAVLSKEDTETLNLCRSMMERGECPPLMVVYDPVEGFTIEADKSIKDLTIITEYVGDV 237
G VL KED ETL C+SM RGECPPL+VV+DP+EG+T+EAD IKDLT I EY GDV
Sbjct: 218 GGMQVLCKEDLETLEQCQSMYRRGECPPLVVVFDPLEGYTVEADGPIKDLTFIAEYTGDV 277
Query: 238 DFLKNREHDDGDSIMTLLSASNPSQSLVICPDKRSNIARFINGINNHTPEGKKKQNLKCV 297
D+LKNRE DD DSIMTLL + +PS++LVICPDK NI+RFINGINNH P KKKQN KCV
Sbjct: 278 DYLKNREKDDCDSIMTLLLSEDPSKTLVICPDKFGNISRFINGINNHNPVAKKKQNCKCV 337
Query: 298 RYNVDGECRVLLIANRDIAKGERLYYDYNGLEHEYPTEHF 337
RY+++GECRVLL+A RDI+KGERLYYDYNG EHEYPT HF
Sbjct: 338 RYSINGECRVLLVATRDISKGERLYYDYNGYEHEYPTHHF 377
>UniRef100_Q9LXE2 Hypothetical protein F17I14_20 [Arabidopsis thaliana]
Length = 379
Score = 305 bits (781), Expect = 1e-81
Identities = 157/341 (46%), Positives = 215/341 (63%), Gaps = 7/341 (2%)
Query: 2 VSPLRRRRIPAPKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILP 61
++ + + +P +++ ++ +V C+KC SG+ +LLLCD CD G+H+ CL PI+
Sbjct: 40 MAEIMAKSVPVVEQEEEEDEDSYSNVTCEKCGSGEGDDELLLCDKCDRGFHMKCLRPIVV 99
Query: 62 SVPKSSWFCPSCSHNPKIPKFPLVQTKIIDFFKIQR-TSDASQILNHEEEKAE**LGCF- 119
VP +W C CS + + L Q KI+ FF+I++ T ++ +EE + C
Sbjct: 100 RVPIGTWLCVDCSDQRPVRR--LSQKKILHFFRIEKHTHQTDKLELSQEETRKRRRSCSL 157
Query: 120 ---KKEKKVVGICAK**FEEEIRTDGVVGDRVDSY*NRVQ**AYVYAWNGAKGC*FSCS* 176
K+ +K++ + ++ + G + + + + + S
Sbjct: 158 TVKKRRRKLLPLVPSEDPDQRLAQMGTLASALTALGIKYSDGLNYVPGMAPRSANQSKLE 217
Query: 177 TWGNAVLSKEDTETLNLCRSMMERGECPPLMVVYDPVEGFTIEADKSIKDLTIITEYVGD 236
G VL KED ETL C+SM RGECPPL+VV+DP+EG+T+EAD IKDLT I EY GD
Sbjct: 218 KGGMQVLCKEDLETLEQCQSMYRRGECPPLVVVFDPLEGYTVEADGPIKDLTFIAEYTGD 277
Query: 237 VDFLKNREHDDGDSIMTLLSASNPSQSLVICPDKRSNIARFINGINNHTPEGKKKQNLKC 296
VD+LKNRE DD DSIMTLL + +PS++LVICPDK NI+RFINGINNH P KKKQN KC
Sbjct: 278 VDYLKNREKDDCDSIMTLLLSEDPSKTLVICPDKFGNISRFINGINNHNPVAKKKQNCKC 337
Query: 297 VRYNVDGECRVLLIANRDIAKGERLYYDYNGLEHEYPTEHF 337
VRY+++GECRVLL+A RDI+KGERLYYDYNG EHEYPT HF
Sbjct: 338 VRYSINGECRVLLVATRDISKGERLYYDYNGYEHEYPTHHF 378
>UniRef100_Q8VZJ1 AT5g09790/F17I14_20 [Arabidopsis thaliana]
Length = 352
Score = 250 bits (638), Expect = 4e-65
Identities = 116/159 (72%), Positives = 133/159 (82%)
Query: 179 GNAVLSKEDTETLNLCRSMMERGECPPLMVVYDPVEGFTIEADKSIKDLTIITEYVGDVD 238
G VL KED ETL C+SM RGECPPL+VV+DP+EG+T+EAD IKDLT I EY GDVD
Sbjct: 193 GMQVLCKEDLETLEQCQSMYRRGECPPLVVVFDPLEGYTVEADGPIKDLTFIAEYTGDVD 252
Query: 239 FLKNREHDDGDSIMTLLSASNPSQSLVICPDKRSNIARFINGINNHTPEGKKKQNLKCVR 298
+LKNRE DD DSIMTLL + +PS++LVICPDK NI+RFINGINNH P KKKQN KCVR
Sbjct: 253 YLKNREKDDCDSIMTLLLSEDPSKTLVICPDKFGNISRFINGINNHNPVAKKKQNCKCVR 312
Query: 299 YNVDGECRVLLIANRDIAKGERLYYDYNGLEHEYPTEHF 337
Y+++GECRVLL+A RDI+KGERLYYDYNG EHEYPT HF
Sbjct: 313 YSINGECRVLLVATRDISKGERLYYDYNGYEHEYPTHHF 351
Score = 69.7 bits (169), Expect = 1e-10
Identities = 27/80 (33%), Positives = 46/80 (56%)
Query: 2 VSPLRRRRIPAPKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILP 61
++ + + +P +++ ++ +V C+KC SG+ +LLLCD CD G+H+ CL PI+
Sbjct: 40 MAEIMAKSVPVVEQEEEEDEDSYSNVTCEKCGSGEGDDELLLCDKCDRGFHMKCLRPIVV 99
Query: 62 SVPKSSWFCPSCSHNPKIPK 81
VP +W C CS + K
Sbjct: 100 RVPIGTWLCVDCSDQRPVRK 119
>UniRef100_UPI00003C19B7 UPI00003C19B7 UniRef100 entry
Length = 2289
Score = 72.0 bits (175), Expect = 2e-11
Identities = 29/74 (39%), Positives = 45/74 (60%), Gaps = 6/74 (8%)
Query: 6 RRRRIPAPKKQSTTF------NNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPI 59
RRR+ +P ++ ++ N ++ +C+ C G+ +LLCD C+ GYH++CL P
Sbjct: 513 RRRKGVSPHLEADSYLRAQAGNQAQEEQMCEICLRGEDGPNMLLCDECNRGYHMYCLQPA 572
Query: 60 LPSVPKSSWFCPSC 73
L S+PKS WFCP C
Sbjct: 573 LTSIPKSQWFCPPC 586
>UniRef100_Q9HFW4 Regulator Ustilago maydis 1 protein [Ustilago maydis]
Length = 2289
Score = 72.0 bits (175), Expect = 2e-11
Identities = 29/74 (39%), Positives = 45/74 (60%), Gaps = 6/74 (8%)
Query: 6 RRRRIPAPKKQSTTF------NNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPI 59
RRR+ +P ++ ++ N ++ +C+ C G+ +LLCD C+ GYH++CL P
Sbjct: 513 RRRKGVSPHLEADSYLRAQAGNQAQEEQMCEICLRGEDGPNMLLCDECNRGYHMYCLQPA 572
Query: 60 LPSVPKSSWFCPSC 73
L S+PKS WFCP C
Sbjct: 573 LTSIPKSQWFCPPC 586
>UniRef100_UPI0000438D8A UPI0000438D8A UniRef100 entry
Length = 1237
Score = 71.6 bits (174), Expect = 3e-11
Identities = 28/54 (51%), Positives = 33/54 (60%)
Query: 20 FNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
FNN+ D VC+ C G KL+LCD CD+ YH FCL P L PK +W CP C
Sbjct: 258 FNNHVDSFVCRMCGRGDEDEKLMLCDGCDDNYHTFCLIPPLTDPPKGNWRCPKC 311
>UniRef100_UPI00003A9969 UPI00003A9969 UniRef100 entry
Length = 1380
Score = 71.6 bits (174), Expect = 3e-11
Identities = 33/67 (49%), Positives = 37/67 (54%), Gaps = 7/67 (10%)
Query: 14 KKQSTTFNN-------NDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKS 66
+K TT+NN D VC C G + KLLLCD CD+ YH FCL P LP VPK
Sbjct: 276 RKPVTTYNNIYVYFSPQVDLYVCLFCGRGNNEDKLLLCDGCDDSYHTFCLIPPLPDVPKG 335
Query: 67 SWFCPSC 73
W CP C
Sbjct: 336 DWRCPKC 342
>UniRef100_UPI00003685B7 UPI00003685B7 UniRef100 entry
Length = 1584
Score = 71.2 bits (173), Expect = 4e-11
Identities = 31/61 (50%), Positives = 34/61 (54%)
Query: 13 PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPS 72
PK +S N D VC C SG +LLLCD CD+ YH FCL P L VPK W CP
Sbjct: 336 PKSRSKKATNAVDLYVCLLCGSGNDEDRLLLCDGCDDSYHTFCLIPPLHDVPKGDWRCPK 395
Query: 73 C 73
C
Sbjct: 396 C 396
>UniRef100_UPI00003685B6 UPI00003685B6 UniRef100 entry
Length = 1682
Score = 71.2 bits (173), Expect = 4e-11
Identities = 31/61 (50%), Positives = 34/61 (54%)
Query: 13 PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPS 72
PK +S N D VC C SG +LLLCD CD+ YH FCL P L VPK W CP
Sbjct: 434 PKSRSKKATNAVDLYVCLLCGSGNDEDRLLLCDGCDDSYHTFCLIPPLHDVPKGDWRCPK 493
Query: 73 C 73
C
Sbjct: 494 C 494
>UniRef100_Q9Y3Q5 PLU-1 protein [Homo sapiens]
Length = 1544
Score = 71.2 bits (173), Expect = 4e-11
Identities = 31/61 (50%), Positives = 34/61 (54%)
Query: 13 PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPS 72
PK +S N D VC C SG +LLLCD CD+ YH FCL P L VPK W CP
Sbjct: 296 PKSRSKKATNAVDLYVCLLCGSGNDEDRLLLCDGCDDSYHTFCLIPPLHDVPKGDWRCPK 355
Query: 73 C 73
C
Sbjct: 356 C 356
>UniRef100_Q9UGL1 RB-binding protein [Homo sapiens]
Length = 1681
Score = 71.2 bits (173), Expect = 4e-11
Identities = 31/61 (50%), Positives = 34/61 (54%)
Query: 13 PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPS 72
PK +S N D VC C SG +LLLCD CD+ YH FCL P L VPK W CP
Sbjct: 433 PKSRSKKATNAVDLYVCLLCGSGNDEDRLLLCDGCDDSYHTFCLIPPLHDVPKGDWRCPK 492
Query: 73 C 73
C
Sbjct: 493 C 493
>UniRef100_Q9UFD3 Hypothetical protein DKFZp434N0335 [Homo sapiens]
Length = 1350
Score = 71.2 bits (173), Expect = 4e-11
Identities = 31/61 (50%), Positives = 34/61 (54%)
Query: 13 PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPS 72
PK +S N D VC C SG +LLLCD CD+ YH FCL P L VPK W CP
Sbjct: 102 PKSRSKKATNAVDLYVCLLCGSGNDEDRLLLCDGCDDSYHTFCLIPPLHDVPKGDWRCPK 161
Query: 73 C 73
C
Sbjct: 162 C 162
>UniRef100_O95811 Retinoblastoma binding protein 2 homolog 1 [Homo sapiens]
Length = 1580
Score = 71.2 bits (173), Expect = 4e-11
Identities = 31/61 (50%), Positives = 34/61 (54%)
Query: 13 PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPS 72
PK +S N D VC C SG +LLLCD CD+ YH FCL P L VPK W CP
Sbjct: 332 PKSRSKKATNAVDLYVCLLCGSGNDEDRLLLCDGCDDSYHTFCLIPPLHDVPKGDWRCPK 391
Query: 73 C 73
C
Sbjct: 392 C 392
>UniRef100_UPI0000428D78 UPI0000428D78 UniRef100 entry
Length = 1476
Score = 70.5 bits (171), Expect = 6e-11
Identities = 30/60 (50%), Positives = 35/60 (58%)
Query: 14 KKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+++ T N D VC C G + KLLLCD CD+ YH FCL P LP VPK W CP C
Sbjct: 129 QRKGTLSVNFVDLYVCMFCGRGNNEDKLLLCDGCDDSYHTFCLLPPLPDVPKGDWRCPKC 188
>UniRef100_UPI000021E349 UPI000021E349 UniRef100 entry
Length = 621
Score = 70.5 bits (171), Expect = 6e-11
Identities = 30/60 (50%), Positives = 35/60 (58%)
Query: 14 KKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+++ T N D VC C G + KLLLCD CD+ YH FCL P LP VPK W CP C
Sbjct: 281 QRKGTLSVNFVDLYVCMFCGRGNNEDKLLLCDGCDDSYHTFCLLPPLPDVPKGDWRCPKC 340
>UniRef100_UPI00001CF564 UPI00001CF564 UniRef100 entry
Length = 1513
Score = 70.5 bits (171), Expect = 6e-11
Identities = 30/60 (50%), Positives = 35/60 (58%)
Query: 14 KKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+++ T N D VC C G + KLLLCD CD+ YH FCL P LP VPK W CP C
Sbjct: 104 QRKGTLSVNFVDLYVCMFCGRGNNEDKLLLCDGCDDSYHTFCLLPPLPDVPKGDWRCPKC 163
>UniRef100_UPI00001CE950 UPI00001CE950 UniRef100 entry
Length = 1448
Score = 70.5 bits (171), Expect = 6e-11
Identities = 27/52 (51%), Positives = 32/52 (60%)
Query: 24 DDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSH 75
DDD C+KC P +LLCD+CD+GYH CL P L +P WFCP C H
Sbjct: 893 DDDEPCKKCGLPNHPELILLCDSCDSGYHTACLRPPLMIIPDGEWFCPPCQH 944
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.333 0.146 0.480
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 577,257,079
Number of Sequences: 2790947
Number of extensions: 23709179
Number of successful extensions: 74162
Number of sequences better than 10.0: 1283
Number of HSP's better than 10.0 without gapping: 736
Number of HSP's successfully gapped in prelim test: 547
Number of HSP's that attempted gapping in prelim test: 72203
Number of HSP's gapped (non-prelim): 2080
length of query: 337
length of database: 848,049,833
effective HSP length: 128
effective length of query: 209
effective length of database: 490,808,617
effective search space: 102579000953
effective search space used: 102579000953
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 75 (33.5 bits)
Medicago: description of AC141323.10


