

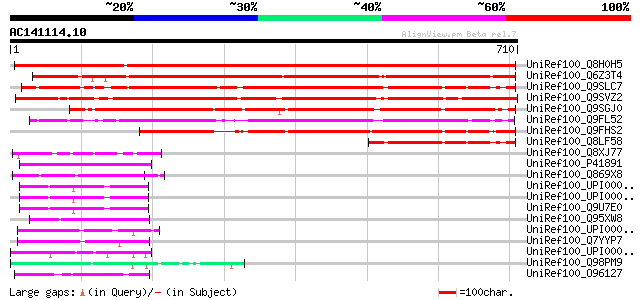
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141114.10 + phase: 0
(710 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8H0H5 Hypothetical protein A20 [Nicotiana tabacum] 855 0.0
UniRef100_Q6Z3T4 Hypothetical protein OSJNBa0025J22.9 [Oryza sat... 704 0.0
UniRef100_Q9SLC7 Hypothetical protein At2g42320 [Arabidopsis tha... 696 0.0
UniRef100_Q9SVZ2 Hypothetical protein F15B8.30 [Arabidopsis thal... 659 0.0
UniRef100_Q9SGJ0 F28J7.14 protein [Arabidopsis thaliana] 499 e-140
UniRef100_Q9FL52 Gb|AAD23715.1 [Arabidopsis thaliana] 462 e-128
UniRef100_Q9FHS2 Gb|AAF03435.1 [Arabidopsis thaliana] 432 e-119
UniRef100_Q8LF58 Hypothetical protein [Arabidopsis thaliana] 218 5e-55
UniRef100_Q8XJ77 Hypothetical protein CPE1884 [Clostridium perfr... 60 2e-07
UniRef100_P41891 Protein gar2 [Schizosaccharomyces pombe] 57 2e-06
UniRef100_Q869X8 Similar to Homo sapiens (Human). dentin sialoph... 56 3e-06
UniRef100_UPI0000060E64 UPI0000060E64 UniRef100 entry 54 2e-05
UniRef100_UPI0000060E63 UPI0000060E63 UniRef100 entry 54 2e-05
UniRef100_Q9U7E0 Transcriptional regulator ATRX homolog [Caenorh... 54 2e-05
UniRef100_Q95XW8 Hypothetical protein Y55B1BR.3 [Caenorhabditis ... 53 4e-05
UniRef100_UPI00004996EE UPI00004996EE UniRef100 entry 51 1e-04
UniRef100_Q7YYP7 Hypothetical garp protein, possible [Cryptospor... 51 1e-04
UniRef100_UPI00004307A7 UPI00004307A7 UniRef100 entry 50 2e-04
UniRef100_Q98PM9 MEMBRANE NUCLEASE, LIPOPROTEIN [Mycoplasma pulm... 50 2e-04
UniRef100_O96127 Hypothetical protein PFB0115w [Plasmodium falci... 50 2e-04
>UniRef100_Q8H0H5 Hypothetical protein A20 [Nicotiana tabacum]
Length = 717
Score = 855 bits (2209), Expect = 0.0
Identities = 446/709 (62%), Positives = 537/709 (74%), Gaps = 11/709 (1%)
Query: 7 IDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLE 66
I++HS+D+E E KEG E+ D DT DS+SSQGD ED VE+AS +K A S+
Sbjct: 10 INDHSSDMESEPKEGLEEESDIDTINDSVSSQGDPQIAEDENVERASTVKVSKKLAKSVA 69
Query: 67 SNRG--SRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSS 124
+N R +SD+K N Q K + S K+ K PS VT+KN + N+K +KV K S
Sbjct: 70 NNSSPAQRAKSDQKENNSQIKATKSTANKAKTPKKDPSTVTSKNVNDNSKNMKVHPKSIS 129
Query: 125 ESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKE 184
+SSE DEK V EV+ ++ILD +S AQSVGS+DE VN EE+ E ED A++ KI +
Sbjct: 130 DSSEEGDEKLVKEVEPVDILDETSVSAQSVGSDDET---VNTEESCEQEDKTALEQKIGQ 186
Query: 185 MESRIENLEEELREVAALEVSLYSIVPEHGSSAHKVHTPARRLSRLYIHACKHWTPKRKA 244
MESR+E LEEELREVAALE++LYS+V EHGSSAHK+HTPARRLSRLY+HACK+W+ ++A
Sbjct: 187 MESRLEKLEEELREVAALEIALYSVVSEHGSSAHKLHTPARRLSRLYLHACKYWSQDKRA 246
Query: 245 TIAKNAVSGLILVAKSCGNDVSRLTFWLSNTIVLREIISQAFGNSGQVSPIMRLAGSNGS 304
T+AKN VSGL+LVAKSCGNDV+RLTFWLSN +VLR IISQAFG+S S ++++ SNG
Sbjct: 247 TVAKNTVSGLVLVAKSCGNDVARLTFWLSNAVVLRVIISQAFGSSCSPSSLVKITESNGG 306
Query: 305 VKRNDGKSASLKWKGIPNGK--SGNGFMQTGEDWQETGTFTLALERVESWIFSRLVESVW 362
K+ + K +S KWK P K S + +Q +DWQET TFT ALERVESWIFSR+VES+W
Sbjct: 307 GKKTESKISSFKWKTHPGNKQSSKHDLLQFFDDWQETRTFTAALERVESWIFSRIVESIW 366
Query: 363 WQALTPYMQSSVGDSCSNKSAGRLLGPALGDHNQGNFSINLWRNAFQDAFQRLCPLRAGG 422
WQ LTP MQS D + KS GRLLGPALGD QGNFSINLW++AFQ+A +RLCP+RAG
Sbjct: 367 WQTLTPNMQSPADDPMARKSVGRLLGPALGDQQQGNFSINLWKHAFQEALKRLCPVRAGS 426
Query: 423 HECGCLPVMARMVMEQCIDRLDVAMFNAILRESALEIPTDPISDPIVDSKVLPIPAGNLS 482
HECGCLPV+AR V+EQC+ RLDVAMFNAILRESA EIPTDPISDPIVDSKVLPIPAG+LS
Sbjct: 427 HECGCLPVLARRVIEQCVARLDVAMFNAILRESAHEIPTDPISDPIVDSKVLPIPAGDLS 486
Query: 483 FGSGAQLKNSVGNWSRLLTDMFGIDAEDCSEEYPENSENDERRGGPGEQKSFALLNDLSD 542
FGSGAQLKNSVGNWSR LTD+FG+DAED S + E S D+ R G + + F LLN LSD
Sbjct: 487 FGSGAQLKNSVGNWSRCLTDLFGMDAED-SGQNDEGSFGDDHRKGGNQPEHFHLLNALSD 545
Query: 543 LLMLPKDMLMDRQVSQEVCPSISLSLIIRVLCNFTPDEFCPDAVPGAVLEALNGETIVER 602
LLMLPKDMLMDR + EVCPSISL L+ R+LCNF+PDEFCPD VPG VLEALN E I+ R
Sbjct: 546 LLMLPKDMLMDRTIRMEVCPSISLPLVKRILCNFSPDEFCPDPVPGTVLEALNAECIIAR 605
Query: 603 RMS-AESIRSFPYSAAPVVYMPPSSVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDEELE 661
R+S +S SFPY AAPVVY PP + +VAEKVAE GK L+R+ SA+QR+GYTSDEELE
Sbjct: 606 RLSGGDSSGSFPYPAAPVVYKPPVAADVAEKVAEVDGKSLLSRSASAIQRKGYTSDEELE 665
Query: 662 ELDSPLSSIIDKVPSSPTVATNGN--GNHEEQGSQTTTNARYQLLREVW 708
E+DSPL+ IIDK+PSSP A NG+ +E+ +N RY LLREVW
Sbjct: 666 EIDSPLACIIDKMPSSPASAQNGSRKAKQKEETGSIGSNKRYDLLREVW 714
>UniRef100_Q6Z3T4 Hypothetical protein OSJNBa0025J22.9 [Oryza sativa]
Length = 687
Score = 704 bits (1816), Expect = 0.0
Identities = 377/694 (54%), Positives = 482/694 (69%), Gaps = 31/694 (4%)
Query: 32 KDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESN-RGSRERSDRKT-NKLQSKVSGS 89
KD+ + + D + + S K +R ++ E++ R ++ R+ NKLQ K S +
Sbjct: 4 KDNGEVRDEKGVGSDYEPARVSGVSKKLMRKDTRENSPRMAKSSGSRQVQNKLQHKASNN 63
Query: 90 NQKKSINSNKGPSRVTNKNTSTNTK---PVKVPAKVSSESSEGVDE-----------KPV 135
Q +S P +V N S + V+VP++ SE SE D+ K
Sbjct: 64 IQSRSPK----PRKVVNAAKSAEVRRSDTVRVPSRAPSELSEETDDIVSEAGTVDDSKGN 119
Query: 136 IEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMESRIENLEEE 195
E KEI++LD + + QS G++DEI EI EE ++ + + +E++S+I+ LE+E
Sbjct: 120 EEAKEIDVLDEAPHCDQSTGTDDEIPEI---EEKIVDDEKPVVYQRNEELQSKIDKLEQE 176
Query: 196 LREVAALEVSLYSIVPEHGSSAHKVHTPARRLSRLYIHACKHWTPKRKATIAKNAVSGLI 255
LREVAALEVSLYS++PEHGSSAHK+HTPARRLSR+YIHA K W+ + A++AK+ VSGL+
Sbjct: 177 LREVAALEVSLYSVLPEHGSSAHKLHTPARRLSRMYIHASKFWSSDKIASVAKSTVSGLV 236
Query: 256 LVAKSCGNDVSRLTFWLSNTIVLREIISQAFGNSGQVSPIMRLAGSNGSVKRNDGKSASL 315
LVAKSC ND SRLTFWLSNT+VLREII+Q G S Q S + NGS K DG+S +
Sbjct: 237 LVAKSCSNDASRLTFWLSNTVVLREIIAQTIGISCQSSSTITAINMNGSAKSLDGRSMPM 296
Query: 316 KWKGIPNGKSGNGF-MQTGEDWQETGTFTLALERVESWIFSRLVESVWWQALTPYMQSSV 374
W +GK MQ +DW ET T ALE++ESWIFSR+VE+VWWQALTP+MQ+ V
Sbjct: 297 LWTNSSSGKQTKFTGMQVPDDWHETSTLLAALEKIESWIFSRIVETVWWQALTPHMQTPV 356
Query: 375 GDSCSNKSAGRLLGPALGDHNQGNFSINLWRNAFQDAFQRLCPLRAGGHECGCLPVMARM 434
S + K+ GR+LGP+LGD QG FS+NLW+ AF DAF R+CPLRAGGHECGCLPV+A++
Sbjct: 357 EGSSTPKT-GRVLGPSLGDQQQGTFSVNLWKAAFHDAFNRICPLRAGGHECGCLPVLAKL 415
Query: 435 VMEQCIDRLDVAMFNAILRESALEIPTDPISDPIVDSKVLPIPAGNLSFGSGAQLKNSVG 494
VMEQC+ RLDVAMFNAILRESA EIPTDPISDPIVD KVLPIPAG+LSFGSGAQLKNS+G
Sbjct: 416 VMEQCVGRLDVAMFNAILRESASEIPTDPISDPIVDPKVLPIPAGDLSFGSGAQLKNSIG 475
Query: 495 NWSRLLTDMFGIDAEDCSEEYPENSENDERRGGPGEQKSFALLNDLSDLLMLPKDMLMDR 554
NWSR LTD FGIDA+D E+ + ER E KSF LLN+LSDLLMLPKDML+++
Sbjct: 476 NWSRWLTDNFGIDADDSEEDGTDT--GSER--SAAESKSFQLLNELSDLLMLPKDMLIEK 531
Query: 555 QVSQEVCPSISLSLIIRVLCNFTPDEFCPDAVPGAVLEALNGETIVERRMSAESIRSFPY 614
+ +E+CPSI L L+ R+LCNFTPDEFCPD VP VLE LN E+++ER + +FP
Sbjct: 532 SIRKEICPSIGLPLVTRILCNFTPDEFCPDPVPSIVLEELNSESLLERCTDKSATSAFPC 591
Query: 615 SAAPVVYMPPSSVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDEELEELDSPLSSIIDKV 674
AAPVVY PPS ++VAEKVA+ GG L R S VQRRGYTSD++L++LDSPL+S+IDK
Sbjct: 592 IAAPVVYRPPSLLDVAEKVADTGGNAKLDRRASMVQRRGYTSDDDLDDLDSPLASLIDK- 650
Query: 675 PSSPTVATNGNGNHEEQGSQTTTNARYQLLREVW 708
S+P + + G+ + Q + NARY LREVW
Sbjct: 651 -SAPPLLSKGSAHFTAQRGVSMENARYTFLREVW 683
>UniRef100_Q9SLC7 Hypothetical protein At2g42320 [Arabidopsis thaliana]
Length = 669
Score = 696 bits (1797), Expect = 0.0
Identities = 373/693 (53%), Positives = 485/693 (69%), Gaps = 56/693 (8%)
Query: 17 EQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRGSRERSD 76
++K E D DT ++S + S D ++ + V L+ + G +D
Sbjct: 31 QKKTEQEKHKDLDTKEESSNI---STVASDSTIQSDPSESYETVDVRYLDDDNGVIASAD 87
Query: 77 RKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEGVDEKPVI 136
S V +K+ S + + + + K A + ++ EG+
Sbjct: 88 AHQESDSSSVVDKTEKEHNLSGS----ICDLEKDVDEQECK-DANIDKDAREGISA---- 138
Query: 137 EVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMESRIENLEEEL 196
++ + +SNGA S GSE+E ++ E NG + + + KI+ +E+RIE LEEEL
Sbjct: 139 -----DVWEDASNGALSAGSENEAADVT--ENNGGNFEDGSSEEKIERLETRIEKLEEEL 191
Query: 197 REVAALEVSLYSIVPEHGSSAHKVHTPARRLSRLYIHACKHWTPKRKATIAKNAVSGLIL 256
REVAALE+SLYS+VP+H SSAHK+HTPARR+SR+YIHACKH+T ++ATIA+N+VSGL+L
Sbjct: 192 REVAALEISLYSVVPDHCSSAHKLHTPARRISRIYIHACKHFTQGKRATIARNSVSGLVL 251
Query: 257 VAKSCGNDVSRLTFWLSNTIVLREIISQAFGNSGQVSPIMRLAGSNGSVKRNDGKSASLK 316
VAKSCGNDVSRLTFWLSN I LR+IISQAFG S I +++ N S + GK +L+
Sbjct: 252 VAKSCGNDVSRLTFWLSNIIALRQIISQAFGRSR----ITQISEPNESGNSDSGKKTNLR 307
Query: 317 WKGIPNGKSGNGFMQTGEDWQETGTFTLALERVESWIFSRLVESVWWQALTPYMQSSVGD 376
WK NGF Q EDWQET TFT ALE++E W+FSR+VESVWWQ TP+MQS D
Sbjct: 308 WK--------NGFQQLLEDWQETETFTTALEKIEFWVFSRIVESVWWQVFTPHMQSPEDD 359
Query: 377 SCSNKSAGRLLGPALGDHNQGNFSINLWRNAFQDAFQRLCPLRAGGHECGCLPVMARMVM 436
S ++KS G+L+GP+LGD NQG FSI+LW+NAF+DA QR+CP+R GHECGCLPV+ARMVM
Sbjct: 360 SSASKSNGKLMGPSLGDQNQGTFSISLWKNAFRDALQRICPMRGAGHECGCLPVLARMVM 419
Query: 437 EQCIDRLDVAMFNAILRESALEIPTDPISDPIVDSKVLPIPAGNLSFGSGAQLKNSVGNW 496
++CI R DVAMFNAILRES +IPTDP+SDPI+DSKVLPIPAG+LSFGSGAQLKN++GNW
Sbjct: 420 DKCIGRFDVAMFNAILRESEHQIPTDPVSDPILDSKVLPIPAGDLSFGSGAQLKNAIGNW 479
Query: 497 SRLLTDMFGIDAEDCSEEYPENSENDERRGGPGEQKSFALLNDLSDLLMLPKDMLMDRQV 556
SR LT+MFG++++D S + NSE+D E K+F LLN+LSDLLMLPKDMLM+ +
Sbjct: 480 SRCLTEMFGMNSDDSSAKEKRNSEDDH-----VESKAFVLLNELSDLLMLPKDMLMEISI 534
Query: 557 SQEVCPSISLSLIIRVLCNFTPDEFCPDAVPGAVLEALN-GETIVERRMSAESIRSFPYS 615
+E+CPSISL LI R+LCNFTPDEFCPD VPGAVLE LN E+I +R++S SFPY+
Sbjct: 535 REEICPSISLPLIKRILCNFTPDEFCPDQVPGAVLEELNAAESIGDRKLSE---ASFPYA 591
Query: 616 AAPVVYMPPSSVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDEELEELDSPLSSIIDKVP 675
A+ V YMPPS++++AEKVAEA K L+RNVS +QR+GYTSDEELEELDSPL+SI+DK
Sbjct: 592 ASSVSYMPPSTMDIAEKVAEASAK--LSRNVSMIQRKGYTSDEELEELDSPLTSIVDK-- 647
Query: 676 SSPTVATNGNGNHEEQGSQTTTNARYQLLREVW 708
A++ G+ T+NARY+LLR+VW
Sbjct: 648 -----ASDFTGS-------ATSNARYKLLRQVW 668
>UniRef100_Q9SVZ2 Hypothetical protein F15B8.30 [Arabidopsis thaliana]
Length = 670
Score = 659 bits (1701), Expect = 0.0
Identities = 372/707 (52%), Positives = 462/707 (64%), Gaps = 48/707 (6%)
Query: 9 NHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSF--TNEDVKVEKASKDPKNKVRANSLE 66
N+S + K G D T SS+ + T E+ V D + + + +
Sbjct: 7 NNSVQSNRSTKNGKRDQKPQKTNGAKRSSEQERLKATKEESNVSVVVDDTTTQSKLSDDD 66
Query: 67 SNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSES 126
+ + S KT K + ++G K S+ + T V P E+
Sbjct: 67 DH--AVNDSSEKTEK-EETINGLACDDEDEEEKEESKELDAIAHEKTDSVSSP-----ET 118
Query: 127 SEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEME 186
EGV+ V+EV D +SNG S GSE+E ++ EN E ED +K ++ +E
Sbjct: 119 CEGVNVDKVVEV-----WDDASNGGLSGGSENEAGDVKEKNENFE-EDEEMLKQMVETLE 172
Query: 187 SRIENLEEELREVAALEVSLYSIVPEHGSSAHKVHTPARRLSRLYIHACKHWTPKRKATI 246
+R+E LEEELREVAALE+SLYS+VP+H SSAHK+HTPARR+SR+YIHACKHW+ ++AT+
Sbjct: 173 TRVEKLEEELREVAALEISLYSVVPDHSSSAHKLHTPARRISRIYIHACKHWSQGKRATV 232
Query: 247 AKNAVSGLILVAKSCGNDVSRLTFWLSNTIVLREIISQAFGNSGQVSPIMRLAGSNGSVK 306
A+N+VSGLIL AKSCGNDVSRLTFWLSN I LREII QAFG + S + SNGS
Sbjct: 233 ARNSVSGLILAAKSCGNDVSRLTFWLSNIISLREIILQAFGKTSVPSHFTETSASNGSEH 292
Query: 307 RNDGKSASLK--WKGIPNGKSGNGFMQTGEDWQETGTFTLALERVESWIFSRLVESVWWQ 364
GK K W K NGF Q EDWQE+ TFT ALE+VE WIFSR+VESVWWQ
Sbjct: 293 NVLGKVRRKKNQWT-----KQSNGFKQVFEDWQESQTFTAALEKVEFWIFSRIVESVWWQ 347
Query: 365 ALTPYMQSSVGDSCSNKSAGRLLGPALGDHNQGNFSINLWRNAFQDAFQRLCPLRAGGHE 424
TP+MQS ++ G+ LGD QG+FSI+LW+NAF+ RLCP+R HE
Sbjct: 348 VFTPHMQSP-------ENGGKTKEHILGDIEQGSFSISLWKNAFKVTLSRLCPMRGARHE 400
Query: 425 CGCLPVMARMVMEQCIDRLDVAMFNAILRESALEIPTDPISDPIVDSKVLPIPAGNLSFG 484
CGCLP++A+MVME+CI R+DVAMFNAILRES +IPTDP+SDPI+DSKVLPI +GNLSFG
Sbjct: 401 CGCLPILAKMVMEKCIARIDVAMFNAILRESEHQIPTDPVSDPILDSKVLPILSGNLSFG 460
Query: 485 SGAQLKNSVGNWSRLLTDMFGIDAEDCSEEYPENSENDERRGGPGEQKSFALLNDLSDLL 544
SGAQLKN++GNWSR L +MF I+ D E END +KSF+LLN+LSDLL
Sbjct: 461 SGAQLKNAIGNWSRCLAEMFSINTRDSVE------ENDPIE----SEKSFSLLNELSDLL 510
Query: 545 MLPKDMLMDRQVSQEVCPSISLSLIIRVLCNFTPDEFCPDAVPGAVLEALNGETIVERRM 604
MLPKDMLMDR +EVCPSISL+LI R+LCNFTPDEFCPD VPGAVLE LN E+I E+++
Sbjct: 511 MLPKDMLMDRSTREEVCPSISLALIKRILCNFTPDEFCPDDVPGAVLEELNNESISEQKL 570
Query: 605 SAESIRSFPYSAAPVVYMPPSSVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDEELEELD 664
S SFPY+A+PV Y PPSS N VAE G ++RNVS +QR+GYTSD+ELEELD
Sbjct: 571 SGV---SFPYAASPVSYTPPSSTN----VAEVGDISRMSRNVSMIQRKGYTSDDELEELD 623
Query: 665 SPLSSIIDKVPSSPTVATNGNGNHE-EQGSQTTTNARYQLLREVWSM 710
SPL+SII+ V SP A N E E+ T +RY+LLREVWSM
Sbjct: 624 SPLTSIIENVSLSPISAQGRNVKQEAEKIGPGVTISRYELLREVWSM 670
>UniRef100_Q9SGJ0 F28J7.14 protein [Arabidopsis thaliana]
Length = 921
Score = 499 bits (1286), Expect = e-140
Identities = 293/639 (45%), Positives = 392/639 (60%), Gaps = 47/639 (7%)
Query: 84 SKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSE--SSEGVDEKPVIEVKEI 141
SK++ + Q + + P ++ N S N P A V+S+ +S+ E V + +++
Sbjct: 312 SKLASNGQHNNGEAKSVPLQIDN--LSENASP---RASVNSQDLTSDQEPESIVEKSRKV 366
Query: 142 EILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMESRIENLEEELREVAA 201
+ + S + +S E A+ ++SKIK +ESR++ LE EL E AA
Sbjct: 367 KSVRSSLDINRSNSRLSLFSERKEAKVYPNSTHDTTLESKIKNLESRVKKLEGELCEAAA 426
Query: 202 LEVSLYSIVPEHGSSAHKVHTPARRLSRLYIHACKHWTPKRKATIAKNAVSGLILVAKSC 261
+E +LYS+V EHGSS+ KVH PARRL RLY+HAC+ R+A A++AVSGL+LVAK+C
Sbjct: 427 IEAALYSVVAEHGSSSSKVHAPARRLLRLYLHACRETHLSRRANAAESAVSGLVLVAKAC 486
Query: 262 GNDVSRLTFWLSNTIVLREIISQAFGNSGQVSPIMRLAGSNGS-VKRNDGKSASLKWKGI 320
GNDV RLTFWLSNTIVLR IIS ++ + P+ G +R K +SLKWK
Sbjct: 487 GNDVPRLTFWLSNTIVLRTIISDT--SAEEELPVSAGPGPRKQKAERETEKRSSLKWKDS 544
Query: 321 PNGKSGNGFMQTGEDWQETGTFTLALERVESWIFSRLVESVWWQALTPYMQSSVGD---- 376
P K +++ W + TF ALE+VE+WIFSR+VES+WWQ LTP MQSS
Sbjct: 545 PLSKKD---IKSFGAWDDPVTFITALEKVEAWIFSRVVESIWWQTLTPRMQSSAASTREF 601
Query: 377 -----SCSNKSAGRLLGPALGDHNQGNFSINLWRNAFQDAFQRLCPLRAGGHECGCLPVM 431
S S K+ GR P+ + G+FS+ LW+ AF++A +RLCPLR GHECGCLP+
Sbjct: 602 DKGNGSASKKTFGRT--PSSTNQELGDFSLELWKKAFREAHERLCPLRGSGHECGCLPIP 659
Query: 432 ARMVMEQCIDRLDVAMFNAILRESALEIPTDPISDPIVDSKVLPIPAGNLSFGSGAQLKN 491
AR++MEQC+ RLDVAMFNAILR+S PTDP+SDPI D +VLPIP+ SFGSGAQLKN
Sbjct: 660 ARLIMEQCVARLDVAMFNAILRDSDDNFPTDPVSDPIADLRVLPIPSRTSSFGSGAQLKN 719
Query: 492 SVGNWSRLLTDMFGIDAEDCSEEYPENSENDERRGGPGEQKSFALLNDLSDLLMLPKDML 551
S+GNWSR LTD+FGID ED + +DE K+F LL LSDL+MLPKDML
Sbjct: 720 SIGNWSRWLTDLFGIDDED-------DDSSDENSYVEKSFKTFNLLKALSDLMMLPKDML 772
Query: 552 MDRQVSQEVCPSISLSLIIRVLCNFTPDEFCPDAVPGAVLEALNGETIVERRMSAESIRS 611
++ V +EVCP LI RVL NF PDEFCPD VP AVL++L E E+ + I S
Sbjct: 773 LNSSVRKEVCPMFGAPLIKRVLNNFVPDEFCPDPVPDAVLKSLESEEEAEKSI----ITS 828
Query: 612 FPYSAAPVVYMPPSSVNVAEKVAEAG--GKCHLTRNVSAVQRRGYTSDEELEELDSPLSS 669
+P +A VY PPS +++ + G L+R S++ R+ YTSD+EL+EL SPL+
Sbjct: 829 YPCTAPSPVYCPPSRTSISTIIGNFGQPQAPQLSRIRSSITRKAYTSDDELDELSSPLAV 888
Query: 670 IIDKVPSSPTVATNGNGNHEEQGSQTTTNARYQLLREVW 708
++ + S + NG+ +E RYQLLRE W
Sbjct: 889 VVLQQAGSKKI---NNGDADE-------TIRYQLLRECW 917
>UniRef100_Q9FL52 Gb|AAD23715.1 [Arabidopsis thaliana]
Length = 657
Score = 462 bits (1188), Expect = e-128
Identities = 299/681 (43%), Positives = 385/681 (55%), Gaps = 91/681 (13%)
Query: 28 TDTTKDSLSSQGDSFTNEDVK-VEKASKDPKNKVRANSLESNRGSRERSDRKTNKLQSKV 86
TD+T S SS+ + N +V ++ A++ + E G+ + SD +TN V
Sbjct: 65 TDSTTGSESSE--VYENVNVHYMDDANEKSRTDGNLVGCEEEEGNGDESDTETNN--GSV 120
Query: 87 SGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEGVDEKPVIEVKEIEILDG 146
S S + K R P+ VS +D P E KE + +
Sbjct: 121 SWSQCELFSPEEKKSER---------------PSMVSK-----IDSTPFEEGKEDDEFED 160
Query: 147 SSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMESRIENLEEELREVAALEVSL 206
+ N SV + + +E + +E + + KI+ ME+RIE LEEELREVAALE+SL
Sbjct: 161 ALN---SVHNNESDNETLVYKEKKRSDVEKVLAQKIETMEARIEKLEEELREVAALEMSL 217
Query: 207 YSIVPEHGSSAHKVHTPARRLSRLYIHACKHWTPKRKATIAKNAVSGLILVAKSCGNDVS 266
YS+ PEHGSS+HK+H PAR LSRLY A K+ + + ++ KN VSGL L+ KSCG+DVS
Sbjct: 218 YSVFPEHGSSSHKLHKPARNLSRLYALARKNQSENKIISVTKNIVSGLSLLLKSCGSDVS 277
Query: 267 RLTFWLSNTIVLREIISQAFGNSGQVSPIMRLAGSNGSVKRNDGKSASLKWKGIPNGKSG 326
RLT+WLSNT++LREIIS FG+S +L G N SLK
Sbjct: 278 RLTYWLSNTVMLREIISLDFGSS-------KLNGLN-----------SLK---------- 309
Query: 327 NGFMQTGEDWQETGTFTLALERVESWIFSRLVESVWWQALTPYMQSSVGDSCSNKSAGRL 386
EDW + T AL RVES F++ VES+W Q + +M DS + G
Sbjct: 310 -------EDWGDVRTLIAALRRVESCFFTQAVESIWSQVMMVHMIPQGVDSTMGEMIGNF 362
Query: 387 LGPALGDHNQGNFSINLWRNAFQDAFQRLCPLRAGGHECGCLPVMARMVMEQCIDRLDVA 446
PA D Q +FS+NLW+ AF++A QRLCP++A +CGCL V+ RMVMEQCI RLDVA
Sbjct: 363 SEPATCDRLQESFSVNLWKEAFEEALQRLCPVQATRRQCGCLHVLTRMVMEQCIVRLDVA 422
Query: 447 MFNAILRESALEIPTDPISDPIVDSKVLPIPAGNLSFGSGAQLKNSVGNWSRLLTDMFGI 506
MFNAILRESA IPTD SDPI DS+VLPIPAG LSF SG +LKN+V WSRLLTD+FGI
Sbjct: 423 MFNAILRESAHHIPTDSASDPIADSRVLPIPAGVLSFESGVKLKNTVSYWSRLLTDIFGI 482
Query: 507 DAEDCSEEYPENSENDERRGGPGEQKSFALLNDLSDLLMLPKDMLMDRQVSQEVCPSISL 566
D E + + G K F LLN+LSDLLMLPK+M +D EVCPSI L
Sbjct: 483 DVE------------QKMQRGDETFKPFHLLNELSDLLMLPKEMFVDSSTRDEVCPSIGL 530
Query: 567 SLIIRVLCNFTPDEFCPDAVPGAVLEALNGETIVERR-MSAESIRSFPYSAAPVVYMPPS 625
SLI R++CNFTPDEFCP VPG VLE LN ++I+E R +S ++ R FP PV Y PPS
Sbjct: 531 SLIKRIVCNFTPDEFCPYPVPGTVLEELNAQSILENRSLSRDTARGFPRQVNPVSYSPPS 590
Query: 626 SVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDEELEELDSPLSSIIDKVPSSPTVATNGN 685
++ + VAE K L S + GY+S+E++E SP + K N
Sbjct: 591 CSHLTDIVAEFSVKLKL----SMTHKNGYSSNEKVETPRSPPYYNVIK-----GAVAKDN 641
Query: 686 GNHEEQGSQTTTNARYQLLRE 706
N E TN RY+LL E
Sbjct: 642 LNLSE------TNERYRLLGE 656
>UniRef100_Q9FHS2 Gb|AAF03435.1 [Arabidopsis thaliana]
Length = 628
Score = 432 bits (1110), Expect = e-119
Identities = 245/530 (46%), Positives = 329/530 (61%), Gaps = 51/530 (9%)
Query: 182 IKEMESRIENLEEELREVAALEVSLYSIVPEHGSSAHKVHTPARRLSRLYIHACKH--WT 239
I +S+ E LE+EL+E A LE ++YS+V EH SS KVH PARRL+R Y+HACK
Sbjct: 141 ITPQDSKTETLEDELKEAAVLEAAIYSVVAEHTSSMSKVHAPARRLARFYLHACKGNGSD 200
Query: 240 PKRKATIAKNAVSGLILVAKSCGNDVSRLTFWLSNTIVLREIISQAFGNSGQVSPIMRLA 299
++AT A+ AVSGLILV+K+CGNDV RLTFWLSN+IVLR I+S+
Sbjct: 201 HSKRATAARAAVSGLILVSKACGNDVPRLTFWLSNSIVLRAILSRGME------------ 248
Query: 300 GSNGSVKRNDGKSASLKWKGIPNGKSGNGFMQTGEDWQETGTFTLALERVESWIFSRLVE 359
K K +P K+G+ ++W++ F ALE+ ESWIFSR+V+
Sbjct: 249 ----------------KMKIVPE-KAGS------DEWEDPRAFLAALEKFESWIFSRVVK 285
Query: 360 SVWWQALTPYMQS-SVGDSCSNKSAGRLLGPALGDHNQGNFSINLWRNAFQDAFQRLCPL 418
SVWWQ++TP+MQS +V S + K +G+ LG NQG ++I LW+NAF+ A +RLCPL
Sbjct: 286 SVWWQSMTPHMQSPAVKGSIARKVSGKR---RLGHRNQGLYAIELWKNAFRAACERLCPL 342
Query: 419 RAGGHECGCLPVMARMVMEQCIDRLDVAMFNAILRESALEIPTDPISDPIVDSKVLPIPA 478
R ECGCLP++A++VMEQ I RLDVAMFNAILRESA E+PTDP+SDPI D VLPIPA
Sbjct: 343 RGSRQECGCLPMLAKLVMEQLISRLDVAMFNAILRESAGEMPTDPVSDPISDINVLPIPA 402
Query: 479 GNLSFGSGAQLKNSVGNWSRLLTDMFGIDAEDCSEEYPENSENDERRGGPGEQKSFALLN 538
G SFG+GAQLKN++G WSR L D F ED S + ND+ + + F LLN
Sbjct: 403 GKASFGAGAQLKNAIGTWSRWLEDQFE-QKEDKSGRNKDEDNNDKEKPECEHFRLFHLLN 461
Query: 539 DLSDLLMLPKDMLMDRQVSQEVCPSISLSLIIRVLCNFTPDEFCPDAVPGAVLEALNGET 598
L DL+MLP ML D+ +EVCP++ +I RVL NF PDEF P +P + + LN E
Sbjct: 462 SLGDLMMLPFKMLADKSTRKEVCPTLGPPIIKRVLRNFVPDEFNPHRIPRRLFDVLNSEG 521
Query: 599 IVERRMSAESIRSFPYSAAPVVYMPPSSVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDE 658
+ E I FP +A+P VY+ PS+ ++ + E ++ S+V ++ YTSD+
Sbjct: 522 LTEEDNGC--ITVFPSAASPTVYLMPSTDSIKRFIGELNNP-SISETGSSVFKKQYTSDD 578
Query: 659 ELEELDSPLSSIIDKVPSSPTVATNGNGNHEEQGSQTTTNARYQLLREVW 708
EL++LD+ ++SI S+P T + +G RYQLLRE+W
Sbjct: 579 ELDDLDTSINSIF----SAP--GTTNSSEWMPKGYGRRKTVRYQLLREIW 622
>UniRef100_Q8LF58 Hypothetical protein [Arabidopsis thaliana]
Length = 184
Score = 218 bits (555), Expect = 5e-55
Identities = 120/207 (57%), Positives = 154/207 (73%), Gaps = 25/207 (12%)
Query: 503 MFGIDAEDCSEEYPENSENDERRGGPGEQKSFALLNDLSDLLMLPKDMLMDRQVSQEVCP 562
MFG++++D S + NSE+D E K+F LLN+LSDLLMLPKDMLM+ + +E+CP
Sbjct: 1 MFGMNSDDSSAKEKRNSEDDHV-----ESKAFVLLNELSDLLMLPKDMLMEISIREEICP 55
Query: 563 SISLSLIIRVLCNFTPDEFCPDAVPGAVLEALN-GETIVERRMSAESIRSFPYSAAPVVY 621
SISL LI R+LCNFTPDEFCPD VPGAVLE LN E+I +R++S SFPY+A+ V Y
Sbjct: 56 SISLPLIKRILCNFTPDEFCPDQVPGAVLEELNAAESIGDRKLSE---ASFPYAASSVSY 112
Query: 622 MPPSSVNVAEKVAEAGGKCHLTRNVSAVQRRGYTSDEELEELDSPLSSIIDKVPSSPTVA 681
MPPS++++AEKVAEA K L+RNVS +QR+GYTSDEELEELDSPL+SI+DK A
Sbjct: 113 MPPSTMDIAEKVAEASAK--LSRNVSMIQRKGYTSDEELEELDSPLTSIVDK-------A 163
Query: 682 TNGNGNHEEQGSQTTTNARYQLLREVW 708
++ G+ T+NARY+LLR+VW
Sbjct: 164 SDFTGS-------ATSNARYKLLRQVW 183
>UniRef100_Q8XJ77 Hypothetical protein CPE1884 [Clostridium perfringens]
Length = 451
Score = 60.1 bits (144), Expect = 2e-07
Identities = 59/214 (27%), Positives = 104/214 (48%), Gaps = 25/214 (11%)
Query: 4 NEVIDNHS---TDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKV 60
N++ID + ++LEK +KE ED+ + + SL + D T+E +EK + K+K
Sbjct: 70 NQLIDYYEKVKSELEKSKKE-IEDLKELEGESVSLKEKLDKITSEKEALEKNLNELKDKK 128
Query: 61 RANSLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPA 120
E SRE + K NKL S+ SN K+ + + +NK + K ++
Sbjct: 129 -----EEIEKSREELNNKFNKLNSE--NSNLKEELKNTNNRMNNSNKEIANLKKEIE-RL 180
Query: 121 KVSSESSEGVDEKPVIEVKEI--EILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAM 178
K + S + +K EV+++ E+ + SN A E+++ + N E +
Sbjct: 181 KSENNSLKSAKDKNSHEVEKLSKELKEVKSNNA-------ELNKTIEISRNKEKN----L 229
Query: 179 KSKIKEMESRIENLEEELREVAALEVSLYSIVPE 212
++I ++S+ N+E+ELR++ SL SIV E
Sbjct: 230 SNEINNLKSKNNNVEKELRDLKEKNNSLSSIVTE 263
Score = 38.9 bits (89), Expect = 0.53
Identities = 44/205 (21%), Positives = 92/205 (44%), Gaps = 15/205 (7%)
Query: 2 KVNEVIDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVR 61
K E ++N L E E++ +T+ ++ + + + E +++ + K+
Sbjct: 133 KSREELNNKFNKLNSENSNLKEELKNTNNRMNNSNKEIANLKKEIERLKSENNSLKSAKD 192
Query: 62 ANSLESNRGSRERSDRKTNKLQ-SKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPA 120
NS E + S+E + K+N + +K ++ K N + + + +KN + + +
Sbjct: 193 KNSHEVEKLSKELKEVKSNNAELNKTIEISRNKEKNLSNEINNLKSKNNNVEKELRDLKE 252
Query: 121 KVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKS 180
K +S SS + K K +E+L+ N S+ ++ N + EGE+ +K
Sbjct: 253 KNNSLSSIVTEAK-----KNLELLNKEIN---SLKERNKTQREENEKLTLEGEN---LKI 301
Query: 181 KIKEMESRIENLEEE---LREVAAL 202
KE+ ++E L +E L+E + L
Sbjct: 302 NCKEIGEKLEGLNKENGQLKETSEL 326
Score = 37.0 bits (84), Expect = 2.0
Identities = 49/232 (21%), Positives = 104/232 (44%), Gaps = 36/232 (15%)
Query: 4 NEVIDNHSTDLEKEQKEGNEDVYDTDTTK---DSLSSQGDSFTNEDVKVEKASKDPKNKV 60
N+ I N ++E+ + E N D + LS + + + ++ K + +NK
Sbjct: 167 NKEIANLKKEIERLKSENNSLKSAKDKNSHEVEKLSKELKEVKSNNAELNKTIEISRNKE 226
Query: 61 R-----ANSLESNRGSRERSDR----KTNKLQSKVSGSNQK-----KSINSNKGPS---R 103
+ N+L+S + E+ R K N L S V+ + + K INS K + R
Sbjct: 227 KNLSNEINNLKSKNNNVEKELRDLKEKNNSLSSIVTEAKKNLELLNKEINSLKERNKTQR 286
Query: 104 VTNKNTSTNTKPVKVPAKVSSESSEGVDEK--------PVIEVKEIEILDGSSNGAQSVG 155
N+ + + +K+ K E EG++++ ++ ++I I D +S + +
Sbjct: 287 EENEKLTLEGENLKINCKEIGEKLEGLNKENGQLKETSELLNKEKIWIKDQNSGLKKQIL 346
Query: 156 SEDEIHEIVNAEENGEGE--------DGAAMKSKIKEMESRIENLEEELREV 199
+E ++ E++ G+ + A+K + +E+++ +E LEEE +++
Sbjct: 347 ELEENLQLALEEKDALGKKISEDMEVEMKALKEEAEEVKAEMEILEEEAKKL 398
>UniRef100_P41891 Protein gar2 [Schizosaccharomyces pombe]
Length = 500
Score = 56.6 bits (135), Expect = 2e-06
Identities = 48/187 (25%), Positives = 80/187 (42%), Gaps = 4/187 (2%)
Query: 15 EKEQKEGNEDVYDTDTTKDSLS--SQGDSFTNEDVK-VEKASKDPKNKVRANSLESNRGS 71
+K++ + V +T + K ++ S+ T E K + K S + + E+ R S
Sbjct: 3 KKDKTSVKKSVKETASKKGAIEKPSKSKKITKEAAKEIAKQSSKTDVSPKKSKKEAKRAS 62
Query: 72 RERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEGVD 131
+K+ K Q K + S + ++ S + ++S + SSESS
Sbjct: 63 SPEPSKKSVKKQKKSKKKEESSSESESESSSSESESSSSESESSSSESESSSSESSSSES 122
Query: 132 EKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMESRIEN 191
E+ VI VK E + SS + S SE+E +V EE E ++ +S E ES +
Sbjct: 123 EEEVI-VKTEEKKESSSESSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESESESSS 181
Query: 192 LEEELRE 198
E E E
Sbjct: 182 SESEEEE 188
Score = 40.8 bits (94), Expect = 0.14
Identities = 34/173 (19%), Positives = 80/173 (45%), Gaps = 8/173 (4%)
Query: 14 LEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRGSRE 73
++K++K ++ +++ +S SS+ +S ++E E +S + ++ +S S+ E
Sbjct: 71 VKKQKKSKKKEESSSESESESSSSESESSSSES---ESSSSESESSSSESS--SSESEEE 125
Query: 74 RSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVS---SESSEGV 130
+ K +S S+ +S + ++ K S++ + + S S SSE
Sbjct: 126 VIVKTEEKKESSSESSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESESESSSSESE 185
Query: 131 DEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIK 183
+E+ V+E E + S + + S S D E +++ + + E ++ + + K
Sbjct: 186 EEEEVVEKTEEKKEGSSESSSDSESSSDSSSESGDSDSSSDSESESSSEDEKK 238
Score = 37.0 bits (84), Expect = 2.0
Identities = 42/176 (23%), Positives = 69/176 (38%), Gaps = 14/176 (7%)
Query: 15 EKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRGSRER 74
E ++ DV + K++ + + + VK +K SK + + ES+ E
Sbjct: 39 EIAKQSSKTDVSPKKSKKEAKRASSPEPSKKSVKKQKKSKKKEESSSESESESSSSESES 98
Query: 75 SDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEGVDEKP 134
S ++ S+ S+ + S S++ V K T K SS SE +E
Sbjct: 99 SSSESESSSSESESSSSESS--SSESEEEVIVK---TEEKKESSSESSSSSESEEEEEAV 153
Query: 135 V-IEVKEIEILDGSSNGAQSVG--------SEDEIHEIVNAEENGEGEDGAAMKSK 181
V IE K+ D SS + S SE+E + EE EG ++ S+
Sbjct: 154 VKIEEKKESSSDSSSESSSSESESESSSSESEEEEEVVEKTEEKKEGSSESSSDSE 209
>UniRef100_Q869X8 Similar to Homo sapiens (Human). dentin sialophosphoprotein
[Dictyostelium discoideum]
Length = 1206
Score = 56.2 bits (134), Expect = 3e-06
Identities = 49/214 (22%), Positives = 92/214 (42%), Gaps = 8/214 (3%)
Query: 4 NEVIDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRAN 63
N N ++ E E + + +++ + SSQG + NE E +++ N+ N
Sbjct: 678 NNESSNQGSNNESSHNETSNQGSNNESSHNESSSQGSN--NESAHNESSNQGSNNESSHN 735
Query: 64 SLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVS 123
S + E S+ +N S SN + +SN+ + +++ N S+N + +
Sbjct: 736 ESSSQGSNNESSNNDSNNQNSNNEPSNNE---SSNQSSNNLSSNNESSNNESSNQGSNNE 792
Query: 124 SESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGED-GAAMKSKI 182
S SE + + + E + S G+ S S + ++ +N+E N + G+ +S
Sbjct: 793 SSHSESSNNESSNQSLNSEYNNEPSQGSNSESSNESSNQSLNSESNNDSSSHGSNSESSN 852
Query: 183 KEMESRIENLEEELREVAALEVSLYSIVPEHGSS 216
E S N E E ++ + S YS +GSS
Sbjct: 853 NESSSHGSNNESSNNESSSQDSSNYS--SNNGSS 884
Score = 48.1 bits (113), Expect = 9e-04
Identities = 45/188 (23%), Positives = 79/188 (41%), Gaps = 7/188 (3%)
Query: 4 NEVIDNHSTDLEKEQKEGNEDVYDTDTTKDSLS--SQGDSFTNEDVKVEKASKDPKNKVR 61
NE +N S++ + N + + + +S S S +S NE E +S+ N
Sbjct: 956 NESSNNESSNQGSNNESSNNESNNESSNNESSSQGSNNESSNNESSNNESSSQSSNNG-S 1014
Query: 62 ANSLESNRGS-RERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPA 120
+N+ SN+GS E S +N S +N+ S SN S + N S+N + +
Sbjct: 1015 SNNESSNQGSNNESSSHGSNNESSSKGSNNESSSHGSNNESSSQGSNNESSNNESSNQNS 1074
Query: 121 KVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKS 180
S ++E + E E SS G+ + S +E + + E+ E+ +
Sbjct: 1075 NNESSNNESSSKGSNNESSNNE---SSSQGSNNESSNNESNNQNSNNESSHKEESESNSH 1131
Query: 181 KIKEMESR 188
+ E ES+
Sbjct: 1132 QSSETESK 1139
Score = 45.8 bits (107), Expect = 0.004
Identities = 36/195 (18%), Positives = 79/195 (40%), Gaps = 11/195 (5%)
Query: 4 NEVIDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRAN 63
NE + S + Q NE + + +S S ++ ++ + + S + + +N
Sbjct: 975 NESNNESSNNESSSQGSNNESSNNESSNNESSSQSSNNGSSNNESSNQGSNNESSSHGSN 1034
Query: 64 SLESNRGSRERSDRKTNKLQSKVSGSNQKKSIN-----------SNKGPSRVTNKNTSTN 112
+ S++GS S + +S GSN + S N SN S + N S+N
Sbjct: 1035 NESSSKGSNNESSSHGSNNESSSQGSNNESSNNESSNQNSNNESSNNESSSKGSNNESSN 1094
Query: 113 TKPVKVPAKVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEG 172
+ + S ++E ++ E E + +S+ + S+D++ + ++ E
Sbjct: 1095 NESSSQGSNNESSNNESNNQNSNNESSHKEESESNSHQSSETESKDQVSNSGSMNQSSES 1154
Query: 173 EDGAAMKSKIKEMES 187
+ +++ + ES
Sbjct: 1155 SESSSLSEQSSAFES 1169
Score = 42.0 bits (97), Expect = 0.063
Identities = 45/197 (22%), Positives = 81/197 (40%), Gaps = 28/197 (14%)
Query: 11 STDLEKEQKEGNEDVYDTDTTKDSLSSQGDS-------FTNEDVKVEKASKDPKNKVRAN 63
S++ K Q NE D +T + SSQG + +N+ E + + N+ N
Sbjct: 545 SSETLKIQNSNNESSDDHSSTNNESSSQGSNNESSNNESSNQGSNNESSHNETSNQGSNN 604
Query: 64 SLESNRGSRERSDRKTNKLQSKVSGSNQKKSIN--SNKGPSRVTNKNTSTNTKPVKVPAK 121
N S + S+ ++ +S GSN + S N SN+G + ++ N S+N + +
Sbjct: 605 ESSHNESSSQGSNNESAHNESSNQGSNNESSHNESSNQGSNNESSNNESSN-QGSNNESS 663
Query: 122 VSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSK 181
+ S++G + + S+N + + GS +E + N G+ +S
Sbjct: 664 HNESSNQGSNN------------ESSNNESSNQGSNNE------SSHNETSNQGSNNESS 705
Query: 182 IKEMESRIENLEEELRE 198
E S+ N E E
Sbjct: 706 HNESSSQGSNNESAHNE 722
Score = 40.4 bits (93), Expect = 0.18
Identities = 44/208 (21%), Positives = 80/208 (38%), Gaps = 17/208 (8%)
Query: 8 DNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDS-------FTNEDVKVEKASKDPKNKV 60
+N S++ E ++ + + ++ + SSQG + NE E +S+ N+
Sbjct: 861 NNESSNNESSSQDSSNYSSNNGSSNNESSSQGSNNESSNQGSNNESSNNESSSQGSNNES 920
Query: 61 RAN-----SLESNRGSRERSDRKTNKLQSKVSGSNQKKSIN--SNKGPSRVTNKNTSTNT 113
N S + + E S+ +++ +S GSN + S N SN+G + ++ N S N
Sbjct: 921 SHNEPSNQSSNNESSNNESSNNESSNNESSSKGSNNESSNNESSNQGSNNESSNNESNNE 980
Query: 114 KPVKVPAKVSSESSEGVDEKPVIEVKEIEILDGSSNGA---QSVGSEDEIHEIVNAEENG 170
+ S + +E E +GSSN Q +E H N +
Sbjct: 981 SSNNESSSQGSNNESSNNESSNNESSSQSSNNGSSNNESSNQGSNNESSSHGSNNESSSK 1040
Query: 171 EGEDGAAMKSKIKEMESRIENLEEELRE 198
+ ++ E S+ N E E
Sbjct: 1041 GSNNESSSHGSNNESSSQGSNNESSNNE 1068
Score = 40.0 bits (92), Expect = 0.24
Identities = 48/224 (21%), Positives = 91/224 (40%), Gaps = 10/224 (4%)
Query: 4 NEVIDNHSTDLEKE-QKEGNEDVYDTDTTKDSLS-SQGDSFTNEDVKVEKASKDPKNKVR 61
N+ +N S+ E Q NE ++ + + S + S + +N+ E ++ + N+
Sbjct: 599 NQGSNNESSHNESSSQGSNNESAHNESSNQGSNNESSHNESSNQGSNNESSNNESSNQGS 658
Query: 62 ANSLESNRGSRERSDRKTNKLQSKVSGSNQKKSIN--SNKGPSRVTNKNTSTNTKPVKVP 119
N N S + S+ +++ +S GSN + S N SN+G + ++ N S++
Sbjct: 659 NNESSHNESSNQGSNNESSNNESSNQGSNNESSHNETSNQGSNNESSHNESSSQGSNNES 718
Query: 120 AKVSSESSEGVDEKPVIEVKEIEILDGSSN---GAQSVGSEDEIHEIVNAEENGEGEDGA 176
A S + +E E + SSN Q+ +E +E N N +
Sbjct: 719 AHNESSNQGSNNESSHNESSSQGSNNESSNNDSNNQNSNNEPSNNESSNQSSNNLSSNNE 778
Query: 177 AMKSKIKEMESRIENLEEELREVAALEVSL---YSIVPEHGSSA 217
+ ++ S E+ E + SL Y+ P GS++
Sbjct: 779 SSNNESSNQGSNNESSHSESSNNESSNQSLNSEYNNEPSQGSNS 822
Score = 38.1 bits (87), Expect = 0.91
Identities = 40/194 (20%), Positives = 85/194 (43%), Gaps = 15/194 (7%)
Query: 4 NEVIDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRAN 63
NE +N S++ + + + + +++ SL+S+ ++ ++ E +++ + N
Sbjct: 777 NESSNNESSNQGSNNESSHSESSNNESSNQSLNSEYNNEPSQGSNSESSNESSNQSL--N 834
Query: 64 SLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVS 123
S +N S S+ +++ +S GSN + S N + S + N S+N + +
Sbjct: 835 SESNNDSSSHGSNSESSNNESSSHGSNNESSNNES---SSQDSSNYSSNN-----GSSNN 886
Query: 124 SESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEI--HEIVNAEENGEGEDGAAMKSK 181
SS+G + + + E S+N + S GS +E +E N N E + + ++
Sbjct: 887 ESSSQGSNNESSNQGSNNE---SSNNESSSQGSNNESSHNEPSNQSSNNESSNNESSNNE 943
Query: 182 IKEMESRIENLEEE 195
ES + E
Sbjct: 944 SSNNESSSKGSNNE 957
Score = 37.4 bits (85), Expect = 1.5
Identities = 39/194 (20%), Positives = 78/194 (40%), Gaps = 9/194 (4%)
Query: 4 NEVIDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDS--FTNEDVKVEKASKDPKNKVR 61
N +H ++ E E + + +++ + SSQ S +N ++S N
Sbjct: 838 NNDSSSHGSNSESSNNESSSHGSNNESSNNESSSQDSSNYSSNNGSSNNESSSQGSNNES 897
Query: 62 ANSLESNRGSR-ERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPA 120
+N +N S E S + +N S SNQ + N+ + ++ N S+N + +
Sbjct: 898 SNQGSNNESSNNESSSQGSNNESSHNEPSNQSSN---NESSNNESSNNESSNNESSSKGS 954
Query: 121 KVSSESSEGVDEKPVIEVKEIEILDGSSN---GAQSVGSEDEIHEIVNAEENGEGEDGAA 177
S ++E ++ E E + SSN +Q +E +E N E + + + +
Sbjct: 955 NNESSNNESSNQGSNNESSNNESNNESSNNESSSQGSNNESSNNESSNNESSSQSSNNGS 1014
Query: 178 MKSKIKEMESRIEN 191
++ S E+
Sbjct: 1015 SNNESSNQGSNNES 1028
Score = 36.2 bits (82), Expect = 3.4
Identities = 43/215 (20%), Positives = 88/215 (40%), Gaps = 27/215 (12%)
Query: 14 LEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNK-------VRANSLE 66
++ +E + ++ + + S S +S +NED + + P N +N+
Sbjct: 474 IQSPTEESSSHNSESSSGQTSSSESNNSQSNEDSNHQSSGNHPTNDSSNQGSGSHSNNHS 533
Query: 67 SNRGSR--ERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSS 124
SN+GS + S +T K+Q+ + S+ S +N+ S+ +N +S N + SS
Sbjct: 534 SNQGSSSGQDSSSETLKIQNSNNESSDDHSSTNNESSSQGSNNESSNNESSNQGSNNESS 593
Query: 125 E---SSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSK 181
S++G + + S N + S GS +E + E + +G + + ++
Sbjct: 594 HNETSNQGSNN------------ESSHNESSSQGSN---NESAHNESSNQGSNNESSHNE 638
Query: 182 IKEMESRIENLEEELREVAALEVSLYSIVPEHGSS 216
S E+ E + S ++ GS+
Sbjct: 639 SSNQGSNNESSNNESSNQGSNNESSHNESSNQGSN 673
>UniRef100_UPI0000060E64 UPI0000060E64 UniRef100 entry
Length = 585
Score = 53.5 bits (127), Expect = 2e-05
Identities = 50/186 (26%), Positives = 80/186 (42%), Gaps = 15/186 (8%)
Query: 15 EKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRGSRER 74
EK ++EG K SS + +E+ K+SK K++ RA S + S E
Sbjct: 41 EKREREGKPPPKKRPAKKRKASSSEEDDDDEEESPRKSSK--KSRKRAKSESESDESDEE 98
Query: 75 SDRKTNKLQSKVSG-----SNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEG 129
DRK +K + KV S +K++ +S++ + + K K + SSESSE
Sbjct: 99 EDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDEEREQKSKKKSKKTKKQTSSESSEE 158
Query: 130 VDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIK-EMESR 188
+E E ++ N +SV E E + +E + +K K K E ES
Sbjct: 159 SEE-------ERKVKKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKGLKKKAKSESESE 211
Query: 189 IENLEE 194
E+ +E
Sbjct: 212 SEDEKE 217
Score = 43.5 bits (101), Expect = 0.022
Identities = 47/191 (24%), Positives = 79/191 (40%), Gaps = 22/191 (11%)
Query: 8 DNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLES 67
++ +D E+EQK + + TK SS+ + E+ KV+K+ KNK + S++
Sbjct: 128 EDEDSDEEREQKSKKK----SKKTKKQTSSESSEESEEERKVKKSK---KNKEK--SVKK 178
Query: 68 NRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESS 127
+ E SD ++ SK S KK S K + K K K SES
Sbjct: 179 RAETSEESDE--DEKPSKKSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESE 236
Query: 128 EGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMES 187
+ EK E ++ +S S +E E ++E E ++ + K K +
Sbjct: 237 DEAPEKKKTEKRK-----------RSKTSSEESSESEKSDEEEEEKESSPKPKKKKPLAV 285
Query: 188 RIENLEEELRE 198
+ + +EE E
Sbjct: 286 KKLSSDEESEE 296
>UniRef100_UPI0000060E63 UPI0000060E63 UniRef100 entry
Length = 1359
Score = 53.5 bits (127), Expect = 2e-05
Identities = 50/186 (26%), Positives = 80/186 (42%), Gaps = 15/186 (8%)
Query: 15 EKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRGSRER 74
EK ++EG K SS + +E+ K+SK K++ RA S + S E
Sbjct: 41 EKREREGKPPPKKRPAKKRKASSSEEDDDDEEESPRKSSK--KSRKRAKSESESDESDEE 98
Query: 75 SDRKTNKLQSKVSG-----SNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEG 129
DRK +K + KV S +K++ +S++ + + K K + SSESSE
Sbjct: 99 EDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDEEREQKSKKKSKKTKKQTSSESSEE 158
Query: 130 VDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIK-EMESR 188
+E E ++ N +SV E E + +E + +K K K E ES
Sbjct: 159 SEE-------ERKVKKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKGLKKKAKSESESE 211
Query: 189 IENLEE 194
E+ +E
Sbjct: 212 SEDEKE 217
Score = 43.5 bits (101), Expect = 0.022
Identities = 47/191 (24%), Positives = 79/191 (40%), Gaps = 22/191 (11%)
Query: 8 DNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLES 67
++ +D E+EQK + + TK SS+ + E+ KV+K+ KNK + S++
Sbjct: 128 EDEDSDEEREQKSKKK----SKKTKKQTSSESSEESEEERKVKKSK---KNKEK--SVKK 178
Query: 68 NRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESS 127
+ E SD ++ SK S KK S K + K K K SES
Sbjct: 179 RAETSEESDE--DEKPSKKSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESE 236
Query: 128 EGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMES 187
+ EK E ++ +S S +E E ++E E ++ + K K +
Sbjct: 237 DEAPEKKKTEKRK-----------RSKTSSEESSESEKSDEEEEEKESSPKPKKKKPLAV 285
Query: 188 RIENLEEELRE 198
+ + +EE E
Sbjct: 286 KKLSSDEESEE 296
>UniRef100_Q9U7E0 Transcriptional regulator ATRX homolog [Caenorhabditis elegans]
Length = 1359
Score = 53.5 bits (127), Expect = 2e-05
Identities = 50/186 (26%), Positives = 80/186 (42%), Gaps = 15/186 (8%)
Query: 15 EKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRGSRER 74
EK ++EG K SS + +E+ K+SK K++ RA S + S E
Sbjct: 41 EKREREGKPPPKKRPAKKRKASSSEEDDDDEEESPRKSSK--KSRKRAKSESESDESDEE 98
Query: 75 SDRKTNKLQSKVSG-----SNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEG 129
DRK +K + KV S +K++ +S++ + + K K + SSESSE
Sbjct: 99 EDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDEEREQKSKKKSKKTKKQTSSESSEE 158
Query: 130 VDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIK-EMESR 188
+E E ++ N +SV E E + +E + +K K K E ES
Sbjct: 159 SEE-------ERKVKKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKGLKKKAKSESESE 211
Query: 189 IENLEE 194
E+ +E
Sbjct: 212 SEDEKE 217
Score = 43.5 bits (101), Expect = 0.022
Identities = 47/191 (24%), Positives = 79/191 (40%), Gaps = 22/191 (11%)
Query: 8 DNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLES 67
++ +D E+EQK + + TK SS+ + E+ KV+K+ KNK + S++
Sbjct: 128 EDEDSDEEREQKSKKK----SKKTKKQTSSESSEESEEERKVKKSK---KNKEK--SVKK 178
Query: 68 NRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESS 127
+ E SD ++ SK S KK S K + K K K SES
Sbjct: 179 RAETSEESDE--DEKPSKKSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESE 236
Query: 128 EGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMES 187
+ EK E ++ +S S +E E ++E E ++ + K K +
Sbjct: 237 DEAPEKKKTEKRK-----------RSKTSSEESSESEKSDEEEEEKESSPKPKKKKPLAV 285
Query: 188 RIENLEEELRE 198
+ + +EE E
Sbjct: 286 KKLSSDEESEE 296
>UniRef100_Q95XW8 Hypothetical protein Y55B1BR.3 [Caenorhabditis elegans]
Length = 679
Score = 52.8 bits (125), Expect = 4e-05
Identities = 42/168 (25%), Positives = 71/168 (42%), Gaps = 3/168 (1%)
Query: 28 TDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRGSRERSDRKTNKLQSKVS 87
++ K +++ +E+ K EK SK K V +S E E+S +K +K K S
Sbjct: 273 SEKRKRAVNDSSSDDEDEEEKPEKRSKKSKKAVIDSSSEDEE--EEKSSKKRSKKSKKES 330
Query: 88 GSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEGVDEKPVIEVKEIEILDGS 147
Q+ S +S + V + S P K K SE S G +E+ V++ K+ ++
Sbjct: 331 DEEQQAS-DSEEEVVEVKKNSKSPKKTPKKTAVKEESEESSGDEEEEVVKKKKSSKINKR 389
Query: 148 SNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMESRIENLEEE 195
S E+E+ E + + +K + E +N EEE
Sbjct: 390 KAKESSSDEEEEVEESPKKKTKSPRKSSKKSAAKEESEEESSDNEEEE 437
Score = 45.4 bits (106), Expect = 0.006
Identities = 58/214 (27%), Positives = 89/214 (41%), Gaps = 27/214 (12%)
Query: 6 VIDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNED-VKVEKASKDP-----KNK 59
VID+ S D E+E+ + K+S Q S + E+ V+V+K SK P K
Sbjct: 305 VIDSSSEDEEEEKSSKKRS---KKSKKESDEEQQASDSEEEVVEVKKNSKSPKKTPKKTA 361
Query: 60 VRANSLESNRGSRE-----RSDRKTNKLQSKVSGSNQKKSINSN-----KGPSRVTNKNT 109
V+ S ES+ E + K NK ++K S S++++ + + K P + + K+
Sbjct: 362 VKEESEESSGDEEEEVVKKKKSSKINKRKAKESSSDEEEEVEESPKKKTKSPRKSSKKSA 421
Query: 110 STNTKPVKVPAKVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEEN 169
+ K +E E VD P +VK + S A V SE+ N EE
Sbjct: 422 A---KEESEEESSDNEEEEEVDYSPKKKVKSPK--KSSKKPAAKVESEEPSD---NEEEE 473
Query: 170 GEGEDGAAMKSKIKEMESRIENLEEELREVAALE 203
E E+ K K SR + E E + E
Sbjct: 474 EEVEESPIKKDKTPRKYSRKSAAKVESTESSGNE 507
Score = 42.4 bits (98), Expect = 0.048
Identities = 41/189 (21%), Positives = 75/189 (38%), Gaps = 8/189 (4%)
Query: 11 STDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRG 70
S D E+ + E + + K +S DS ++ D +K K KNK + + + +
Sbjct: 191 SDDSEESEDEEEKSPRKRSSKKRRAASVSDSDSDSDSDFDK-KKWKKNKAKRSKRDDSSD 249
Query: 71 SRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAK----VSSES 126
+R+ K + KKS + + ++ + KP K K V S
Sbjct: 250 DDSEMERRRKKSKKSKKSKKFKKSEKRKRAVNDSSSDDEDEEEKPEKRSKKSKKAVIDSS 309
Query: 127 SEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEME 186
SE +E+ + + + S Q+ SE+E+ E+ ++N + K+ +KE
Sbjct: 310 SEDEEEEKSSKKRSKKSKKESDEEQQASDSEEEVVEV---KKNSKSPKKTPKKTAVKEES 366
Query: 187 SRIENLEEE 195
EEE
Sbjct: 367 EESSGDEEE 375
Score = 38.9 bits (89), Expect = 0.53
Identities = 35/189 (18%), Positives = 67/189 (34%), Gaps = 8/189 (4%)
Query: 7 IDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLE 66
+++ + +E++E E T S + + D + E + PK +
Sbjct: 498 VESTESSGNEEEEEVEESPKKKGKTPRKSSKKSAAVEESDNEEEDVEESPKKRTSPRKSS 557
Query: 67 SNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSES 126
R ++E S+ ++ +V S +K K P + K V+ +E
Sbjct: 558 KKRAAKEESEESSDNEVEEVEDSPKKNDKTLRKSPRKPAAK--------VESEESFGNEE 609
Query: 127 SEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEME 186
E V+E P + K S + + E AEE + KSK E +
Sbjct: 610 EEEVEESPKKKGKTPRKSSKKSAAKEESEESSDDEEEEQAEETNGSHTESPTKSKTDESD 669
Query: 187 SRIENLEEE 195
+ + E++
Sbjct: 670 NSTSSDEDD 678
Score = 38.5 bits (88), Expect = 0.69
Identities = 42/192 (21%), Positives = 85/192 (43%), Gaps = 23/192 (11%)
Query: 15 EKEQKEGNED---VYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRGS 71
E E+ G+E+ V ++K + +S ++E+ +VE++ K R +S +S
Sbjct: 365 ESEESSGDEEEEVVKKKKSSKINKRKAKESSSDEEEEVEESPKKKTKSPRKSSKKSAAKE 424
Query: 72 RERSDRKTNKLQSKVSGSNQKK----SINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESS 127
+ N+ + +V S +KK +S K ++V ++ S N E
Sbjct: 425 ESEEESSDNEEEEEVDYSPKKKVKSPKKSSKKPAAKVESEEPSDN-----------EEEE 473
Query: 128 EGVDEKPVIEVKEIEILDGSS----NGAQSVGSEDEIHEIVNAEENGEGEDGAAMKS-KI 182
E V+E P+ + K S +S G+E+E + ++ G+ ++ KS +
Sbjct: 474 EEVEESPIKKDKTPRKYSRKSAAKVESTESSGNEEEEEVEESPKKKGKTPRKSSKKSAAV 533
Query: 183 KEMESRIENLEE 194
+E ++ E++EE
Sbjct: 534 EESDNEEEDVEE 545
>UniRef100_UPI00004996EE UPI00004996EE UniRef100 entry
Length = 497
Score = 50.8 bits (120), Expect = 1e-04
Identities = 47/206 (22%), Positives = 90/206 (42%), Gaps = 22/206 (10%)
Query: 11 STDLEKEQKEGN-EDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNR 69
+T+L KE ++ E++ D+ T++ + + + V +EK+ K + K+ E +
Sbjct: 211 NTELTKECQDNTIEEMLDSQNTENPKNEKKSEIEQKQVSIEKSYKTNEGKLLEKKTELTQ 270
Query: 70 GSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEG 129
+D + K Q + + Q K N+ +T+ +TNTK + + SS
Sbjct: 271 IQLNTNDIQKEKYQMQETQLTQNKLFNN------ITHSEGTTNTKTITTIPNIEINSSTN 324
Query: 130 VDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEG-----EDGAAMKSKIKE 184
+ ++ I + E L ++ E E E+ E N +G ED ++ +KI+E
Sbjct: 325 LQQEEAINNQLNEEL-------ETTEGEIENTEVQKKESNSDGMMELEEDSISLINKIEE 377
Query: 185 MESRIENLEEELREVAALEVSLYSIV 210
L EL+ L+VSL ++
Sbjct: 378 KNQYSFRLPHELKN---LQVSLCLLI 400
>UniRef100_Q7YYP7 Hypothetical garp protein, possible [Cryptosporidium parvum]
Length = 789
Score = 50.8 bits (120), Expect = 1e-04
Identities = 45/197 (22%), Positives = 85/197 (42%), Gaps = 21/197 (10%)
Query: 12 TDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSL--ESNR 69
TDL +E + T + SQ E +K+EK K K + L +
Sbjct: 182 TDLSNTNQENQSENVPVSTL-NLTKSQKKKLEKERLKLEKKEKKLKKQKEKERLRLDKKE 240
Query: 70 GSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEG 129
RER ++K + + K N+KK+ N NK +KN + K ++ P + S
Sbjct: 241 KKREREEKKRLEAEKKKQLKNEKKNKNKNK------SKNEGSGEKSIESPKEKKSSCFSK 294
Query: 130 VDEKPVIEVKEIEILDGSSNGAQ-----------SVGSEDEIHEIVNAEENGEGEDGAAM 178
++ ++++ + L+ N + +V SE+E+ + +++ + GE + A
Sbjct: 295 KKKQNYVQIESHDSLEEKLNSQEVVPEDINDEVSTVASENEVFDNISSIDEGE-QTEAED 353
Query: 179 KSKIKEMESRIENLEEE 195
K+++ E + E EEE
Sbjct: 354 DIKVEKSEDQFEFDEEE 370
Score = 40.4 bits (93), Expect = 0.18
Identities = 50/223 (22%), Positives = 86/223 (38%), Gaps = 48/223 (21%)
Query: 4 NEVIDNHSTDLEKEQKEGNEDVY---------------DTDTTKDSLSSQGDSFTNEDVK 48
NEV DN S+ E EQ E +D+ D++ L+ F E +
Sbjct: 334 NEVFDNISSIDEGEQTEAEDDIKVEKSEDQFEFDEEEEDSEIKARILAGNESDFKTERIS 393
Query: 49 VEKASKDPKNK-VRANSLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNK 107
E+ +D + + + + + R R +++ L +K + I +G S ++
Sbjct: 394 SEEVLQDSDSDYLEGDEYQEDSNERYRFSQESEDLIAKEKAVKPLEGIEI-EGISEGEDE 452
Query: 108 NTSTNTKPVKVPAKVSSESSEGVDEK-----------------------PVIEVKEIEIL 144
+TS N + V + S + E DE+ +I EIE+L
Sbjct: 453 DTS-NISDIPVVSNYSLDHEEEKDEENKNEDSGIQELSSGIELSNDNDNDIISNNEIEVL 511
Query: 145 DGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMES 187
+ S +D+++EI+ EE ED SK+ E ES
Sbjct: 512 N-------SADDDDDVNEIIVEEEKKVDEDFTNKDSKLDERES 547
>UniRef100_UPI00004307A7 UPI00004307A7 UniRef100 entry
Length = 386
Score = 50.4 bits (119), Expect = 2e-04
Identities = 62/223 (27%), Positives = 95/223 (41%), Gaps = 29/223 (13%)
Query: 1 MKVNEVIDNHSTDLEKEQKEGNEDVYDTD--TTKDSLSSQGDSFTNEDVKVEKASKD--- 55
+K E D S++ E+E + T + + S DS + E+ K +K
Sbjct: 78 VKKKESTDESSSEEEEETPKTQPKAVSTPKISKEQKSSDDSDSESEEESKTNITAKTNTK 137
Query: 56 ----PKNKVRANSLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTST 111
K KV AN ES+ S+ +TN + K S + S +S++ VT T +
Sbjct: 138 SIITSKTKVGANQKESSSEDSSESEDETNTVIKKTESSTED-SDDSDENEKSVTKSTTKS 196
Query: 112 NTKPVKVPAKVSSESSEGVDEKPV-------IEVKEIEILDGSSNGAQSVGSEDEIHE-I 163
K + S +SSE DEKPV + K + SS + S SE+E + I
Sbjct: 197 IGKKDE---SSSEDSSESEDEKPVPTKTPVPTKTKADTKKESSSEDSDSDSSEEEKPKTI 253
Query: 164 VNAEENGE-----GEDGAAMKSKIKEMESR---IENLEEELRE 198
V+ + E GE + K +I E ESR + LEEE ++
Sbjct: 254 VSKSKTTEIKQDTGEKKDSRKREINENESRKRSKKELEEETKK 296
Score = 36.6 bits (83), Expect = 2.6
Identities = 32/166 (19%), Positives = 69/166 (41%), Gaps = 10/166 (6%)
Query: 29 DTTKDSLSSQGDSFTNEDVKVEKASKDPKNKV-RANSLESNRGSRERSDRKTNKLQSKVS 87
DT K+S S DS ++E+ K PK V ++ + E + + E+ D + ++ S
Sbjct: 230 DTKKESSSEDSDSDSSEEEK-------PKTIVSKSKTTEIKQDTGEKKDSRKREINENES 282
Query: 88 GSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSESSEGVDEKPVIEVKEIEILDGS 147
KK + S+ + K++S+ + K+ + + + + + E
Sbjct: 283 RKRSKKELEEETKKSKRSRKDSSSGDEDQKIKRNKTEDDRSRLRKSKKDSSSDDE--PKK 340
Query: 148 SNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKEMESRIENLE 193
+ + SEDE + ++ ED ++ K + +S ++ E
Sbjct: 341 TRKRRDSSSEDEKSRTRKSRKDSTSEDEGXVRLKKSKRDSSTDDEE 386
Score = 36.6 bits (83), Expect = 2.6
Identities = 38/160 (23%), Positives = 64/160 (39%), Gaps = 18/160 (11%)
Query: 57 KNKVRANSLESNRGSRERSDRKTNKLQSKVSGS-------NQKKSINSNKGPSR------ 103
K V+A + +S+ + S+ +T K S+++G N KK+ +S++ S
Sbjct: 1 KLNVKAQAKDSSSDTSSESEEETPKKLSQINGKTTVNAVINNKKNESSSEDTSSENEKAL 60
Query: 104 --VTNKNTSTNTK---PVKVPAKVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSED 158
VT T+ T PVK SSE +E P + K + S S S+
Sbjct: 61 KVVTKAKTAVKTSKASPVKKKESTDESSSEEEEETPKTQPKAVSTPKISKEQKSSDDSDS 120
Query: 159 EIHEIVNAEENGEGEDGAAMKSKIKEMESRIENLEEELRE 198
E E + + + SK K ++ E+ E+ E
Sbjct: 121 ESEEESKTNITAKTNTKSIITSKTKVGANQKESSSEDSSE 160
>UniRef100_Q98PM9 MEMBRANE NUCLEASE, LIPOPROTEIN [Mycoplasma pulmonis]
Length = 1125
Score = 50.4 bits (119), Expect = 2e-04
Identities = 70/358 (19%), Positives = 142/358 (39%), Gaps = 46/358 (12%)
Query: 2 KVNEVIDNHS-TDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKV 60
K+++ N+S T EK+QK ++D + K + Q DS + + SK+ ++
Sbjct: 507 KISDASQNNSNTTNEKDQKLDSQDESKNNAIKSQQNDQKDSNLSSSKNDTQPSKESSPQI 566
Query: 61 RANSLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPA 120
N + S + + + S S Q K+ ++ +T KN S+N+ V
Sbjct: 567 NPNLENNQEISHSNNGENDDSKEQNTSNSRQTKNDLRSEQKQNLTTKNPSSNSS--NVET 624
Query: 121 KVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENG------EGED 174
K ++++E K + D +++ ++ +++ N E N + E+
Sbjct: 625 KNETQNNENSSTKKDEIDTSAKTQDSTNSNLKNEEKTNQVETKTNTESNNSNSTNKQEEN 684
Query: 175 GAAMKSKIKEMESRI----------ENLEEELREVAALEVSLYSIVPEHGSSAHKVHTPA 224
+ K +I + ES + EN++ + E++ E ++ + + ++ TP
Sbjct: 685 SSTKKEEISKSESNVNNSNSTNKQEENIDNKKEEISKSESNVNN---SNSTNTQNQETPE 741
Query: 225 RRLSRLYIHACKHWTPKRKATIAKNAVSGLILVAKSCGNDVSRLTFWLSNTIVLREIISQ 284
S+ + K+ ++ +NA+ + AK ++ +W N V + S+
Sbjct: 742 TNESQNNVIIGKN---PNNQSLNQNAID---VSAKKV-----KIGYWNINESVGKSSASK 790
Query: 285 AFGNSGQVS-PIMRLAGSNGSVKRND-----------GKSASLKW-KGIPNGKSGNGF 329
AF + + + L G G V K +S KW + I K G GF
Sbjct: 791 AFAVAKVIDHNKLDLVGIGGLVHEETLTKIVEEMNKLSKDSSDKWVQVISEKKQGEGF 848
>UniRef100_O96127 Hypothetical protein PFB0115w [Plasmodium falciparum]
Length = 1192
Score = 50.4 bits (119), Expect = 2e-04
Identities = 44/191 (23%), Positives = 81/191 (42%), Gaps = 22/191 (11%)
Query: 7 IDNHSTDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLE 66
+ + D EKE KEG +D DTD +D+ + D+ ED E+ + D +NK
Sbjct: 367 VGEETKDDEKEDKEGTDDEEDTDDEEDT-DDEEDTDDEEDTSDEETTGDQENKEET---- 421
Query: 67 SNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAKVSSES 126
E ++KT K + ++ K+ +K S + + + + + + + E
Sbjct: 422 ------EVDEKKTEKAEEEL--EEDKEESEKDKEESEKDKEESEKDKEESEKDKEKTEED 473
Query: 127 SEGVDEKPVIEV--KEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGAAMKSKIKE 184
E +++ EV KE ++ + G G++DE E+ + ED K + K+
Sbjct: 474 EEKTEDEKGTEVYKKETDVDEKKEKGEYGEGTDDE-------EDKEKEEDDEETKVEEKK 526
Query: 185 MESRIENLEEE 195
E E + E
Sbjct: 527 TEKDEEGTDYE 537
Score = 43.5 bits (101), Expect = 0.022
Identities = 48/202 (23%), Positives = 79/202 (38%), Gaps = 13/202 (6%)
Query: 2 KVNEVIDNHSTDLEKE--QKEGNEDVYDTDTTKDSLSSQ--GDSFTNEDVKVEKASKDPK 57
K +E D TD E++ +E +D DTD +D+ + GD E+ +V++ +
Sbjct: 372 KDDEKEDKEGTDDEEDTDDEEDTDDEEDTDDEEDTSDEETTGDQENKEETEVDEKKTEKA 431
Query: 58 NKVRANSLESNRGSRERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVK 117
+ E + +E S++ + + S + K K T K
Sbjct: 432 EEELEEDKEESEKDKEESEKDKEESEKDKEESEKDKEKTEEDEEKTEDEKGTEVYKKETD 491
Query: 118 V-PAKVSSESSEGVDEKPVIEVKEIEILDGSSNGAQSVGSEDEIHEIVNAEENGEGEDGA 176
V K E EG D++ E KE E D + + +DE E + EE+ + D
Sbjct: 492 VDEKKEKGEYGEGTDDE---EDKEKEEDDEETKVEEKKTEKDE--EGTDYEEDTDDSD-- 544
Query: 177 AMKSKIKEMESRIENLEEELRE 198
+ K E + E EEE E
Sbjct: 545 -KDEETKVEEKKTERDEEETEE 565
Score = 38.5 bits (88), Expect = 0.69
Identities = 31/130 (23%), Positives = 54/130 (40%), Gaps = 9/130 (6%)
Query: 12 TDLEKEQKEGNEDVYDTDTTKDSLSSQGDSFTNEDVKVEKASKDPKNKVRANSLESNRGS 71
TDL+ ++++G ED D D KD + D EK +D K K + + E ++
Sbjct: 626 TDLDDQEEDGEEDKED-DKEKDKEDDKEDD-------KEKDKEDDKEKYKEDDKEDDKED 677
Query: 72 RERSDRKTNKLQSKVSGSNQKKSINSNKGPSRVTNKNTSTNTKPVKVPAK-VSSESSEGV 130
+ D++ NK + K + K + K K+ N + K K E +
Sbjct: 678 DKEKDKEDNKEKDKEDNKEKDKEDDKEKDKEDDKEKDKEDNKEKDKEDNKEKDKEDDKEK 737
Query: 131 DEKPVIEVKE 140
+K V +K+
Sbjct: 738 HDKHVRRIKK 747
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.128 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,196,366,221
Number of Sequences: 2790947
Number of extensions: 51775236
Number of successful extensions: 195258
Number of sequences better than 10.0: 2697
Number of HSP's better than 10.0 without gapping: 170
Number of HSP's successfully gapped in prelim test: 2668
Number of HSP's that attempted gapping in prelim test: 182176
Number of HSP's gapped (non-prelim): 10176
length of query: 710
length of database: 848,049,833
effective HSP length: 135
effective length of query: 575
effective length of database: 471,271,988
effective search space: 270981393100
effective search space used: 270981393100
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC141114.10


