

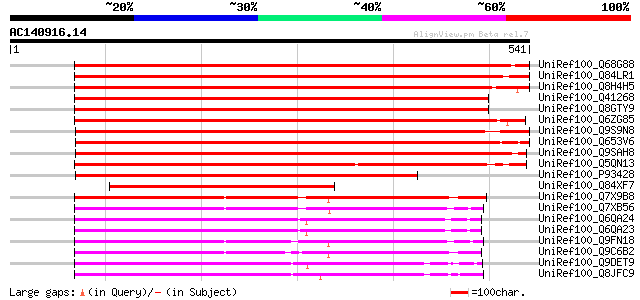
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140916.14 - phase: 0 /pseudo
(541 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q68G88 Root-specific metal transporter [Malus baccata] 746 0.0
UniRef100_Q84LR1 Root-specific metal transporter [Lycopersicon e... 728 0.0
UniRef100_Q8H4H5 Putative Nramp1 protein [Oryza sativa] 670 0.0
UniRef100_Q41268 OsNramp1 [Oryza sativa] 645 0.0
UniRef100_Q8GTY9 Nramp1 protein [Oryza sativa] 644 0.0
UniRef100_Q6ZG85 Putative root-specific metal transporter [Oryza... 570 e-161
UniRef100_Q9S9N8 Metal transporter Nramp6 [Arabidopsis thaliana] 563 e-159
UniRef100_Q653V6 Putative NRAMP metal ion transporter 1 [Oryza s... 560 e-158
UniRef100_Q9SAH8 Metal transporter Nramp1 [Arabidopsis thaliana] 558 e-157
UniRef100_Q5QN13 Putative root-specific metal transporter [Oryza... 492 e-137
UniRef100_P93428 Integral membrane protein OsNramp3 [Oryza sativa] 416 e-115
UniRef100_Q84XF7 Integral membrane protein Nramp1 [Malus xiaojin... 308 3e-82
UniRef100_Q7X9B8 Ferrous ion membrane transport protein DMT1 [Gl... 302 1e-80
UniRef100_Q7XB56 Nramp metal transporter homolog [Thlaspi japoni... 299 1e-79
UniRef100_Q6QA24 Putative divalent metal transporter DMT1A [Schi... 298 2e-79
UniRef100_Q6QA23 Putative divalent metal transporter DMT1B [Schi... 298 2e-79
UniRef100_Q9FN18 Metal transporter Nramp4 [Arabidopsis thaliana] 298 3e-79
UniRef100_Q9C6B2 Metal transporter Nramp2 [Arabidopsis thaliana] 298 4e-79
UniRef100_Q9DET9 Natural resistance-associated macrophage protei... 295 3e-78
UniRef100_Q8JFC9 Natural resistance-associated macrophage protei... 293 7e-78
>UniRef100_Q68G88 Root-specific metal transporter [Malus baccata]
Length = 551
Score = 746 bits (1927), Expect = 0.0
Identities = 376/475 (79%), Positives = 422/475 (88%), Gaps = 4/475 (0%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+ETDLQAGANH YELLW++LIGLIFALIIQSL+ANLGV TGKHLSE+C+ EYP FVKYCL
Sbjct: 79 LETDLQAGANHRYELLWVILIGLIFALIIQSLSANLGVITGKHLSELCKAEYPPFVKYCL 138
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLI 187
WLLAEVAVIAADIPEVIGTAFALNILFNIP+W GVLLTGFSTLLLL LQ++GVRKLE+LI
Sbjct: 139 WLLAEVAVIAADIPEVIGTAFALNILFNIPVWTGVLLTGFSTLLLLGLQKYGVRKLEMLI 198
Query: 188 TILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHS 247
+LVFVMA CFF EMSYV PPASGVLKGMF+PKL+G+GA DAIALLGALIMPHNLFLHS
Sbjct: 199 AVLVFVMAACFFGEMSYVKPPASGVLKGMFIPKLSGQGATGDAIALLGALIMPHNLFLHS 258
Query: 248 ALVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVER 307
ALVLSRK+P SVRG+N+ACRYFL ESGFALFVAFLINVA+ISVSGTVC ++LS N +
Sbjct: 259 ALVLSRKIPNSVRGMNDACRYFLIESGFALFVAFLINVAIISVSGTVCHQSNLSDKNNDT 318
Query: 308 CNDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWK 367
C+DLTLNSASFLL+NVLGRSS +YAIALLASGQSSTITGTYAGQ++MQGFLDI+MK+W
Sbjct: 319 CSDLTLNSASFLLQNVLGRSSKILYAIALLASGQSSTITGTYAGQFVMQGFLDIKMKKWA 378
Query: 368 RNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPH 427
RNLMTRCIAI PSL V+IIGGSSG+ RLIIIASMILSFELPFALIPLLKFS S+TKMGPH
Sbjct: 379 RNLMTRCIAITPSLIVSIIGGSSGAGRLIIIASMILSFELPFALIPLLKFSGSATKMGPH 438
Query: 428 KNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIAS 487
KNS+ II ISWI+G II IN+YYLST FV WIIHSSLPKVA VFIGI+VFP+MAIYI +
Sbjct: 439 KNSIYIIVISWIIGMAIIGINIYYLSTGFVGWIIHSSLPKVATVFIGILVFPIMAIYILA 498
Query: 488 VIYLTFRKDTVEMFIE-TKNDTVMQNQVEKGIVDNGQLELSHVPYREDLADIPLP 541
V+YL RKD+V F+E TKND QNQ+E G+ + G+ VP+REDLAD+ LP
Sbjct: 499 VLYLALRKDSVVTFVEPTKNDPAAQNQMENGLPNYGE---PPVPFREDLADVSLP 550
>UniRef100_Q84LR1 Root-specific metal transporter [Lycopersicon esculentum]
Length = 530
Score = 728 bits (1879), Expect = 0.0
Identities = 368/474 (77%), Positives = 414/474 (86%), Gaps = 6/474 (1%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+ETDLQAGANH YELLW++LIGLIFALIIQSLAANLGV+TGKHLSE+C EYP+FVKYCL
Sbjct: 62 LETDLQAGANHRYELLWVILIGLIFALIIQSLAANLGVSTGKHLSELCRAEYPVFVKYCL 121
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLI 187
WLLAEVAVIAADIPEVIGTAFALNILF+IP+WAGVL TG STLL + LQR+GVRKLELLI
Sbjct: 122 WLLAEVAVIAADIPEVIGTAFALNILFHIPVWAGVLCTGVSTLLFIGLQRYGVRKLELLI 181
Query: 188 TILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHS 247
ILVFVMA CFF EMSYV PPA+ + KGMF+PKL G+GA ADAIALLGAL+MPHNLFLHS
Sbjct: 182 AILVFVMAACFFGEMSYVKPPANELFKGMFIPKLNGDGATADAIALLGALVMPHNLFLHS 241
Query: 248 ALVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVER 307
ALVLSRK+P SVRGIN+AC++FL ESGFALFV+FLINVA++SVSGTVC A++LS N E
Sbjct: 242 ALVLSRKIPNSVRGINDACKFFLIESGFALFVSFLINVAVVSVSGTVCGADNLSDQNKES 301
Query: 308 CNDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWK 367
C+D+TLNSASFLLKNVLG+SSST+YAIALLASGQSSTITGTYAGQ+IMQGFLD++MK W
Sbjct: 302 CSDITLNSASFLLKNVLGKSSSTVYAIALLASGQSSTITGTYAGQFIMQGFLDLKMKTWL 361
Query: 368 RNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPH 427
RNL TR IAI PSL V+IIGGSSG+ RLIIIASMILSFELPFALIPLLKFSSSSTK+GPH
Sbjct: 362 RNLFTRLIAITPSLVVSIIGGSSGAGRLIIIASMILSFELPFALIPLLKFSSSSTKLGPH 421
Query: 428 KNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIAS 487
KNS+ II ISWILG GII IN+YYLSTAFV W+I ++LPKV NV IGI+VFPLMAIYI +
Sbjct: 422 KNSIYIIVISWILGLGIIGINIYYLSTAFVGWLISNNLPKVGNVLIGIVVFPLMAIYILA 481
Query: 488 VIYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDLADIPLP 541
VIYL FRKD V +I+ D M+N + N + VPYREDLADIPLP
Sbjct: 482 VIYLMFRKDKVVTYIDPIKDDHMENGI------NSMELVDRVPYREDLADIPLP 529
>UniRef100_Q8H4H5 Putative Nramp1 protein [Oryza sativa]
Length = 538
Score = 670 bits (1729), Expect = 0.0
Identities = 336/477 (70%), Positives = 401/477 (83%), Gaps = 6/477 (1%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+ETDLQAGANH YELLW++LIGLIFALIIQSLAANLGV TG+HL+E+C+ EYP FVK L
Sbjct: 64 LETDLQAGANHRYELLWVILIGLIFALIIQSLAANLGVVTGRHLAEICKSEYPKFVKIFL 123
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLI 187
WLLAE+AVIAADIPEVIGTAFA NILF+IP+W GVL+TG STLLLL LQ++GVRKLE LI
Sbjct: 124 WLLAELAVIAADIPEVIGTAFAFNILFHIPVWVGVLITGTSTLLLLGLQKYGVRKLEFLI 183
Query: 188 TILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHS 247
++LVFVMA CFF E+S V PPA V+KG+F+P+L G+GA ADAIALLGAL+MPHNLFLHS
Sbjct: 184 SMLVFVMAACFFGELSIVKPPAKEVMKGLFIPRLNGDGATADAIALLGALVMPHNLFLHS 243
Query: 248 ALVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVER 307
ALVLSRK P SVRGI + CR+FLYESGFALFVA LIN+A++SVSGT CS+ +LS ++ ++
Sbjct: 244 ALVLSRKTPASVRGIKDGCRFFLYESGFALFVALLINIAVVSVSGTACSSANLSQEDADK 303
Query: 308 CNDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWK 367
C +L+L+++SFLLKNVLG+SS+ +Y +ALLASGQSSTITGTYAGQYIMQGFLDIRM++W
Sbjct: 304 CANLSLDTSSFLLKNVLGKSSAIVYGVALLASGQSSTITGTYAGQYIMQGFLDIRMRKWL 363
Query: 368 RNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPH 427
RNLMTR IAIAPSL V+IIGGS G+ RLIIIASMILSFELPFALIPLLKFSSS +KMGPH
Sbjct: 364 RNLMTRTIAIAPSLIVSIIGGSRGAGRLIIIASMILSFELPFALIPLLKFSSSKSKMGPH 423
Query: 428 KNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIAS 487
KNS+ II SW LG II IN+Y+LST+FV W+IH+ LPK ANV +G VFP M +YI +
Sbjct: 424 KNSIYIIVFSWFLGLLIIGINMYFLSTSFVGWLIHNDLPKYANVLVGAAVFPFMLVYIVA 483
Query: 488 VIYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSH---VPYREDLADIPLP 541
V+YLT RKD+V F+ D+ + V+ D G L + +PYR+DLADIPLP
Sbjct: 484 VVYLTIRKDSVVTFVA---DSSLAAVVDAEKADAGDLAVDDDEPLPYRDDLADIPLP 537
>UniRef100_Q41268 OsNramp1 [Oryza sativa]
Length = 517
Score = 645 bits (1663), Expect = 0.0
Identities = 310/432 (71%), Positives = 381/432 (87%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+ETDLQAGANH YELLW++LIGLIFALIIQSL+ANLGV TG+HL+E+C+ EYP++VK CL
Sbjct: 54 METDLQAGANHKYELLWVILIGLIFALIIQSLSANLGVVTGRHLAELCKTEYPVWVKTCL 113
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLI 187
WLLAE+AVIA+DIPEVIGT FA N+LF+IP+W GVL+ G STLLLL LQR+GVRKLE+++
Sbjct: 114 WLLAELAVIASDIPEVIGTGFAFNLLFHIPVWTGVLIAGSSTLLLLGLQRYGVRKLEVVV 173
Query: 188 TILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHS 247
+LVFVMAGCFF EMS V PP + VL+G+F+P+L+G GA D+IALLGAL+MPHNLFLHS
Sbjct: 174 ALLVFVMAGCFFVEMSIVKPPVNEVLQGLFIPRLSGPGATGDSIALLGALVMPHNLFLHS 233
Query: 248 ALVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVER 307
ALVLSR P S +G+ +ACR+FL+ESG ALFVA L+N+A+ISVSGTVC+A +LS ++ +
Sbjct: 234 ALVLSRNTPASAKGMKDACRFFLFESGIALFVALLVNIAIISVSGTVCNATNLSPEDAVK 293
Query: 308 CNDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWK 367
C+DLTL+S+SFLL+NVLG+SS+T+Y +ALLASGQSSTITGTYAGQY+MQGFLDI+MK+W
Sbjct: 294 CSDLTLDSSSFLLRNVLGKSSATVYGVALLASGQSSTITGTYAGQYVMQGFLDIKMKQWL 353
Query: 368 RNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPH 427
RNLMTR IAI PSL V+IIGGSSG+ RLI+IASMILSFELPFALIPLLKFSSSS KMG +
Sbjct: 354 RNLMTRSIAIVPSLIVSIIGGSSGAGRLIVIASMILSFELPFALIPLLKFSSSSNKMGEN 413
Query: 428 KNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIAS 487
KNS+ I+ SW+LGF II IN+Y+LST V WI+H++LP ANV IGI++FPLM +Y+ +
Sbjct: 414 KNSIYIVGFSWVLGFVIIGINIYFLSTKLVGWILHNALPTFANVLIGIVLFPLMLLYVVA 473
Query: 488 VIYLTFRKDTVE 499
VIYLTFRKDTV+
Sbjct: 474 VIYLTFRKDTVK 485
>UniRef100_Q8GTY9 Nramp1 protein [Oryza sativa]
Length = 519
Score = 644 bits (1660), Expect = 0.0
Identities = 310/432 (71%), Positives = 379/432 (86%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
VETDLQAGANH YELLW++LIGLIFALIIQSL+ANLGV TG HL+E+C+ EYP++VK CL
Sbjct: 56 VETDLQAGANHKYELLWVILIGLIFALIIQSLSANLGVVTGSHLAELCKTEYPVWVKTCL 115
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLI 187
WLLAE+AVIA+DIPEVIGT FA N+LF+IP+W GVL+ G STLLLL LQR+GVRKLE+++
Sbjct: 116 WLLAELAVIASDIPEVIGTGFAFNLLFHIPVWTGVLIAGSSTLLLLGLQRYGVRKLEVVV 175
Query: 188 TILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHS 247
+LVFVMAGCFF EMS V PP + VL+G+F+P+L+G GA D+IALLGAL+MPHNLFLHS
Sbjct: 176 ALLVFVMAGCFFVEMSIVKPPVNEVLQGLFIPRLSGPGATGDSIALLGALVMPHNLFLHS 235
Query: 248 ALVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVER 307
ALVLSR P S +G+ + CR+FL+ESG ALFVA L+N+A+ISVSGTVC+A +LS ++ +
Sbjct: 236 ALVLSRNTPASAKGMKDVCRFFLFESGIALFVALLVNIAIISVSGTVCNATNLSPEDAVK 295
Query: 308 CNDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWK 367
C+DLTL+S+SFLL+NVLG+SS+T+Y +ALLASGQSSTITGTYAGQY+MQGFLDI+MK+W
Sbjct: 296 CSDLTLDSSSFLLRNVLGKSSATVYGVALLASGQSSTITGTYAGQYVMQGFLDIKMKQWL 355
Query: 368 RNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPH 427
RNLMTR IAI PSL V+IIGGSSG+ RLI+IASMILSFELPFALIPLLKFSSSS KMG +
Sbjct: 356 RNLMTRSIAIVPSLIVSIIGGSSGAGRLIVIASMILSFELPFALIPLLKFSSSSNKMGEN 415
Query: 428 KNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIAS 487
KNS+ I+ SW+LGF II IN+Y+LST V WI+H++LP ANV IGI++FPLM +Y+ +
Sbjct: 416 KNSIYIVGFSWVLGFVIIGINIYFLSTKLVGWILHNALPTFANVLIGIVLFPLMLLYVVA 475
Query: 488 VIYLTFRKDTVE 499
VIYLTFRKDTV+
Sbjct: 476 VIYLTFRKDTVK 487
>UniRef100_Q6ZG85 Putative root-specific metal transporter [Oryza sativa]
Length = 545
Score = 570 bits (1468), Expect = e-161
Identities = 285/474 (60%), Positives = 365/474 (76%), Gaps = 6/474 (1%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+ETD+QAGA+ YELLW++L+G++FAL+IQ+LAANLGV TG+HL+E+C EYP +V L
Sbjct: 71 LETDMQAGADFKYELLWVILVGMVFALLIQTLAANLGVKTGRHLAELCREEYPHYVNIFL 130
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLI 187
W++AE+AVI+ DIPEV+GTAFA NIL IP+WAGV+LT FSTLLLL +QRFG RKLE +I
Sbjct: 131 WIIAELAVISDDIPEVLGTAFAFNILLKIPVWAGVILTVFSTLLLLGVQRFGARKLEFII 190
Query: 188 TILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHS 247
+F MA CFF E+SY+ P A V+KGMFVP L G+GA A+AIAL GA+I P+NLFLHS
Sbjct: 191 AAFMFTMAACFFGELSYLRPSAGEVVKGMFVPSLQGKGAAANAIALFGAIITPYNLFLHS 250
Query: 248 ALVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVER 307
ALVLSRK P+S + I ACRYFL E A VAFLINV+++ V+G++C+AN+LS +
Sbjct: 251 ALVLSRKTPRSDKSIRAACRYFLIECSLAFIVAFLINVSVVVVAGSICNANNLSPADANT 310
Query: 308 CNDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWK 367
C DLTL S LL+NVLGRSSS +YA+ALLASGQS+TI+ T+AGQ IMQGFLD++MK W
Sbjct: 311 CGDLTLQSTPLLLRNVLGRSSSVVYAVALLASGQSTTISCTFAGQVIMQGFLDMKMKNWV 370
Query: 368 RNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPH 427
RNL+TR IAIAPSL V+I+ G SG+ +LII++SMILSFELPFALIPLLKF +SS K+GP
Sbjct: 371 RNLITRVIAIAPSLIVSIVSGPSGAGKLIILSSMILSFELPFALIPLLKFCNSSKKVGPL 430
Query: 428 KNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIAS 487
K S+ + I+WIL F +I +N Y+L +V W++H++LPK AN I ++VF LMA Y+ +
Sbjct: 431 KESIYTVVIAWILSFALIVVNTYFLVWTYVDWLVHNNLPKYANGLISVVVFALMAAYLVA 490
Query: 488 VIYLTFRKDTVEMFIETKNDTVMQNQVEKG---IVD-NGQLELSHVPYREDLAD 537
V+YLTFRKDTV ++ Q QVE G +VD + E PYR+DLAD
Sbjct: 491 VVYLTFRKDTVATYVPVPERA--QAQVEAGGTPVVDASAADEDQPAPYRKDLAD 542
>UniRef100_Q9S9N8 Metal transporter Nramp6 [Arabidopsis thaliana]
Length = 527
Score = 563 bits (1450), Expect = e-159
Identities = 282/473 (59%), Positives = 362/473 (75%), Gaps = 14/473 (2%)
Query: 69 ETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCLW 128
ETDLQ+GA + YELLW++L+ AL+IQSLAANLGV TGKHL+E C EY + LW
Sbjct: 59 ETDLQSGAQYKYELLWIILVASCAALVIQSLAANLGVVTGKHLAEHCRAEYSKVPNFMLW 118
Query: 129 LLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLIT 188
++AE+AV+A DIPEVIGTAFALN+LFNIP+W GVLLTG STL+LL+LQ++G+RKLE LI
Sbjct: 119 VVAEIAVVACDIPEVIGTAFALNMLFNIPVWIGVLLTGLSTLILLALQQYGIRKLEFLIA 178
Query: 189 ILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHSA 248
LVF +A CFF E+ Y P VL G+FVP+L G GA AI+LLGA++MPHNLFLHSA
Sbjct: 179 FLVFTIALCFFVELHYSKPDPKEVLYGLFVPQLKGNGATGLAISLLGAMVMPHNLFLHSA 238
Query: 249 LVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVERC 308
LVLSRK+P+SV GI EACRY+L ESG AL VAFLINV++ISVSG VC+A+DLS ++ C
Sbjct: 239 LVLSRKIPRSVTGIKEACRYYLIESGLALMVAFLINVSVISVSGAVCNASDLSPEDRASC 298
Query: 309 NDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWKR 368
DL LN ASFLL+NV+G+ SS ++AIALLASGQSSTITGTYAGQY+MQGFLD+R++ W R
Sbjct: 299 QDLDLNKASFLLRNVVGKWSSKLFAIALLASGQSSTITGTYAGQYVMQGFLDLRLEPWLR 358
Query: 369 NLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPHK 428
N +TRC+AI PSL VA+IGGS+G+ +LIIIASMILSFELPFAL+PLLKF+SS TKMG H
Sbjct: 359 NFLTRCLAIIPSLIVALIGGSAGAGKLIIIASMILSFELPFALVPLLKFTSSKTKMGSHA 418
Query: 429 NSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIASV 488
NS++I +++WI+G I+ IN+YYL ++F+K ++HS + VA VF+G++ F +A Y+A++
Sbjct: 419 NSLVISSVTWIIGGLIMGINIYYLVSSFIKLLLHSHMNLVAIVFLGVLGFSGIATYLAAI 478
Query: 489 IYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDLADIPLP 541
YL RK+ + +D + RED+A++ LP
Sbjct: 479 SYLVLRKN--------------RESSSTHFLDFSNSQTEETLPREDIANMQLP 517
>UniRef100_Q653V6 Putative NRAMP metal ion transporter 1 [Oryza sativa]
Length = 550
Score = 560 bits (1444), Expect = e-158
Identities = 288/473 (60%), Positives = 363/473 (75%), Gaps = 2/473 (0%)
Query: 69 ETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCLW 128
ETDLQAGA + YELLW++LI ALIIQSLAA LGV TGKHL+E C EYP + LW
Sbjct: 71 ETDLQAGAQYKYELLWIILIASCAALIIQSLAARLGVVTGKHLAEHCRAEYPKATNFILW 130
Query: 129 LLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLIT 188
+LAE+AV+A DIPEVIGTAFALN+LF IP+W GVL+TG STL+LL LQ++GVRKLE LI
Sbjct: 131 ILAELAVVACDIPEVIGTAFALNMLFKIPVWCGVLITGLSTLMLLLLQQYGVRKLEFLIA 190
Query: 189 ILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHSA 248
ILV ++A CF E+ Y P +S V++G+FVP+L G GA AI+LLGA++MPHNLFLHSA
Sbjct: 191 ILVSLIATCFLVELGYSKPNSSEVVRGLFVPELKGNGATGLAISLLGAMVMPHNLFLHSA 250
Query: 249 LVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVERC 308
LVLSRKVP+SV GI EACR+++ ES FAL +AFLIN+++ISVSG VC +++LS ++ C
Sbjct: 251 LVLSRKVPRSVHGIKEACRFYMIESAFALTIAFLINISIISVSGAVCGSDNLSPEDQMNC 310
Query: 309 NDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWKR 368
+DL LN ASFLLKNVLG SS ++A+ALLASGQSSTITGTYAGQY+MQGFLD+RM W R
Sbjct: 311 SDLDLNKASFLLKNVLGNWSSKLFAVALLASGQSSTITGTYAGQYVMQGFLDLRMTPWIR 370
Query: 369 NLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPHK 428
NL+TR +AI PSL V+IIGGSS + +LIIIASMILSFELPFAL+PLLKF+SS TKMG H
Sbjct: 371 NLLTRSLAILPSLIVSIIGGSSAAGQLIIIASMILSFELPFALVPLLKFTSSRTKMGQHT 430
Query: 429 NSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIASV 488
NS I I+W +G I+ IN Y+L T+FVK ++H+ L V+ VF GI F M IY+A++
Sbjct: 431 NSKAISVITWGIGSFIVVINTYFLITSFVKLLLHNGLSTVSQVFSGIFGFLGMLIYMAAI 490
Query: 489 IYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDLADIPLP 541
+YL FRK+ + D+ ++ V + G+ L H+P RED++ + LP
Sbjct: 491 LYLVFRKNRKATLPLLEGDSTVR-IVGRDTATEGEGSLGHLP-REDISSMQLP 541
>UniRef100_Q9SAH8 Metal transporter Nramp1 [Arabidopsis thaliana]
Length = 532
Score = 558 bits (1438), Expect = e-157
Identities = 281/470 (59%), Positives = 360/470 (75%), Gaps = 5/470 (1%)
Query: 69 ETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCLW 128
ETDLQAGA++ YELLW++L+ AL+IQSLAANLGV TGKHL+E C EY + LW
Sbjct: 67 ETDLQAGAHYKYELLWIILVASCAALVIQSLAANLGVVTGKHLAEQCRAEYSKVPNFMLW 126
Query: 129 LLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLIT 188
++AE+AV+A DIPEVIGTAFALN+LF+IP+W GVLLTG STL+LL+LQ++GVRKLE LI
Sbjct: 127 VVAEIAVVACDIPEVIGTAFALNMLFSIPVWIGVLLTGLSTLILLALQKYGVRKLEFLIA 186
Query: 189 ILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHSA 248
LVF +A CFF E+ Y P VL G+FVP+L G GA AI+LLGA++MPHNLFLHSA
Sbjct: 187 FLVFTIAICFFVELHYSKPDPGEVLHGLFVPQLKGNGATGLAISLLGAMVMPHNLFLHSA 246
Query: 249 LVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVERC 308
LVLSRK+P+S GI EACR++L ESG AL VAFLINV++ISVSG VC+A +LS ++ C
Sbjct: 247 LVLSRKIPRSASGIKEACRFYLIESGLALMVAFLINVSVISVSGAVCNAPNLSPEDRANC 306
Query: 309 NDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWKR 368
DL LN ASFLL+NV+G+ SS ++AIALLASGQSSTITGTYAGQY+MQGFLD+R++ W R
Sbjct: 307 EDLDLNKASFLLRNVVGKWSSKLFAIALLASGQSSTITGTYAGQYVMQGFLDLRLEPWLR 366
Query: 369 NLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPHK 428
NL+TRC+AI PSL VA+IGGS+G+ +LIIIASMILSFELPFAL+PLLKF+S TKMG H
Sbjct: 367 NLLTRCLAIIPSLIVALIGGSAGAGKLIIIASMILSFELPFALVPLLKFTSCKTKMGSHV 426
Query: 429 NSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIASV 488
N M I ++W++G I+ IN+YYL ++F+K +IHS + + VF GI+ F +A+Y+A++
Sbjct: 427 NPMAITALTWVIGGLIMGINIYYLVSSFIKLLIHSHMKLILVVFCGILGFAGIALYLAAI 486
Query: 489 IYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDLADI 538
YL FRK+ V + D+ + + + N QL P R +D+
Sbjct: 487 AYLVFRKNRVATSLLISRDSQNVETLPRQDIVNMQL-----PCRVSTSDV 531
>UniRef100_Q5QN13 Putative root-specific metal transporter [Oryza sativa]
Length = 550
Score = 492 bits (1267), Expect = e-137
Identities = 245/471 (52%), Positives = 337/471 (71%), Gaps = 16/471 (3%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
++TDL AG++H Y LLW++L G IF L +QSLAANLG+ TG+HL+E+C EYP +VKYCL
Sbjct: 92 LQTDLVAGSSHRYSLLWVLLFGFIFVLTVQSLAANLGIITGRHLAELCMGEYPKYVKYCL 151
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLI 187
WLLAE+ VIAA IP V+GTA A N+L +IP WAGVL G T L+L LQ +G RK+E I
Sbjct: 152 WLLAELGVIAATIPGVLGTALAYNMLLHIPFWAGVLACGACTFLILGLQGYGARKMEFTI 211
Query: 188 TILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHS 247
++L+ VMA CFF E+ VNPPA GV++G+F+P+ G+ + +DA+A+ G+L++PHNLFLHS
Sbjct: 212 SVLMLVMATCFFMELGKVNPPAGGVIEGLFIPRPKGDYSTSDAVAMFGSLVVPHNLFLHS 271
Query: 248 ALVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVER 307
+LVL+RK+P + +G +A +FL E+ ALF+A L+NVA++S+SGT+C AN+LS +
Sbjct: 272 SLVLTRKMPYTSKGRKDASTFFLLENALALFIALLVNVAIVSISGTIC-ANNLSFADTST 330
Query: 308 CNDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWK 367
C+ LTLNS LLKN+LG+SSST+Y +ALL SGQS + +YAGQYIMQGF M++
Sbjct: 331 CSSLTLNSTYVLLKNILGKSSSTVYGVALLVSGQSCMVATSYAGQYIMQGFSG--MRKCI 388
Query: 368 RNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPH 427
L+ C + PSL + IGG+ R+I IA+++LSF LPFALIPL+KFSSS T +GP+
Sbjct: 389 IYLVAPCFTLLPSLIICSIGGTLRVHRIINIAAIVLSFVLPFALIPLIKFSSSCTNIGPY 448
Query: 428 KNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIAS 487
KN+ II I+WIL II IN+Y+ T+FV W++HS LP+V N I +VFP MA YIA+
Sbjct: 449 KNATSIIRIAWILSLVIIGINIYFFCTSFVAWLVHSDLPRVVNAIISSLVFPFMAAYIAA 508
Query: 488 VIYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDLADI 538
+IYL FRK + +D N V +G++E+ H+ +E D+
Sbjct: 509 LIYLAFRKVNL-------SDPFPTNSV------SGEIEVQHIQIQEKQEDL 546
>UniRef100_P93428 Integral membrane protein OsNramp3 [Oryza sativa]
Length = 385
Score = 416 bits (1070), Expect = e-115
Identities = 221/359 (61%), Positives = 272/359 (75%), Gaps = 2/359 (0%)
Query: 69 ETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLS-EVCEVEYPLFVKYCL 127
ETDLQAGA + YELLW++LI ALIIQSLAA LGV TGK + EVEYP + L
Sbjct: 19 ETDLQAGAQYKYELLWIILIASCAALIIQSLAARLGVVTGKPPGPSIAEVEYPKATNFIL 78
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLI 187
W+LAE+AV+A DIPEVIGTAFALN+LF +L+TG STL+LL LQ++GVRKLE LI
Sbjct: 79 WILAELAVVACDIPEVIGTAFALNMLFKSLCGVVILITGLSTLMLLLLQQYGVRKLEFLI 138
Query: 188 TILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHS 247
ILV ++A CF E+ Y P +S V++G+FVP+L G GA AI+LLGA++MPHNLFLHS
Sbjct: 139 AILVSLIATCFLVELGYSKPNSSVVVRGLFVPELKGNGATGLAISLLGAMVMPHNLFLHS 198
Query: 248 ALVLSRKVPKSVRGINEACRYFLYESGF-ALFVAFLINVAMISVSGTVCSANDLSGDNVE 306
LVLSRK +SV GI EACR+++ ES F AL +AFLIN+++ISV+ TVC +++LS ++
Sbjct: 199 ELVLSRKEKRSVHGIKEACRFYMIESAFLALTIAFLINISIISVNNTVCGSDNLSPEDQM 258
Query: 307 RCNDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRW 366
C+DL LN ASFLLKNVLG SS ++A+ALLASGQSSTITGTYAGQY+MQGFLD+RM W
Sbjct: 259 NCSDLDLNKASFLLKNVLGNWSSKLFAVALLASGQSSTITGTYAGQYVMQGFLDLRMTPW 318
Query: 367 KRNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMG 425
RNL+TR +AI PSL V+IIGGSS + +LIIIASMIL+FE FA K K G
Sbjct: 319 IRNLLTRSLAILPSLIVSIIGGSSAAGQLIIIASMILTFEPSFASSSTPKIHKQQDKNG 377
>UniRef100_Q84XF7 Integral membrane protein Nramp1 [Malus xiaojinensis]
Length = 234
Score = 308 bits (788), Expect = 3e-82
Identities = 151/234 (64%), Positives = 189/234 (80%)
Query: 105 VTTGKHLSEVCEVEYPLFVKYCLWLLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLL 164
V TGKHL+E C EYP + LW+LAE+AV+A DIPEVIGTAFALN+LF IP+W GVL+
Sbjct: 1 VVTGKHLAEHCRDEYPKVTNFILWILAELAVVACDIPEVIGTAFALNMLFKIPIWCGVLI 60
Query: 165 TGFSTLLLLSLQRFGVRKLELLITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGE 224
TG STL+LL LQ++GVRKLE LI LVF++A CF E+ Y P +S V++G+FVP++ G+
Sbjct: 61 TGLSTLMLLFLQQYGVRKLEFLIAFLVFLIATCFLVELGYSKPNSSEVVRGLFVPEIKGD 120
Query: 225 GAVADAIALLGALIMPHNLFLHSALVLSRKVPKSVRGINEACRYFLYESGFALFVAFLIN 284
GA AI+LLGA++MPHNLFLHSALVLSRKVP+SV GI EACR+++ ES FAL VAFLIN
Sbjct: 121 GATGLAISLLGAMVMPHNLFLHSALVLSRKVPRSVHGIKEACRFYMIESAFALTVAFLIN 180
Query: 285 VAMISVSGTVCSANDLSGDNVERCNDLTLNSASFLLKNVLGRSSSTIYAIALLA 338
+++ISVSG VCSA++L+ ++ CNDL LN ASFLLKNVLG SS ++AIAL A
Sbjct: 181 ISIISVSGAVCSADNLNPEDRMNCNDLDLNKASFLLKNVLGNWSSKVFAIALWA 234
>UniRef100_Q7X9B8 Ferrous ion membrane transport protein DMT1 [Glycine max]
Length = 516
Score = 302 bits (774), Expect = 1e-80
Identities = 180/438 (41%), Positives = 264/438 (60%), Gaps = 24/438 (5%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E DLQAGA GY LLWL++ L+IQ L+A LGV TGKHL+E+C EYP + + L
Sbjct: 77 LEGDLQAGAIAGYSLLWLLMWATAMGLLIQLLSARLGVATGKHLAELCREEYPPWARIVL 136
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
W++AE+A+I +DI EVIG+A A+ IL + +PLWAGV++T + L L+ +GVR LE
Sbjct: 137 WIMAELALIGSDIQEVIGSAIAIRILSHGVVPLWAGVVITALDCFIFLFLENYGVRTLEA 196
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFL 245
IL+ VMA F P +L G+ +PKL+ + + A+ ++G LIMPHN+FL
Sbjct: 197 FFAILIGVMAISFAWMFGEAKPSGKELLLGVLIPKLSSK-TIQQAVGVVGCLIMPHNVFL 255
Query: 246 HSALVLSRKVPKSVRG-INEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDN 304
HSALV SR+V +S +G + EA Y+ ES AL V+F+IN+ + +V ++L+
Sbjct: 256 HSALVQSRQVDRSKKGRVQEALNYYSIESTLALVVSFIINIFVTTVFAKGFYGSELA--- 312
Query: 305 VERCNDLTLNSASFLLKNVLGRSSST---IYAIALLASGQSSTITGTYAGQYIMQGFLDI 361
N + L +A L+ G I+ I LLA+GQSSTITGTYAGQ+IM GFL++
Sbjct: 313 ----NSIGLVNAGQYLEETYGGGLFPILYIWGIGLLAAGQSSTITGTYAGQFIMGGFLNL 368
Query: 362 RMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLII--IASMILSFELPFALIPLLKFSS 419
R+K+W R L+TR AI P++ VA++ +S S ++ +++ S ++PFALIPLL S
Sbjct: 369 RLKKWMRALITRSCAIIPTMIVALLFDTSEESLDVLNEWLNVLQSVQIPFALIPLLCLVS 428
Query: 420 SSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFP 479
MG + ++ T SW++ +I IN Y L+ F +V IG +V
Sbjct: 429 KEQIMGTFRIGAVLKTTSWLVAALVIVINGYLLTEFFSS--------EVNGPMIGTVVGV 480
Query: 480 LMAIYIASVIYLTFRKDT 497
+ A Y+A V+YL ++ T
Sbjct: 481 ITAAYVAFVVYLIWQAIT 498
>UniRef100_Q7XB56 Nramp metal transporter homolog [Thlaspi japonicum]
Length = 510
Score = 299 bits (766), Expect = 1e-79
Identities = 177/435 (40%), Positives = 261/435 (59%), Gaps = 24/435 (5%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E+DLQAGA GY L+WL++ L+IQ L+A LGV TG+HL+E+C EYP + + L
Sbjct: 71 LESDLQAGAIAGYSLIWLLMWATAIGLLIQLLSARLGVATGRHLAELCREEYPTWARMVL 130
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
W++AE+A+I ADI EVIG+A A+ IL N IPLWAGV++T + L L+ +GVRKLE
Sbjct: 131 WIMAEIALIGADIQEVIGSAIAIKILSNGLIPLWAGVVITALDCFIFLILENYGVRKLEA 190
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFL 245
+ +L+ MA F P + +L G VPKL+ + A+ ++G +IMPHN++L
Sbjct: 191 VFAVLIATMALSFAWMFGQTKPSGTELLVGALVPKLSSR-TIKQAVGIVGCIIMPHNVYL 249
Query: 246 HSALVLSRKV-PKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDN 304
HSALV SR++ PK + EA RY+ ES AL V+F+INV + +V D++
Sbjct: 250 HSALVQSREIDPKKRFRVKEALRYYSIESTGALVVSFIINVCVTTVFAKSFYKTDIA--- 306
Query: 305 VERCNDLTLNSASFLLKNVLGRSSSTIY---AIALLASGQSSTITGTYAGQYIMQGFLDI 361
+ + L +A L+ G +Y A+ LLA+GQSSTITGTYAGQ+IM GFL++
Sbjct: 307 ----DTIGLANAGDYLQEKYGGGYFPVYYIWAVGLLAAGQSSTITGTYAGQFIMGGFLNL 362
Query: 362 RMKRWKRNLMTRCIAIAPSLAVAIIGGSSGS--SRLIIIASMILSFELPFALIPLLKFSS 419
+MK+W R +TR AI P++ VAI+ SS + L +++ S ++PFA+IPLL S
Sbjct: 363 KMKKWIRATITRSCAIIPTMIVAIVFNSSATLLDELNEWLNVLQSVQIPFAVIPLLCLVS 422
Query: 420 SSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFP 479
+ MG K ++ ISW++ +I+IN Y + F S K + + +I+F
Sbjct: 423 NERIMGSFKIKPLMQAISWLVAALVIAINAYLMVNFF------SGAAKSVVMLVLVIIF- 475
Query: 480 LMAIYIASVIYLTFR 494
+ Y+ V+YL R
Sbjct: 476 -VVAYVFFVLYLISR 489
>UniRef100_Q6QA24 Putative divalent metal transporter DMT1A [Schistosoma mansoni]
Length = 583
Score = 298 bits (764), Expect = 2e-79
Identities = 175/448 (39%), Positives = 264/448 (58%), Gaps = 32/448 (7%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E+DLQ+GA Y LLWL+L+ + L +Q LA LGV G+HL+EV EYP + L
Sbjct: 84 IESDLQSGAIANYRLLWLLLLATLMGLFLQRLAVRLGVVPGRHLAEVYYDEYPTPARIIL 143
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILF--NIPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
WL+ EVA+I +D+ EVIGTA A+N+L IPLWAGVL+T T L L ++G+RKLE
Sbjct: 144 WLMVEVAIIGSDMQEVIGTAIAINLLSVGYIPLWAGVLITILDTFTFLFLDKYGLRKLEF 203
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEG--AVADAIALLGALIMPHNL 243
+L+ +MA F E V P +L+G+FVP G G + A+ +LGA++MPHN
Sbjct: 204 FFGLLITIMAVTFGYEYIVVKPDQLSLLRGLFVPSCPGCGWPELEQAVGILGAVVMPHNF 263
Query: 244 FLHSALVLSRKVPKSVRG-INEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSG 302
+LHSALV SR + ++ + EA Y+ ES ALFV+ +IN+ ++SV G ++
Sbjct: 264 YLHSALVKSRDIDRNNPFLVREANMYYFIESTIALFVSLMINIFVVSVFGAGFYGKNVK- 322
Query: 303 DNVERC-------------------NDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSS 343
+E C N + L + L G + I+AI LLA+GQSS
Sbjct: 323 QVIENCTREGKIPVRFTNAASSFDPNSVDLYTGGIFLGCEFGLACLFIWAIGLLAAGQSS 382
Query: 344 TITGTYAGQYIMQGFLDIRMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMIL 403
T+TGTY+GQ+ M+GFL+++ KRW+R L+TR IAI P++ + G + + ++++
Sbjct: 383 TMTGTYSGQFAMEGFLNLKWKRWQRVLITRSIAILPTILTTVFRGIEDLTGMNDFLNVLM 442
Query: 404 SFELPFALIPLLKFSSSSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHS 463
S +LPFA+IPLL F+SS + M KNS+ + ++ ++ ++SIN++ S +I +
Sbjct: 443 SVQLPFAVIPLLTFTSSRSIMAEFKNSLPVSIVANLISVAVVSINLFLGSV-----VIKA 497
Query: 464 SLPKVANVFIGIIVFPLMAIYIASVIYL 491
LP V++G+ + L A Y+ V YL
Sbjct: 498 RLPHHWAVYLGMGL--LFACYLGFVFYL 523
>UniRef100_Q6QA23 Putative divalent metal transporter DMT1B [Schistosoma mansoni]
Length = 543
Score = 298 bits (764), Expect = 2e-79
Identities = 175/448 (39%), Positives = 264/448 (58%), Gaps = 32/448 (7%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E+DLQ+GA Y LLWL+L+ + L +Q LA LGV G+HL+EV EYP + L
Sbjct: 44 IESDLQSGAIANYRLLWLLLLATLMGLFLQRLAVRLGVVPGRHLAEVYYDEYPTPARIIL 103
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILF--NIPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
WL+ EVA+I +D+ EVIGTA A+N+L IPLWAGVL+T T L L ++G+RKLE
Sbjct: 104 WLMVEVAIIGSDMQEVIGTAIAINLLSVGYIPLWAGVLITILDTFTFLFLDKYGLRKLEF 163
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEG--AVADAIALLGALIMPHNL 243
+L+ +MA F E V P +L+G+FVP G G + A+ +LGA++MPHN
Sbjct: 164 FFGLLITIMAVTFGYEYIVVKPDQLSLLRGLFVPSCPGCGWPELEQAVGILGAVVMPHNF 223
Query: 244 FLHSALVLSRKVPKSVRG-INEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSG 302
+LHSALV SR + ++ + EA Y+ ES ALFV+ +IN+ ++SV G ++
Sbjct: 224 YLHSALVKSRDIDRNNPFLVREANMYYFIESTIALFVSLMINIFVVSVFGAGFYGKNVK- 282
Query: 303 DNVERC-------------------NDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSS 343
+E C N + L + L G + I+AI LLA+GQSS
Sbjct: 283 QVIENCTREGKIPVRFTNAASSFDPNSVDLYTGGIFLGCEFGLACLFIWAIGLLAAGQSS 342
Query: 344 TITGTYAGQYIMQGFLDIRMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMIL 403
T+TGTY+GQ+ M+GFL+++ KRW+R L+TR IAI P++ + G + + ++++
Sbjct: 343 TMTGTYSGQFAMEGFLNLKWKRWQRVLITRSIAILPTILTTVFRGIEDLTGMNDFLNVLM 402
Query: 404 SFELPFALIPLLKFSSSSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHS 463
S +LPFA+IPLL F+SS + M KNS+ + ++ ++ ++SIN++ S +I +
Sbjct: 403 SVQLPFAVIPLLTFTSSRSIMAEFKNSLPVSIVANLISVAVVSINLFLGSV-----VIKA 457
Query: 464 SLPKVANVFIGIIVFPLMAIYIASVIYL 491
LP V++G+ + L A Y+ V YL
Sbjct: 458 RLPHHWAVYLGMGL--LFACYLGFVFYL 483
>UniRef100_Q9FN18 Metal transporter Nramp4 [Arabidopsis thaliana]
Length = 512
Score = 298 bits (763), Expect = 3e-79
Identities = 180/435 (41%), Positives = 262/435 (59%), Gaps = 24/435 (5%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E+DLQAGA GY L+WL++ L+IQ L+A LGV TG+HL+E+C EYP + + L
Sbjct: 72 LESDLQAGAIAGYSLIWLLMWATAIGLLIQLLSARLGVATGRHLAELCREEYPTWARMVL 131
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
W++AE+A+I ADI EVIG+A A+ IL N +PLWAGV++T + L L+ +G+RKLE
Sbjct: 132 WIMAEIALIGADIQEVIGSAIAIKILSNGLVPLWAGVVITALDCFIFLFLENYGIRKLEA 191
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFL 245
+ IL+ MA F P + +L G VPKL+ + A+ ++G +IMPHN+FL
Sbjct: 192 VFAILIATMALAFAWMFGQTKPSGTELLVGALVPKLSSR-TIKQAVGIVGCIIMPHNVFL 250
Query: 246 HSALVLSRKV-PKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDN 304
HSALV SR+V PK + EA +Y+ ES AL V+F+INV + +V S
Sbjct: 251 HSALVQSREVDPKKRFRVKEALKYYSIESTGALAVSFIINVFVTTVFAK-------SFYG 303
Query: 305 VERCNDLTLNSASFLLKNVLGRSSST---IYAIALLASGQSSTITGTYAGQYIMQGFLDI 361
E + + L +A L++ G I+AI +LA+GQSSTITGTYAGQ+IM GFL++
Sbjct: 304 TEIADTIGLANAGQYLQDKYGGGFFPILYIWAIGVLAAGQSSTITGTYAGQFIMGGFLNL 363
Query: 362 RMKRWKRNLMTRCIAIAPSLAVAIIGGSSGS--SRLIIIASMILSFELPFALIPLLKFSS 419
+MK+W R L+TR AI P++ VA++ SS S L +++ S ++PFA+IPLL S
Sbjct: 364 KMKKWVRALITRSCAIIPTMIVALVFDSSDSMLDELNEWLNVLQSVQIPFAVIPLLCLVS 423
Query: 420 SSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFP 479
+ MG K ++ TISWI+ +I+IN Y + F S + + +I+F
Sbjct: 424 NEQIMGSFKIQPLVQTISWIVAALVIAINGYLMVDFF------SGAATNLILLVPVIIFA 477
Query: 480 LMAIYIASVIYLTFR 494
+ Y+ V+YL R
Sbjct: 478 I--AYVVFVLYLISR 490
>UniRef100_Q9C6B2 Metal transporter Nramp2 [Arabidopsis thaliana]
Length = 530
Score = 298 bits (762), Expect = 4e-79
Identities = 176/432 (40%), Positives = 257/432 (58%), Gaps = 24/432 (5%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E DLQAGA GY LLWL++ L+IQ L+A +GV TG+HL+E+C EYP + +Y L
Sbjct: 88 LEGDLQAGAIAGYSLLWLLMWATAMGLLIQMLSARVGVATGRHLAELCRDEYPTWARYVL 147
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
W +AE+A+I ADI EVIG+A A+ IL +PLWAGV++T L L L+ +GVRKLE
Sbjct: 148 WSMAELALIGADIQEVIGSAIAIQILSRGFLPLWAGVVITASDCFLFLFLENYGVRKLEA 207
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFL 245
+ +L+ M F P ++ G+ +P+L+ + + A+ ++G +IMPHN+FL
Sbjct: 208 VFAVLIATMGLSFAWMFGETKPSGKELMIGILLPRLSSK-TIRQAVGVVGCVIMPHNVFL 266
Query: 246 HSALVLSRKV-PKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDN 304
HSALV SRK+ PK + EA Y+L ES ALF++F+IN+ + T A G
Sbjct: 267 HSALVQSRKIDPKRKSRVQEALNYYLIESSVALFISFMINLFV-----TTVFAKGFYG-- 319
Query: 305 VERCNDLTLNSASFLLKNVLGRSSST---IYAIALLASGQSSTITGTYAGQYIMQGFLDI 361
E+ N++ L +A L+ G I+ I LLA+GQSSTITGTYAGQ+IM GFL++
Sbjct: 320 TEKANNIGLVNAGQYLQEKFGGGLLPILYIWGIGLLAAGQSSTITGTYAGQFIMGGFLNL 379
Query: 362 RMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLII--IASMILSFELPFALIPLLKFSS 419
R+K+W R ++TR AI P++ VAI+ +S +S ++ +++ S ++PFAL+PLL S
Sbjct: 380 RLKKWMRAVITRSCAIVPTMIVAIVFNTSEASLDVLNEWLNVLQSVQIPFALLPLLTLVS 439
Query: 420 SSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFP 479
MG K I+ I+W + ++ IN Y L FV +V G+ V
Sbjct: 440 KEEIMGDFKIGPILQRIAWTVAALVMIINGYLLLDFFVS--------EVDGFLFGVTVCV 491
Query: 480 LMAIYIASVIYL 491
YIA ++YL
Sbjct: 492 WTTAYIAFIVYL 503
>UniRef100_Q9DET9 Natural resistance-associated macrophage protein [Morone saxatilis]
Length = 554
Score = 295 bits (754), Expect = 3e-78
Identities = 175/448 (39%), Positives = 269/448 (59%), Gaps = 38/448 (8%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E+DLQ+GA G++LLW++L+ I L++Q LAA LGV TG HL+EVC +YP + L
Sbjct: 84 IESDLQSGAKAGFKLLWVLLVATIIGLLLQRLAARLGVVTGMHLAEVCNRQYPTVPRVIL 143
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILF--NIPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
WL+ E+A+I +D+ EVIG A ALN+L IPLW GVL+T T + L L ++G+RKLE
Sbjct: 144 WLMVELAIIGSDMQEVIGCAIALNLLSVGRIPLWGGVLITITDTFVFLFLDKYGLRKLEA 203
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAV--ADAIALLGALIMPHNL 243
L+ VMA F E V P +LKGMF+P AG G V A+ ++GA+IMPHN+
Sbjct: 204 FFGFLITVMALSFGYEYVLVKPDQGELLKGMFLPYCAGCGPVQLEQAVGIVGAVIMPHNI 263
Query: 244 FLHSALVLSRKVP-KSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSG 302
+LHSALV SR + K+ + + EA +Y+ ES ALF++FLINV +++V N +
Sbjct: 264 YLHSALVKSRDIDRKNKKEVKEANKYYFIESTIALFISFLINVFVVAVFAE-AFYNKTNM 322
Query: 303 DNVERCN-----------------DLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTI 345
+ E CN ++ + +L V G ++ I+AI +LA+GQSST+
Sbjct: 323 EVHEFCNKTGSPHSDLFPQNNNTLEVDIYKGGVVLGCVFGPAALYIWAIGILAAGQSSTM 382
Query: 346 TGTYAGQYIMQGFLDIRMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSF 405
TGTY+GQ++M+GFL++R R+ R L+TR IAI P+L VAI + + +++ S
Sbjct: 383 TGTYSGQFVMEGFLNLRWSRFARVLLTRSIAITPTLLVAIFQDVQHLTGMNDFLNVLQSM 442
Query: 406 ELPFALIPLLKFSSSSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSL 465
+LPFALIP+L F+S ++ M N ++ W + G++++ V ++ FV +++
Sbjct: 443 QLPFALIPILTFTSLTSIMNDFANGLV-----WKISGGVVTLVVCAINMYFV--VVY--- 492
Query: 466 PKVANVFIGIIVFPLMAIYIASVIYLTF 493
++++ L A++ S+ YL F
Sbjct: 493 ---VTALNSVLLYVLAALF--SIAYLCF 515
>UniRef100_Q8JFC9 Natural resistance-associated macrophage protein large transcript
[Ictalurus punctatus]
Length = 550
Score = 293 bits (751), Expect = 7e-78
Identities = 179/449 (39%), Positives = 271/449 (59%), Gaps = 32/449 (7%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E+DLQ+GA G++LLW++L I L++Q LAA LGV TG HL+EVC +YP + L
Sbjct: 80 IESDLQSGAVAGFKLLWVLLGATIIGLLLQRLAARLGVVTGMHLAEVCHRQYPTVPRIIL 139
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILF--NIPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
WL+ E+A+I +D+ EVIG A A N+L IPLWAGVL+T T + L L ++G+RKLE
Sbjct: 140 WLMVELAIIGSDMQEVIGCAIAFNLLSVGRIPLWAGVLITIVDTFVFLFLDKYGLRKLEA 199
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAV--ADAIALLGALIMPHNL 243
L+ +MA F + V P VLKG+FVP G G V A+ ++GA+IMPHN+
Sbjct: 200 FFGFLITIMAVTFGYQYVRVAPDQGAVLKGLFVPYCQGCGPVQLEQAVGIVGAVIMPHNI 259
Query: 244 FLHSALVLSRKVPK-SVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSG 302
+LHSALV SR+V + + R + EA +YF ES ALF++FLINV +++V N +
Sbjct: 260 YLHSALVKSRQVDRANKREVKEANKYFFIESCIALFISFLINVFVVAVFAE-AFYNKTNM 318
Query: 303 DNVERCNDL-TLNSASFLLKN----------------VLGRSSSTIYAIALLASGQSSTI 345
+ ++CN+ +++S F L N G ++ I+A+ +LA+GQSST+
Sbjct: 319 EVNQQCNETGSIHSNLFPLNNNTLQVDIYKGGVVLGCFFGPAALYIWAVGILAAGQSSTM 378
Query: 346 TGTYAGQYIMQGFLDIRMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSF 405
TGTY+GQ++M+GFL++R R+ R L+TR IAI P+L VAI + + + +++ S
Sbjct: 379 TGTYSGQFVMEGFLNLRWSRFARVLLTRSIAIFPTLLVAIFQDVTHLTGMNDFLNVLQSM 438
Query: 406 ELPFALIPLLKFSSSSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSL 465
+LPFALIP+L F+S + M N + W +G G+ + V ++ FV +++ +
Sbjct: 439 QLPFALIPILTFTSLTPIMNDFANGLF-----WKIGGGLTILLVCAINLYFV--VVYVTA 491
Query: 466 PKVANVFIGIIVFPLMAIYIASVIYLTFR 494
K +V + + L Y+ VIYL +R
Sbjct: 492 LK--SVLLYVFAALLSIAYLCFVIYLAWR 518
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.328 0.141 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 813,453,135
Number of Sequences: 2790947
Number of extensions: 32173739
Number of successful extensions: 273069
Number of sequences better than 10.0: 419
Number of HSP's better than 10.0 without gapping: 309
Number of HSP's successfully gapped in prelim test: 111
Number of HSP's that attempted gapping in prelim test: 270088
Number of HSP's gapped (non-prelim): 1270
length of query: 541
length of database: 848,049,833
effective HSP length: 132
effective length of query: 409
effective length of database: 479,644,829
effective search space: 196174735061
effective search space used: 196174735061
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC140916.14


